A Canine Leptospirosis Clinical Case Due to Leptospira interrogans (Serogroup Icterohaemorrhagiae) in a Dog Kennel in Castelvetrano (Western Sicily, South Italy)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Kennel
2.2. Samples
2.2.1. Suspected Clinical Case
2.2.2. Sampling from the Other Dogs Housed in the Kennel
2.3. Sample Processing and DNA Extraction
2.4. Differential Diagnosis
2.5. Serological Test for Leptospirosis
2.6. Molecular Tests for Leptospirosis
2.7. Multi-Locus Sequence Typing (MLST)
2.8. Prevention Measures at the Kennel
2.9. Ethical Statement
3. Results
3.1. Leptospirosis Clinical Case: From Clinical Suspicion to Laboratory Confirmation
3.2. Investigations on the Other Dogs in the Kennel and Containment Measures
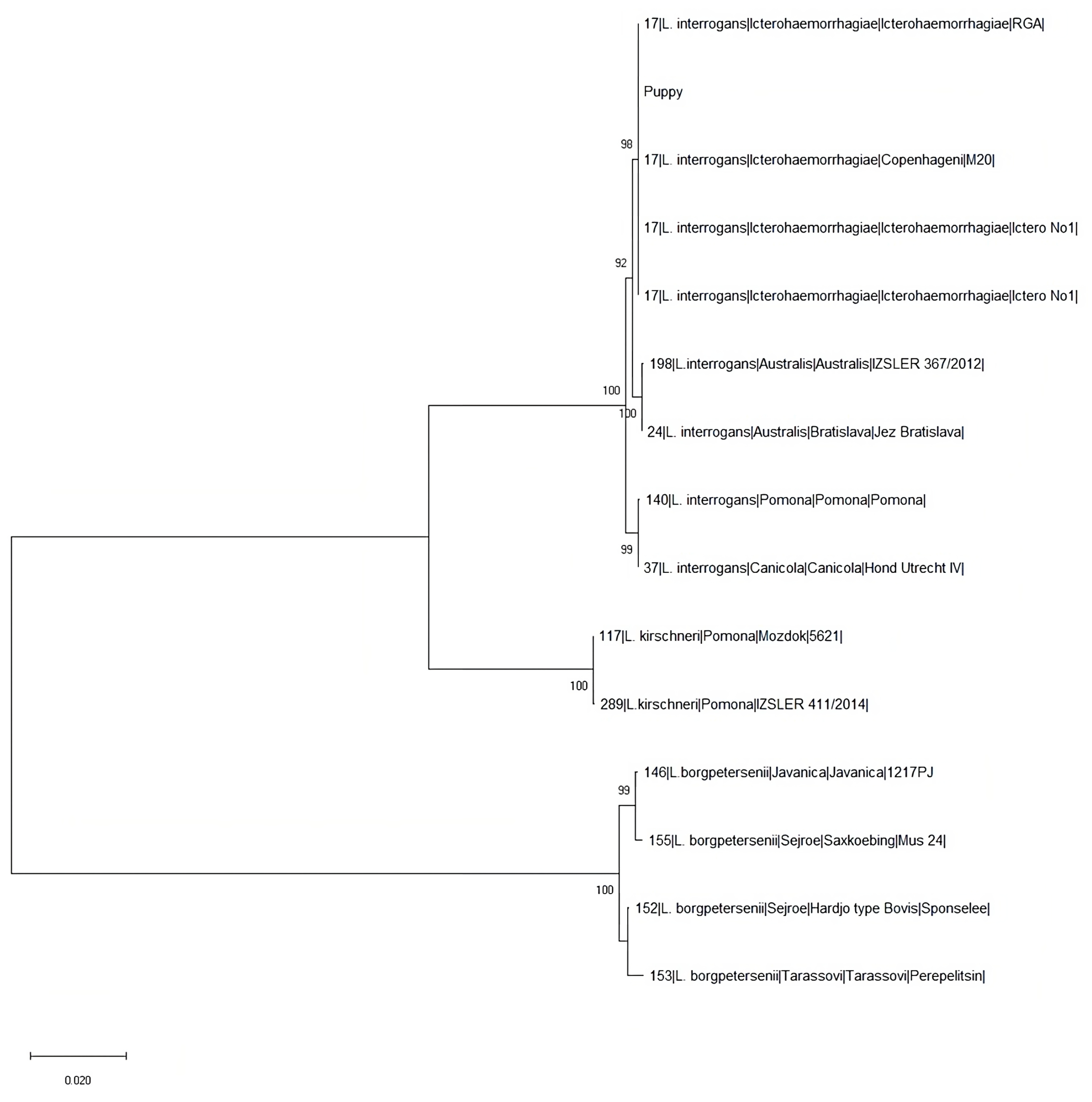
3.3. Genotyping Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pappas, G.; Papadimitriou, P.; Siozopoulou, V.; Christou, L.; Akritidis, N. The globalization of leptospirosis: Worldwide incidence trends. Int. J. Infect. Dis. 2008, 12, 351–357. [Google Scholar] [CrossRef]
- Grippi, F.; Giudice, E.; Pietro, S.D.; Sciacca, C.; Santangelo, F.; Galluzzo, P.; Barreca, S.; Guercio, A. Leptospira Interrogans Serogroup Sejroe Serovar Hardjo in Aborting Cows: Two Herd Cases in Sicily (Italy). J. Vet. Res. 2020, 64, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Bryson, D.G.; Ellis, W.A. Leptospirosis in a British domestic cat. J. Small Anim. Pract. 1976, 17, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A.; O’Brien, J.J.; Cassells, J.A.; Montgomery, J. Leptospiral infection in horses in Northen Ireland: Serological and microbiological findings. Equine Vet. J. 1983, 15, 317–320. [Google Scholar] [CrossRef]
- Ellis, W.A. Bovine leptospirosis in the tropics: Prevalence, pathogenesis and control. Prev. Vet. Med. 1984, 2, 411–421. [Google Scholar] [CrossRef]
- Gravekamp, C.; Korver, H.; Montgomery, J.; Everdard, C.O.R.; Carrington, D.; Ellis, W.A.; Terpstra, W.J. Leptospires isolated from Toads and Frogs on the Island of Barbados. Zentralblatt Bakteriol. 1991, 275, 403–411. [Google Scholar] [CrossRef]
- Shophet, R. A serological survey of leptospirosis in cats. N. Z. Vet. J. 1979, 27, 245–246. [Google Scholar] [CrossRef] [PubMed]
- White, F.H.; Sulzer, K.R.; Engel, R.W. Isolations of Leptospira interrogans serovars hardjo, balcanica, and pomona from cattle at slaughter. Am. J. Vet. Res. 1982, 43, 1172–1173. [Google Scholar]
- Arbour, J.; Blais, M.C.; Carioto, L.; Sylvestre, D. Clinical leptospirosis in three cats (2001–2009). J. Am. Anim. Hosp. Assoc. 2012, 48, 256–260. [Google Scholar] [CrossRef]
- Miotto, B.A.; Guilloux, A.; Tozzi, B.F.; Moreno, L.Z.; da Hora, A.S.; Dias, R.A.; Heinemann, M.B.; Moreno, A.M.; Filho, A.F.D.S.; Lilenbaum, W.; et al. Prospective study of canine leptospirosis in shelter and stray dog populations: Identification of chronic carriers and different Leptospira species infecting dogs. PLoS ONE 2018, 13, e0200384. [Google Scholar] [CrossRef]
- Mohammed, H.; Nozha, C.; Hakim, K.; Abdelaziz, F. Leptospira: Morphology, classification and pathogenesis. J. Bacteriol. Parasitol. 2011, 2, 120. [Google Scholar] [CrossRef]
- Schuller, S.; Francey, T.; Hartman, K.; Hugonnard, M.; Kohn, B.; Nally, J.E.; Sykes, J. European consensus statement on leptospirosis in dogs and cats. J. Small Anim. Pract. 2015, 56, 159–179. [Google Scholar] [CrossRef] [PubMed]
- de Paula Dreer, M.K.; Gonçalves, D.D.; da Silva Caetano, I.C.; Gerônimo, E.; Menegas, P.H.; Bergo, D.; Lopes-Mori, F.M.R.; Benitez, A.; de Freitas, J.C.; Evers, F.; et al. Toxoplasmosis, leptospirosis and brucellosis in stray dogs housed at the shelter in Umuarama municipality, Paraná, Brazil. J. Venom. Anim. Toxins Incl. Trop. Dis. 2013, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.C.; Najera, P.; Pereira, M.M.; Machado, G.; dos Anjos, C.B.; Rodrigues, R.O.; Cavagni, G.M.; Muñoz-Zanzi, C.; Corbellini, L.G.; Leone, M.; et al. Leptospirosis in Rio Grande do Sul, Brazil: An Ecosystem Approach in the Animal-Human Interface. PLoS Negl. Trop. Dis. 2015, 9, e0004095. [Google Scholar] [CrossRef]
- Ward, M.P. Clustering of reported cases of leptospirosis among dogs in the United States and Canada. Prev. Vet. Med. 2002, 56, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Mwachui, M.A.; Crump, L.; Hartskeerl, R.; Zinsstag, J.; Hattendorf, J. Environmental and Behavioural Determinants of Leptospirosis Transmission: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003843. [Google Scholar] [CrossRef]
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef]
- Soo, Z.M.P.; Khan, N.A.; Siddiqui, R. Leptospirosis: Increasing importance in developing countries. Acta Trop. 2020, 201, 105183. [Google Scholar] [CrossRef]
- Goarant, C. Leptospirosis: Risk factors and management challenges in developing countries. Res. Rep. Trop. Med. 2016, 7, 49–62. [Google Scholar] [CrossRef]
- Brightman, C. Leptospirosis: A leisure and occupational hazard. Trends Urol. Men’s Health 2018, 9, 29–31. [Google Scholar] [CrossRef]
- Rojas, P.; Monahan, A.M.; Schuller, S.; Miller, I.S.; Markey, B.K.; Nally, J.E. Detection and quantification of leptospires in urine of dogs: A maintenance host for the zoonotic disease leptospirosis. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1305–1309. [Google Scholar] [CrossRef]
- Wang, Y.; Zhuang, X.; Zhong, Y.; Zhang, C.; Zhang, Y.; Zeng, L.; Zhu, Y.; He, P.; Dong, K.; Pal, U.; et al. Distribution of plasmids in distinct Leptospira pathogenic species. PLoS Negl. Trop. Dis. 2015, 9, e0004220-14. [Google Scholar] [CrossRef] [PubMed]
- Spangler, D.; Kish, D.; Beigel, B.; Morgan, J.; Gruszynski, K.; Naikare, H.; Nahar, V.K.; Coarsey, M.D.; Verma, A. Leptospiral shedding and seropositivity in shelter dogs in the Cumberland Gap Region of Southeastern Appalachia. PLoS ONE 2020, 15, e0228038. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.W.; Couto, C.G. Small Animal Medicine, 3rd ed.; Mosby: St. Louis, MO, USA, 2003. [Google Scholar]
- Pennisi, M.G.; Caprì, A.; Solano-Gallego, L.; Lombardo, G.; Torina, A.; Masucci, M. Prevalence of antibodies against Rickettsia conorii, Babesia canis, Ehrlichia canis, and Anaplasma phagocytophilum antigens in dogs from the Stretto di Messina area (Italy). Ticks Tick. Borne Dis. 2012, 3, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, S.; Figarolli, B.M.; D’Incau, M.; Foschi, G.; Gennero, M.S.; Giordani, R.; Natale, A.; Papa, P.; Ponti, N.; Scaltrito, D.; et al. Serological surveillance of Leptospirosis in Italy: Two-year national data (2010–2011). Vet. Ital. 2016, 52, 129–138. [Google Scholar] [CrossRef]
- Scanziani, E.; Origgi, F.; Giusti, A.M.; Iacchia, G.; Vasino, A.; Pirovano, G.; Scarpa, P.; Tagliabue, S. Serological survey of leptospiral infection in kennelled dogs in Italy. J. Small Anim. Pract. 2002, 43, 154–157. [Google Scholar] [CrossRef]
- Ehricht, R.; Slickers, P.; Goellner, S.; Hotzel, H.; Sachse, K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell Probes. 2006, 20, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Ghalmi, F.; China, B.; Kaidi, R.; Daube, G.; Losson, B. Detection of Neospora caninum in dog organs using real time PCR systems. Vet. Parasitol. 2008, 155, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Schets, F.M.; de Heer, L.; de Roda Husman, A.M. Coxiella burnetii in sewage water at sewage water treatment plants in a Q fever epidemic area. Int. J. Hyg. Environ. Health. 2013, 216, 698–702. [Google Scholar] [CrossRef]
- Briciu, V.T.; Sebah, D.; Coroiu, G.; Lupşe, M.; Cârstina, D.; Ţăţulescu, D.F.; Mihalca, A.D.; Gherman, C.M.; Leucuţa, D.; Meyer, F.; et al. Immunohistochemistry and real-time PCR as diagnostic tools for detection of Borrelia burgdorferi sensu lato in ticks collected from humans. Exp. Appl. Acarol. 2016, 69, 49–60. [Google Scholar] [CrossRef]
- Ivacic, L.; Reed, K.D.; Mitchell, P.D.; Ghebranious, N. A LightCycler TaqMan assay for detection of Borrelia burgdorferi sensu lato in clinical samples. Diagn. Microbiol. Infect. Dis. 2007, 57, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Edvinsson, B.; Lappalainen, M.; Evengård, B.; ESCMID Study Group for Toxoplasmosis. Real-time PCR targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Clin. Microbiol. Infect. 2006, 12, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Burg, J.L.; Grover, C.M.; Pouletty, P.; Boothroyd, J.C. Direct and sensitive detection of a pathogenic protozoan, Toxoplasma gondii, by polymerase chain reaction. J. Clin. Microbiol. 1989, 27, 1787–1792. [Google Scholar] [CrossRef]
- Hu, R.L.; Huang, G.; Qiu, W.; Zhong, Z.H.; Xia, X.Z.; Yin, Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet. Res. Commun. 2001, 25, 77–84. [Google Scholar] [CrossRef]
- Buonavoglia, C.; Martella, V.; Pratelli, A.; Tempesta, M.; Cavalli, A.; Buonavoglia, D.; Bozzo, G.; Elia, G.; Decaro, N.; Carmichael, L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001, 82, 3021–3025. [Google Scholar] [CrossRef] [PubMed]
- Touihri, L.; Bouzid, I.; Daoud, R.; Desario, C.; El Goulli, A.F.; Decaro, N.; Ghorbel, A.; Buonavoglia, C.; Bahloul, C. Molecular characterization of canine parvovirus-2 variants circulating in Tunisia. Virus Genes. 2009, 38, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Grippi, F.; Cannella, V.; Macaluso, G.; Blanda, V.; Emmolo, G.; Santangelo, F.; Vicari, D.; Galluzzo, P.; Sciacca, C.; D’Agostino, R.; et al. Serological and Molecular Evidence of Pathogenic Leptospira spp. in Stray Dogs and Cats of Sicily (South Italy), 2017–2021. Microorganisms 2023, 11, 385. [Google Scholar] [CrossRef]
- Stear, M. O.I.E. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2018, 8th ed.; OIE 2018, Chapter. 3.1.12., Par. B 2.1. 2014; OIE: Paris, France, 2018. [Google Scholar]
- Aliberti, A.; Blanda, V.; Di Marco Lo Presti, V.; Macaluso, G.; Galluzzo, P.; Bertasio, C.; Sciacca, C.; Arcuri, F.; D’Agostino, R.; Ippolito, D.; et al. Leptospira interrogans Serogroup Pomona in a Dairy Cattle Farm in a Multi-Host Zootechnical System. Vet Sci. 2022, 9, 83. [Google Scholar] [CrossRef]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through Taqman polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Galluzzo, P.; Grippi, F.; Di Bella, S.; Santangelo, F.; Sciortino, S.; Castiglia, A.; Sciacca, C.; Arnone, M.; Alduina, R.; Chiarenza, G. Seroprevalence of Borrelia burgdorferi in Stray Dogs from Southern Italy. Microorganisms 2020, 8, 1688. [Google Scholar] [CrossRef]
- Bertasio, C.; Papetti, A.; Scaltriti, E.; Tagliabue, S.; D’Incau, M.; Boniotti, M.B. Serological Survey and Molecular Typing Reveal New Leptospira Serogroup Pomona Strains among Pigs of Northern Italy. Pathogens 2020, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef]
- Salaün, L.; Mérien, F.; Gurianova, S.; Baranton, G.; Picardeau, M. Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J. Clin. Microbiol. 2006, 44, 3954–3962. [Google Scholar] [CrossRef]
- Weiss, S.; Menezes, A.; Woods, K.; Chanthongthip, A.; Dittrich, S.; Opoku-Boateng, A.; Kimuli, M.; Chalker, V. An Extended Multilocus Sequence Typing (MLST) Scheme for Rapid Direct Typing of Leptospira from Clinical Samples. PLoS Negl. Trop. Dis. 2016, 10, e0004996. [Google Scholar] [CrossRef]
- Macaluso, G.; Torina, A.; Blanda, V.; Guercio, A.; Lastra, A.; Giacchino, I.; D’Agostino, R.; Sciacca, C.; D’Incau, M.; Bertasio, C.; et al. Leptospira in Slaughtered Fattening Pigs in Southern Italy: Serological Survey and Molecular Typing. Animals 2022, 12, 585. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Harkin, K.R.; Roshto, Y.M.; Sullivan, J.T.; Purivs, T.J.; Chengappa, M.M. Comparison of polymerase chain reaction assay, bacteriologic culture, and serologic testing in assessment of prevalence of urinary shedding of leptospires in dogs. J. Am. Vet. Med. Assoc. 2003, 222, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, R.; Vieira, A.S.; Oliveira, J.; Lilenbaum, W. Asymptomatic leptospiral infection is associated with canine chronic kidney disease. Comp. Immunol. Microbiol. Infect. Dis. 2019, 62, 64–67. [Google Scholar] [CrossRef]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Peru-United States Leptospirosis Consortium, Leptospirosis: A zoonotic disease of global importance. Lancet. 2003, 12, 757–771. [Google Scholar] [CrossRef]
- Grippi, F.; Vesco, G.; Santangelo, F.; Giannitrapani, V.; Gargano, V.; Randazzo, V.; Francaviglia, F.; Giunta, F.; Lombardo, S.; Giambruno, P. Caso di leptospirosi canina: Focolaio di infezione presso il Canile Municipale di Palermo (Sicilia) e sua gestione. Summa-Anim. Compagnia 2015, 1, 61–68. [Google Scholar]
- Balboni, A.; Mazzotta, E.; Boniotti, M.B.; Bertasio, C.; Bellinati, L.; Lucchese, L.; Battilani, M.; Ceglie, L.; Marchione, S.; Esposito, G.; et al. Outbreak of Leptospira borgpetersenii Serogroup Sejroe Infection in Kennel: The Role of Dogs as Sentinel in Specific Environments. Int. J. Environ. Res. Public Health 2022, 19, 3906. [Google Scholar] [CrossRef]
- Piredda, I.; Ponti, M.N.; Piras, A.; Palmas, B.; Pintore, P.; Pedditzi, A.; Chisu, V. New Insights on Leptospira Infections in a Canine Population from North Sardinia, Italy: A Sero-Epidemiological Study. Biology 2021, 10, 507. [Google Scholar] [CrossRef]
- Ellis, W.A. Control of canine leptospirosis in Europe: Time for a change? Vet. Rec. 2010, 167, 602–605. [Google Scholar] [CrossRef]
- Delaude, A.; Rodriguez-Campos, S.; Dreyfus, A.; Counotte, M.J.; Francey, T.; Schweighauser, A.; Lettry, S.; Schuller, S. Canine leptospirosis in Switzerland—A prospective cross-sectional study examining seroprevalence, risk factors and ur- inary shedding of pathogenic leptospires. Prev. Vet. Med. 2017, 141, 48–60. [Google Scholar] [CrossRef]
- Majed, Z.; Bellenger, E.; Postic, D.; Pourcel, C.; Baranton, G.; Picardeau, M. Identification of variable-number tandem-repeat loci in leptospira interrogans sensu stricto. J. Clin. Microbiol. 2005, 43, 539–545. [Google Scholar] [CrossRef]
- Kmety, E.; Dikken, H. Classification of the Species Leptospira Interrogans and History of its Serovars; University Press Groningen: Groningen, The Netherlands, 1993. [Google Scholar]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Samir, A.; Soliman, R.; El-Hariri, M.; Abdel-Moein, K.; Hatem, M.E. Leptospirosis in animals and human contacts in Egypt: Broad range surveillance. Rev. Soc. Bras. Med. Trop. 2015, 48, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.C.; Jancloes, M.; Buss, D.F.; Aldighieri, S.; Bertherat, E.; Najera, P.; Galan, D.I.; Durski, K.; Espinal, M.A. Leptospirosis: A silent epidemic disease. Int. J. Environ. Res. Public Health 2013, 10, 7229–7234. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Leptospirosis: Risk of Exposure. Available online: https://www.cdc.gov/leptospirosis/exposure/index.html (accessed on 9 February 2023).
- Awosanya, E.J.; Nguku, P.; Oyemakinde, A.; Omobowale, O. Factors Associated with Probable Cluster of Leptospirosis among Kennel Workers in Abuja, Nigeria. Pan Afr. Med. J. 2013, 16, 144. [Google Scholar] [CrossRef]
- André-Fontaine, G. Canine leptospirosis—Do we have a problem? Vet. Microbiol. 2006, 117, 19–24. [Google Scholar] [CrossRef]
- Novak, A.; Hindriks, E.; Hoek, A.; Veraart, C.; Broens, E.M.; Ludwig, I.; Rutten, V.; Sloots, A.; Broere, F. Cellular and humoral immune responsiveness to inactivated Leptospira interrogans in dogs vaccinated with a tetravalent Leptospira vaccine. Vaccine 2023, 41, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.; Stirling, C.; Thomas, A.; King, V.; Plevová, E.; Chromá, L.; Siedek, E.; Illambas, J.; Salt, J.; Sture, G. Duration of immunity of a multivalent (DHPPi/L4R) canine vaccine against four Leptospira serovars. Vaccine 2013, 31, 3126–3130. [Google Scholar] [CrossRef] [PubMed]
| Group | Vaccination Status | Number of Dogs |
|---|---|---|
| A | Never vaccinated | 42/67 |
| B | Vaccination between 1 and 2 years ago | 12/67 |
| C | Vaccinated more than 2 years ago | 13/67 |
| Time | Negative Samples (%) | Positive Samples (%) |
|---|---|---|
| T0 | 58/66 (87.9%) | 8/66 (12.1%) |
| T1 | 62/66 (93.9%) | 4/66 (6.1%) |
| ID Positive Sample (n = 66) | Leptospira spp. Serogroup/Serovar | Titer T0 | Titer T1 | Vaccination Status |
|---|---|---|---|---|
| Dog n. 1 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 1600 | 1600 | |
| L. interrogans serogroup Australis serovar Bratislava | 800 | 200 | Group A | |
| L. borgpetersenii serogroup Ballum serovar Ballum | 100 | <100 | ||
| Dog n. 2 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 400 | <100 | |
| L. interrogans serogroup Australis serovar Bratislava | 100 | <100 | Group A | |
| Dog n. 3 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 200 | 200 | Group A |
| Dog n. 4 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 200 | 200 | Group A |
| Dog n. 5 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 200 | 100 | Group A |
| Dog n. 6 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 100 | <100 | Group B |
| L. interrogans serogroup Australis serovar Bratislava | 100 | <100 | ||
| Dog n. 7 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 100 | <100 | Group B |
| Dog n. 8 | L. interrogans serogroup Icterohaemorragiae serovar Copenhageni | 1100 | <100 | Group B |
| Sample | ST | glmU | pntA | sucA | tpiA | pfkB | mreA | caiB |
|---|---|---|---|---|---|---|---|---|
| Puppy | 17 | 1 | 1 | 2 | 2 | 10 | 4 | 8 |
| Dog n. 1 | 17 (partial) | n.d. | 1 | 2 | 2 | 10 | n.d. | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grippi, F.; Blanda, V.; Galluzzo, P.; Bongiorno, M.; Sciacca, C.; Arcuri, F.; D’Agostino, R.; Giacchino, I.; Gucciardi, F.; D’Incau, M.; et al. A Canine Leptospirosis Clinical Case Due to Leptospira interrogans (Serogroup Icterohaemorrhagiae) in a Dog Kennel in Castelvetrano (Western Sicily, South Italy). Vet. Sci. 2023, 10, 508. https://doi.org/10.3390/vetsci10080508
Grippi F, Blanda V, Galluzzo P, Bongiorno M, Sciacca C, Arcuri F, D’Agostino R, Giacchino I, Gucciardi F, D’Incau M, et al. A Canine Leptospirosis Clinical Case Due to Leptospira interrogans (Serogroup Icterohaemorrhagiae) in a Dog Kennel in Castelvetrano (Western Sicily, South Italy). Veterinary Sciences. 2023; 10(8):508. https://doi.org/10.3390/vetsci10080508
Chicago/Turabian StyleGrippi, Francesca, Valeria Blanda, Paola Galluzzo, Manuel Bongiorno, Carmela Sciacca, Francesca Arcuri, Rosalia D’Agostino, Ilenia Giacchino, Francesca Gucciardi, Mario D’Incau, and et al. 2023. "A Canine Leptospirosis Clinical Case Due to Leptospira interrogans (Serogroup Icterohaemorrhagiae) in a Dog Kennel in Castelvetrano (Western Sicily, South Italy)" Veterinary Sciences 10, no. 8: 508. https://doi.org/10.3390/vetsci10080508
APA StyleGrippi, F., Blanda, V., Galluzzo, P., Bongiorno, M., Sciacca, C., Arcuri, F., D’Agostino, R., Giacchino, I., Gucciardi, F., D’Incau, M., Bertasio, C., Torina, A., & Guercio, A. (2023). A Canine Leptospirosis Clinical Case Due to Leptospira interrogans (Serogroup Icterohaemorrhagiae) in a Dog Kennel in Castelvetrano (Western Sicily, South Italy). Veterinary Sciences, 10(8), 508. https://doi.org/10.3390/vetsci10080508







