Development of a Universal Multi-Epitope Vaccine Candidate against Streptococcus suis Infections Using Immunoinformatics Approaches
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of Protein Sequences
2.2. Prediction of B-Cell (BCL) Epitopes
2.3. Predictions of Cytotoxic T-Lymphocyte (CTL) Epitopes
2.4. Predictions of Helper T-Lymphocyte (HTL) Epitopes
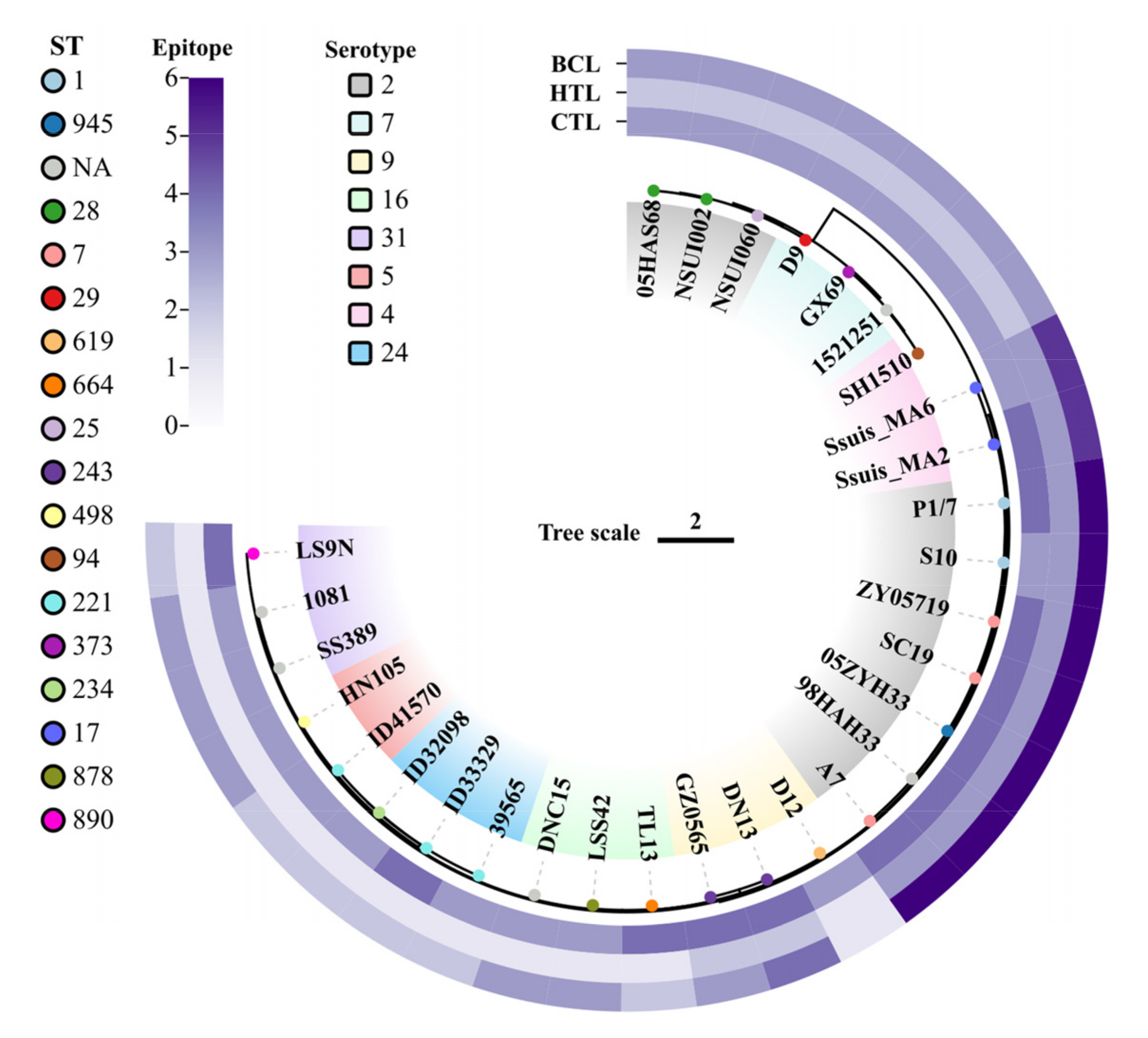
2.5. Predicted Epitopes Conservation Analysis
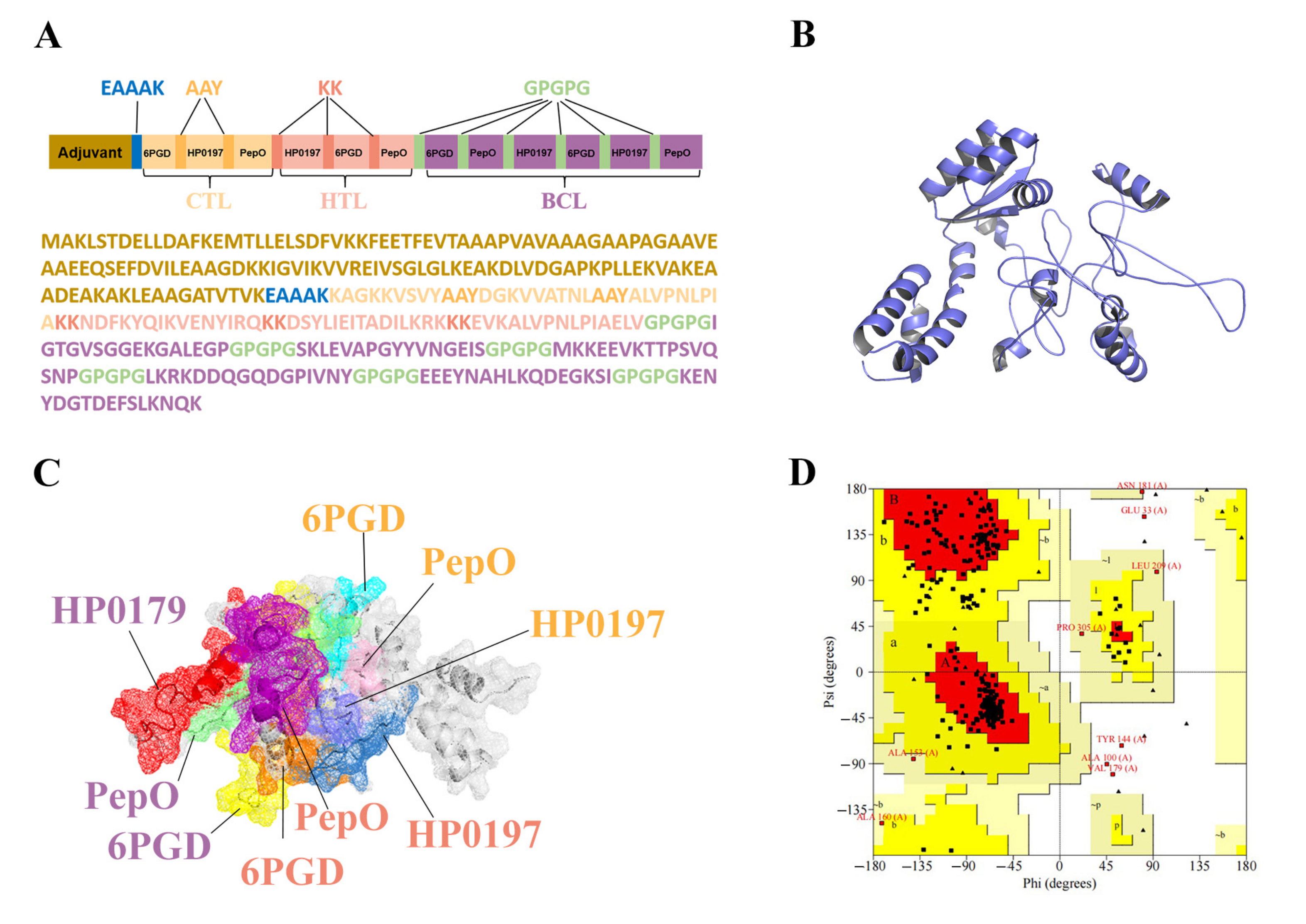
2.6. Multiple-Epitope Vaccine Designing and Processing
2.7. Prediction, Refinement, and Validation of the Tertiary Structure
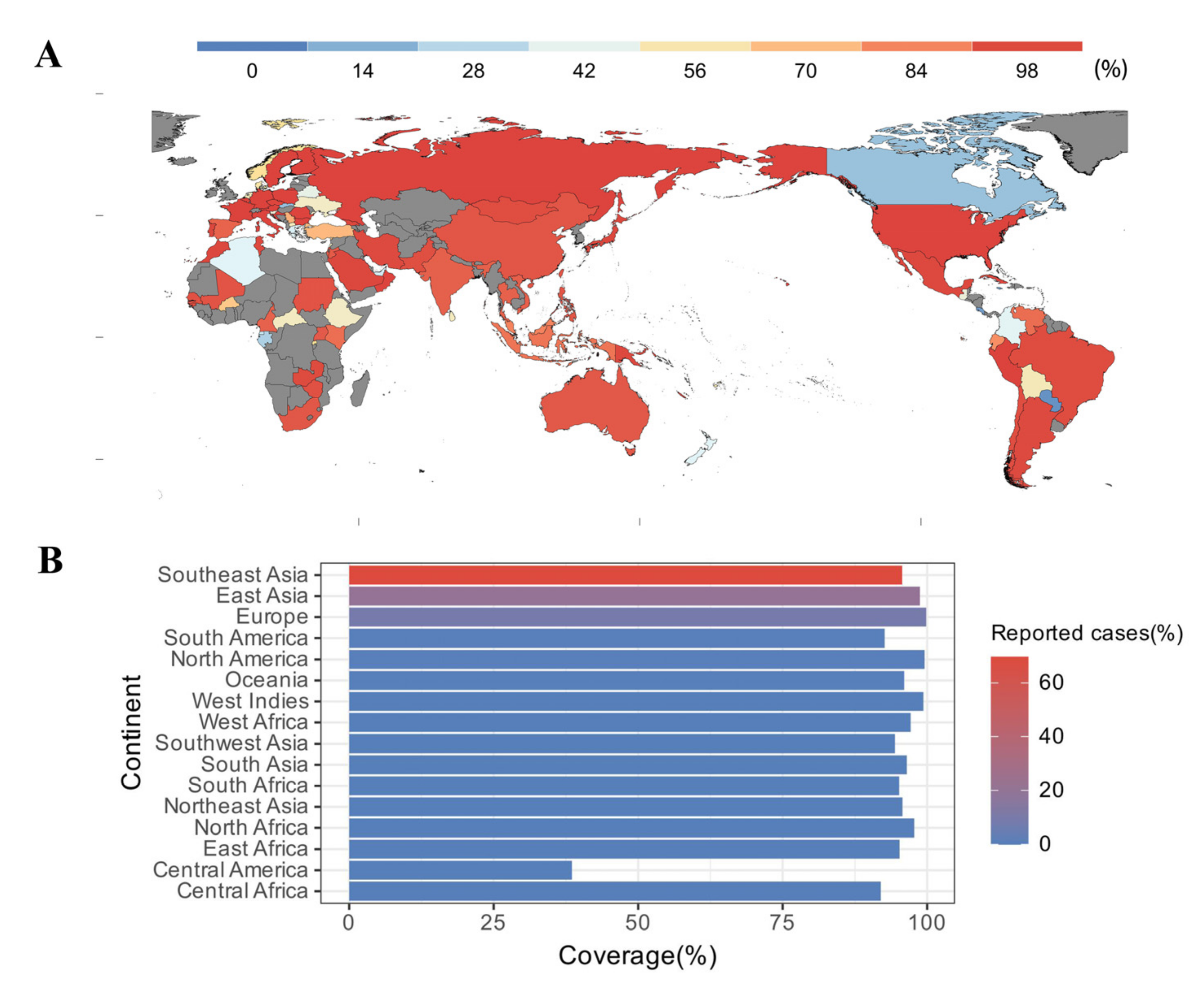
2.8. Population Coverage
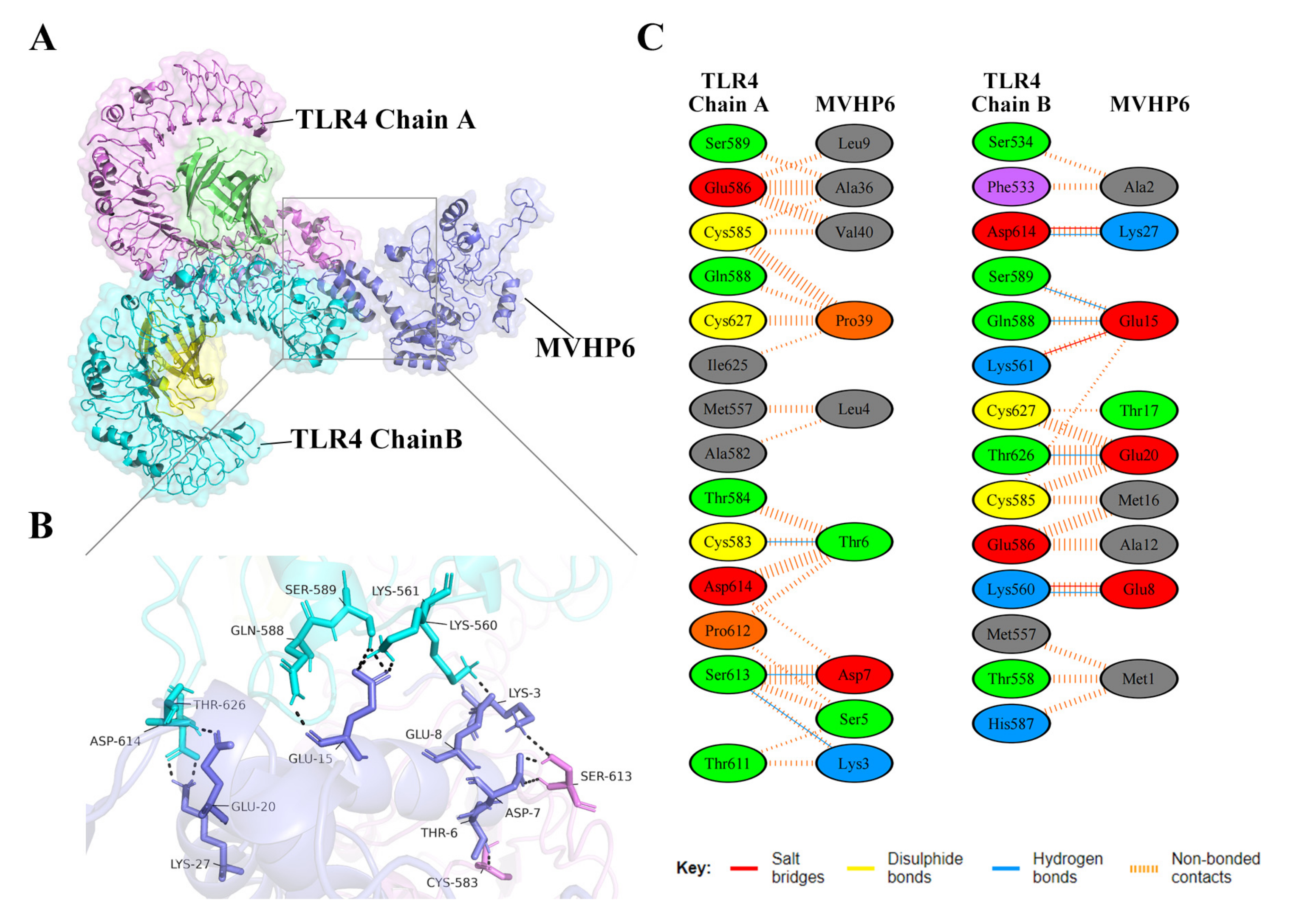
2.9. Molecular Docking and Molecular Dynamics (MD) Simulation
2.10. Immune Response Simulation
2.11. Codon Optimization and In Silico Cloning
3. Results
3.1. Prediction and Analysis of BCL Epitopes
3.2. Prediction of CTL Epitopes
3.3. Prediction of HTL Epitopes
3.4. Analysis of Epitope Conservation
3.5. Estimated Population Coverage
3.6. Construction of the Multi-Epitope Vaccine (MVHP6)
3.7. Tertiary Structure Prediction, Refinement, and Validation of MVHP6
3.8. Molecular Docking of the Constructed Vaccine with the Human Toll-like Receptor, TLR4
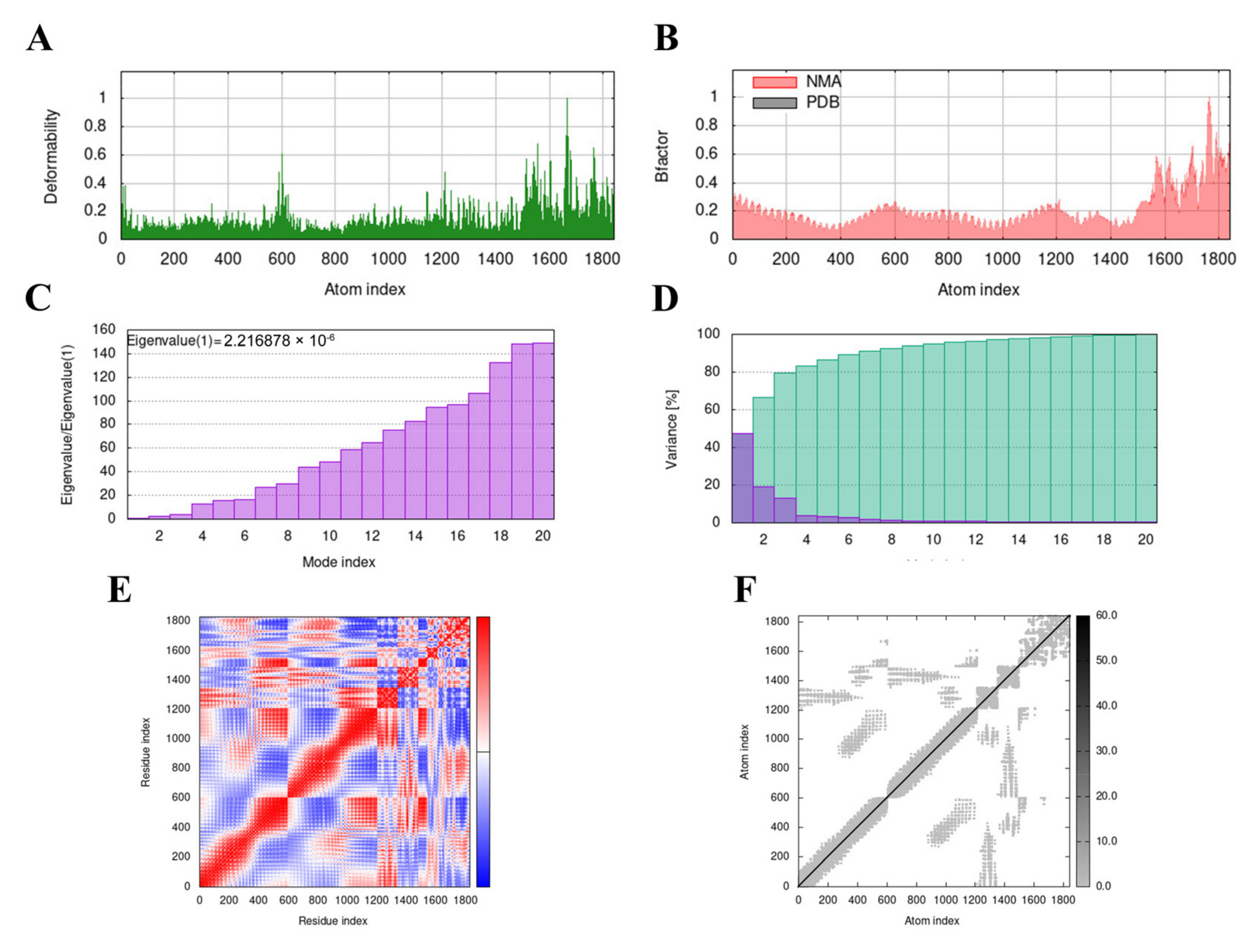
3.9. Molecular Dynamics (MD) Simulation
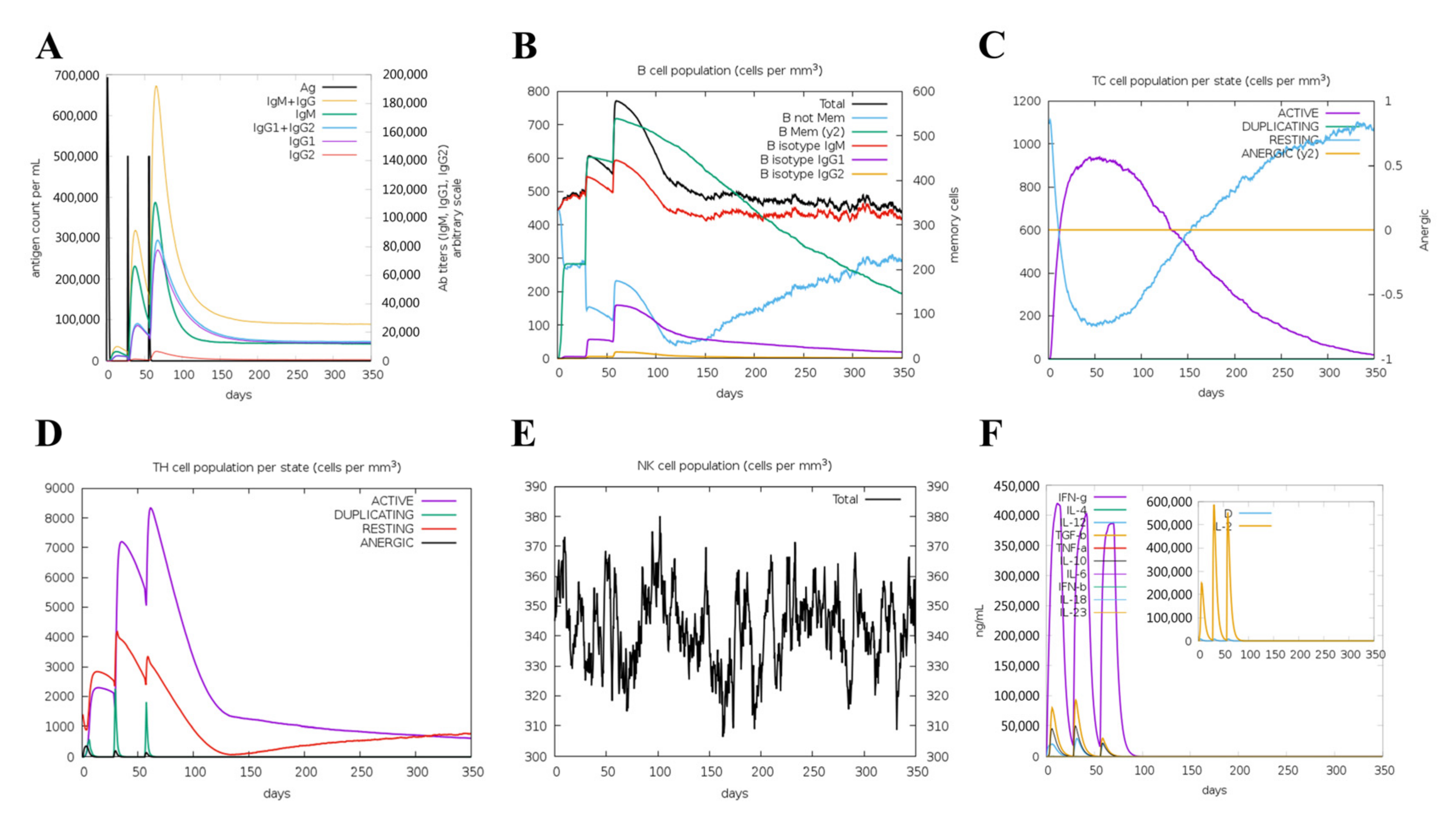
3.10. Immunogenicity Evaluation of the Vaccine
3.11. In Silico Cloning of the Vaccine Candidate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haas, B.; Grenier, D. Understanding the Virulence of Streptococcus Suis: A Veterinary, Medical, and Economic Challenge. Med. Malpract. Infect. 2018, 48, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; de Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus Suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. Suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus Suis: A New Emerging or an Old Neglected Zoonotic Pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Sroka, J.; Zając, V.; Wasiński, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus Suis: A Re-Emerging Pathogen Associated with Occupational Exposure to Pigs or Pork Products. Part I—Epidemiology. Ann. Agric. Environ. Med. 2017, 24, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jing, H.; Chen, Z.; Zheng, H.; Zhu, X.; Wang, H.; Wang, S.; Liu, L.; Zu, R.; Luo, L.; et al. Human Streptococcus Suis Outbreak, Sichuan, China. Emerg. Infect. Dis. 2006, 12, 914–920. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, F.; Wang, H.; Chen, S.; Wang, G.; Sun, J.; Hua, C.; Yang, H. Studies on human streptococcal infectious syndrome caused by infected pigs. Zhonghua Yu Fang Yi Xue Za Zhi 2000, 34, 150–152. [Google Scholar]
- Kerdsin, A.; Segura, M.; Fittipaldi, N.; Gottschalk, M. Sociocultural Factors Influencing Human Streptococcus Suis Disease in Southeast Asia. Foods 2022, 11, 1190. [Google Scholar] [CrossRef]
- Yu, Y.; Fang, J.-T.; Zheng, M.; Zhang, Q.; Walsh, T.R.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Combination Therapy Strategies Against Multiple-Resistant Streptococcus Suis. Front. Pharmacol. 2018, 9, 489. [Google Scholar] [CrossRef] [PubMed]
- Petrocchi-Rilo, M.; Martínez-Martínez, S.; Aguarón-Turrientes, Á.; Roca-Martínez, E.; García-Iglesias, M.-J.; Pérez-Fernández, E.; González-Fernández, A.; Herencia-Lagunar, E.; Gutiérrez-Martín, C.-B. Anatomical Site, Typing, Virulence Gene Profiling, Antimicrobial Susceptibility and Resistance Genes of Streptococcus Suis Isolates Recovered from Pigs in Spain. Antibiotics 2021, 10, 707. [Google Scholar] [CrossRef]
- Rieckmann, K.; Pendzialek, S.-M.; Vahlenkamp, T.; Baums, C.G. A Critical Review Speculating on the Protective Efficacies of Autogenous Streptococcus Suis Bacterins as Used in Europe. Porc. Health Manag. 2020, 6, 12. [Google Scholar] [CrossRef]
- Segura, M. Streptococcus Suis Vaccines: Candidate Antigens and Progress. Expert Rev. Vaccines 2015, 14, 1587–1608. [Google Scholar] [CrossRef]
- Yan, Z.; Yao, X.; Pan, R.; Zhang, J.; Ma, X.; Dong, N.; Wei, J.; Liu, K.; Qiu, Y.; Sealey, K.; et al. Subunit Vaccine Targeting Phosphate ABC Transporter ATP-Binding Protein, PstB, Provides Cross-Protection against Streptococcus Suis Serotype 2, 7, and 9 in Mice. Vet. Sci. 2023, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Du, Y.; Mao, C.; Li, J.; Jin, M.; Sun, L.; Wang, Y. Immunogenicity and Protective Ability of RpoE against Streptococcus Suis Serotype 2. J. Appl. Microbiol. 2021, 130, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Fan, Q.; Wang, Y.; Mao, C.; Li, J.; Jin, M.; Zhang, X.; Ding, K.; Wang, Y. Evaluation of Immune Effect of Streptococcus Suis Biofilm-Associated Protein PDH. Vet. Microbiol. 2021, 263, 109270. [Google Scholar] [CrossRef]
- Wang, H.; Qin, Z.; Li, M. Recent Advances in Pathogenic Streptococcus Vaccine Development. Curr. Issues Mol. Biol. 2019, 32, 645–700. [Google Scholar] [CrossRef] [PubMed]
- Weiße, C.; Dittmar, D.; Jakóbczak, B.; Florian, V.; Schütze, N.; Alber, G.; Klose, K.; Michalik, S.; Valentin-Weigand, P.; Völker, U.; et al. Immunogenicity and Protective Efficacy of a Streptococcus Suis Vaccine Composed of Six Conserved Immunogens. Vet. Res. 2021, 52, 112. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Fu, Y.; Zhang, Y.; Li, Q.; Wang, S.; Shi, H. Salmonella Enterica Serovar Choleraesuis Vector Delivering a Dual-Antigen Expression Cassette Provides Mouse Cross-Protection against Streptococcus Suis Serotypes 2, 7, 9, and 1/2. Vet. Res. 2022, 53, 46. [Google Scholar] [CrossRef]
- Samad, A.; Ahammad, F.; Nain, Z.; Alam, R.; Imon, R.R.; Hasan, M.; Rahman, M.S. Designing a Multi-Epitope Vaccine against SARS-CoV-2: An Immunoinformatics Approach. J. Biomol. Struct. Dyn. 2020, 40, 14–30. [Google Scholar] [CrossRef]
- Majidiani, H.; Dalimi, A.; Ghaffarifar, F.; Pirestani, M. Multi-Epitope Vaccine Expressed in Leishmania Tarentolae Confers Protective Immunity to Toxoplasma Gondii in BALB/c Mice. Microb. Pathog. 2021, 155, 104925. [Google Scholar] [CrossRef]
- Bibi, S.; Ullah, I.; Zhu, B.; Adnan, M.; Liaqat, R.; Kong, W.-B.; Niu, S. In Silico Analysis of Epitope-Based Vaccine Candidate against Tuberculosis Using Reverse Vaccinology. Sci. Rep. 2021, 11, 1249. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A.; et al. Designing Multi-Epitope Vaccine against Staphylococcus Aureus by Employing Subtractive Proteomics, Reverse Vaccinology and Immuno-Informatics Approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Chen, B.; Li, R.; Mu, X.; Han, L.; Zhou, H.; Chen, H.; Meilin, J. Identification of a Surface Protective Antigen, HP0197 of Streptococcus Suis Serotype 2. Vaccine 2009, 27, 5209–5213. [Google Scholar] [CrossRef]
- Li, J.; Xia, J.; Tan, C.; Zhou, Y.; Wang, Y.; Zheng, C.; Chen, H.; Bei, W. Evaluation of the Immunogenicity and the Protective Efficacy of a Novel Identified Immunogenic Protein, SsPepO, of Streptococcus Suis Serotype 2. Vaccine 2011, 29, 6514–6519. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Liu, M.; Liu, J.; Yuan, F.; Fu, S.; Liu, Y.; Jin, M.; Bei, W.; Chen, H. Vaccination with Streptococcus Suis Serotype 2 Recombinant 6PGD Protein Provides Protection against S. Suis Infection in Swine. FEMS Microbiol. Lett. 2009, 296, 78–83. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. Prediction of Continuous B-Cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins: Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A Server for in Silico Prediction of Allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Sharma, N.; Naorem, L.D.; Jain, S.; Raghava, G.P.S. ToxinPred2: An Improved Method for Predicting Toxicity of Proteins. Brief. Bioinform. 2022, 23, bbac174. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped Sequence Alignment Using Artificial Neural Networks: Application to the MHC Class I System. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef]
- Calis, J.J.A.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide Binding Predictions for HLA DR, DP and DQ Molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P. Designing of Interferon-Gamma Inducing MHC Class-II Binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Gupta, S.; Vir, P.; Raghava, G.P.S. Prediction of IL4 Inducing Peptides. Clin. Dev. Immunol. 2013, 2013, 263952. [Google Scholar] [CrossRef]
- Nagpal, G.; Usmani, S.S.; Dhanda, S.K.; Kaur, H.; Singh, S.; Sharma, M.; Raghava, G.P.S. Computer-Aided Designing of Immunosuppressive Peptides Based on IL-10 Inducing Potential. Sci. Rep. 2017, 7, 42851. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.-C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols; Link, A.J., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1999; pp. 531–552. ISBN 978-1-59259-584-6. [Google Scholar]
- Zhang, Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for Checking the Quality of Protein Structures Solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Bui, H.-H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting Population Coverage of T-Cell Epitope-Based Diagnostics and Vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. IMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef] [PubMed]
- Dey, J.; Mahapatra, S.R.; Patnaik, S.; Lata, S.; Kushwaha, G.S.; Panda, R.K.; Misra, N.; Suar, M. Molecular Characterization and Designing of a Novel Multiepitope Vaccine Construct Against Pseudomonas Aeruginosa. Int. J. Pept. Res. 2022, 28, 49. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A Novel Tool to Adapt Codon Usage of a Target Gene to Its Potential Expression Host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus Suis, an Important Pig Pathogen and Emerging Zoonotic Agent—An Update on the Worldwide Distribution Based on Serotyping and Sequence Typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Yuan, Z.; Yan, X.; Zhang, A.; Chen, B.; Shen, Y.; Jin, M. Molecular Mechanism by Which Surface Antigen HP0197 Mediates Host Cell Attachment in the Pathogenic Bacteria Streptococcus Suis. J. Biol. Chem. 2013, 288, 956–963. [Google Scholar] [CrossRef]
- Zhang, A.; Chen, B.; Yuan, Z.; Li, R.; Liu, C.; Zhou, H.; Chen, H.; Jin, M. HP0197 Contributes to CPS Synthesis and the Virulence of Streptococcus Suis via CcpA. PLoS ONE 2012, 7, e50987. [Google Scholar] [CrossRef]
- Geng, H.; Zhu, L.; Yuan, Y.; Zhang, W.; Li, W.; Wang, J.; Zheng, Y.; Wei, K.; Cao, W.; Wang, H.; et al. Identification and Characterization of Novel Immunogenic Proteins of Streptococcus Suis Serotype 2. J. Proteome Res. 2008, 7, 4132–4142. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, K.; Sun, C.; Liu, F.; Peng, W.; Chen, H.; Yuan, F.; Bei, W.; Li, J. Binding of Plasminogen to Streptococcus Suis Protein Endopeptidase O Facilitates Evasion of Innate Immunity in Streptococcus Suis. Front. Microbiol. 2021, 12, 694103. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Yan, K.; Li, H.; Sun, C.; Zhang, S.; Yuan, F.; Wang, X.; Tan, C.; Chen, H.; et al. Binding of Fibronectin to SsPepO Facilitates the Development of Streptococcus Suis Meningitis. J. Infect. Dis. 2018, 217, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Yuan, J.; Wang, H.; Zhang, J.; Li, S.; Zhang, H.; Liu, Y.; Yin, Y.; Zhang, X. Streptococcus Pneumoniae PepO Promotes Host Anti-Infection Defense via Autophagy in a Toll-like Receptor 2/4 Dependent Manner. Virulence 2020, 11, 270–282. [Google Scholar] [CrossRef]
- Bahadori, Z.; Shafaghi, M.; Madanchi, H.; Ranjbar, M.M.; Shabani, A.A.; Mousavi, S.F. In Silico Designing of a Novel Epitope-Based Candidate Vaccine against Streptococcus Pneumoniae with Introduction of a New Domain of PepO as Adjuvant. J. Transl. Med. 2022, 20, 389. [Google Scholar] [CrossRef] [PubMed]
- Daniely, D.; Portnoi, M.; Shagan, M.; Porgador, A.; Givon-Lavi, N.; Ling, E.; Dagan, R.; Nebenzahl, Y.M. Pneumococcal 6-Phosphogluconate-Dehydrogenase, a Putative Adhesin, Induces Protective Immune Response in Mice. Clin. Exp. Immunol. 2006, 144, 254–263. [Google Scholar] [CrossRef]
- Tan, C.; Fu, S.; Liu, M.; Jin, M.; Liu, J.; Bei, W.; Chen, H. Cloning, Expression and Characterization of a Cell Wall Surface Protein, 6-Phosphogluconate-Dehydrogenase, of Streptococcus Suis Serotype 2. Vet. Microbiol. 2008, 130, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Yuan, F.; Zhang, M.; Tan, C.; Chen, H.; Bei, W. Cloning, Expression and Characterization of a Cell Wall Surface Protein, 6-Phosphogluconate Dehydrogenase, of Haemophilus Parasuis. Res. Vet. Sci. 2012, 93, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Shang, J.; Li, Y.; Wang, S.; Shi, H. Live Attenuated Salmonella Enterica Serovar Choleraesuis Vaccine Vector Displaying Regulated Delayed Attenuation and Regulated Delayed Antigen Synthesis to Confer Protection against Streptococcus Suis in Mice. Vaccine 2015, 33, 4858–4867. [Google Scholar] [CrossRef]
- Wu, M.-C.; Wu, H.-C.; Lee, J.-W.; Chang, W.-C.; Chu, C.-Y. A Protein-Based Subunit Vaccine with Biological Adjuvants Provides Effective Protection against Pasteurella Multocida in Pigs. Vet. Res. 2023, 54, 17. [Google Scholar] [CrossRef]
- Ogunniyi, A.D.; Paton, J.C.; Kirby, A.C.; McCullers, J.A.; Cook, J.; Hyodo, M.; Hayakawa, Y.; Karaolis, D.K.R. C-Di-GMP Is an Effective Immunomodulator and Vaccine Adjuvant Against Pneumococcal Infection. Vaccine 2008, 26, 4676–4685. [Google Scholar] [CrossRef]
- Kumar, S.; Sunagar, R.; Gosselin, E. Bacterial Protein Toll-Like-Receptor Agonists: A Novel Perspective on Vaccine Adjuvants. Front. Immunol. 2019, 10, 1144. [Google Scholar] [CrossRef]
- Aziz, S.; Almajhdi, F.N.; Waqas, M.; Ullah, I.; Salim, M.A.; Khan, N.A.; Ali, A. Contriving Multi-Epitope Vaccine Ensemble for Monkeypox Disease Using an Immunoinformatics Approach. Front. Immunol. 2022, 13, 1004804. [Google Scholar] [CrossRef] [PubMed]
- Jang, A.-R.; Choi, J.-H.; Shin, S.J.; Park, J.-H. Mycobacterium Tuberculosis ESAT6 Induces IFN-β Gene Expression in Macrophages via TLRs-Mediated Signaling. Cytokine 2018, 104, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Shin, S.J.; Lee, M.H.; Lee, M.-G.; Kang, T.H.; Park, W.S.; Soh, B.Y.; Park, J.H.; Shin, Y.K.; Kim, H.W.; et al. A Potential Protein Adjuvant Derived from Mycobacterium Tuberculosis Rv0652 Enhances Dendritic Cells-Based Tumor Immunotherapy. PLoS ONE 2014, 9, e104351. [Google Scholar] [CrossRef] [PubMed]

| Epitope | Protein | Sequence | Antigenicity | Hydropathicity |
|---|---|---|---|---|
| BCL | HP0197 | MKKEEVKTTPSVQSNP | 1.1755 | −1.35 |
| EEEYNAHLKQDEGKSI | 1.0451 | −1.744 | ||
| PepO | KENYDGTDEFSLKNQK | 1.288 | −2.05 | |
| SKLEVAPGYYVNGEIS | 0.8605 | −0.156 | ||
| 6PGD | LKRKDDQGQDGPIVNY | 1.4914 | −1.531 | |
| IGTGVSGGEKGALEGP | 1.3547 | −0.131 | ||
| CTL | HP0197 | DGKVVATNL | 1.4692 | 0.222 |
| PepO | ALVPNLPIA | 1.5535 | 1.467 | |
| 6PGD | KAGKKVSVY | 1.3846 | −0.444 | |
| HTL | HP0197 | NDFKYQIKVENYIRQ | 0.8205 | −1.327 |
| PepO | EVKALVPNLPIAELV | 0.6247 | 0.967 | |
| 6PGD | DSYLIEITADILKRK | 0.5099 | −0.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhao, G.; Xiong, Y.; Li, F.; Chen, Y.; Cheng, Y.; Ma, J.; Wang, H.; Yan, Y.; Wang, Z.; et al. Development of a Universal Multi-Epitope Vaccine Candidate against Streptococcus suis Infections Using Immunoinformatics Approaches. Vet. Sci. 2023, 10, 383. https://doi.org/10.3390/vetsci10060383
Zhang Y, Zhao G, Xiong Y, Li F, Chen Y, Cheng Y, Ma J, Wang H, Yan Y, Wang Z, et al. Development of a Universal Multi-Epitope Vaccine Candidate against Streptococcus suis Infections Using Immunoinformatics Approaches. Veterinary Sciences. 2023; 10(6):383. https://doi.org/10.3390/vetsci10060383
Chicago/Turabian StyleZhang, Yumin, Guoqing Zhao, Yangjing Xiong, Feiyu Li, Yifan Chen, Yuqiang Cheng, Jingjiao Ma, Henan Wang, Yaxian Yan, Zhaofei Wang, and et al. 2023. "Development of a Universal Multi-Epitope Vaccine Candidate against Streptococcus suis Infections Using Immunoinformatics Approaches" Veterinary Sciences 10, no. 6: 383. https://doi.org/10.3390/vetsci10060383
APA StyleZhang, Y., Zhao, G., Xiong, Y., Li, F., Chen, Y., Cheng, Y., Ma, J., Wang, H., Yan, Y., Wang, Z., & Sun, J. (2023). Development of a Universal Multi-Epitope Vaccine Candidate against Streptococcus suis Infections Using Immunoinformatics Approaches. Veterinary Sciences, 10(6), 383. https://doi.org/10.3390/vetsci10060383







