Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Origin and Collection
2.2. DNA Extraction, Sequencing
2.3. Bioinformatics Analysis
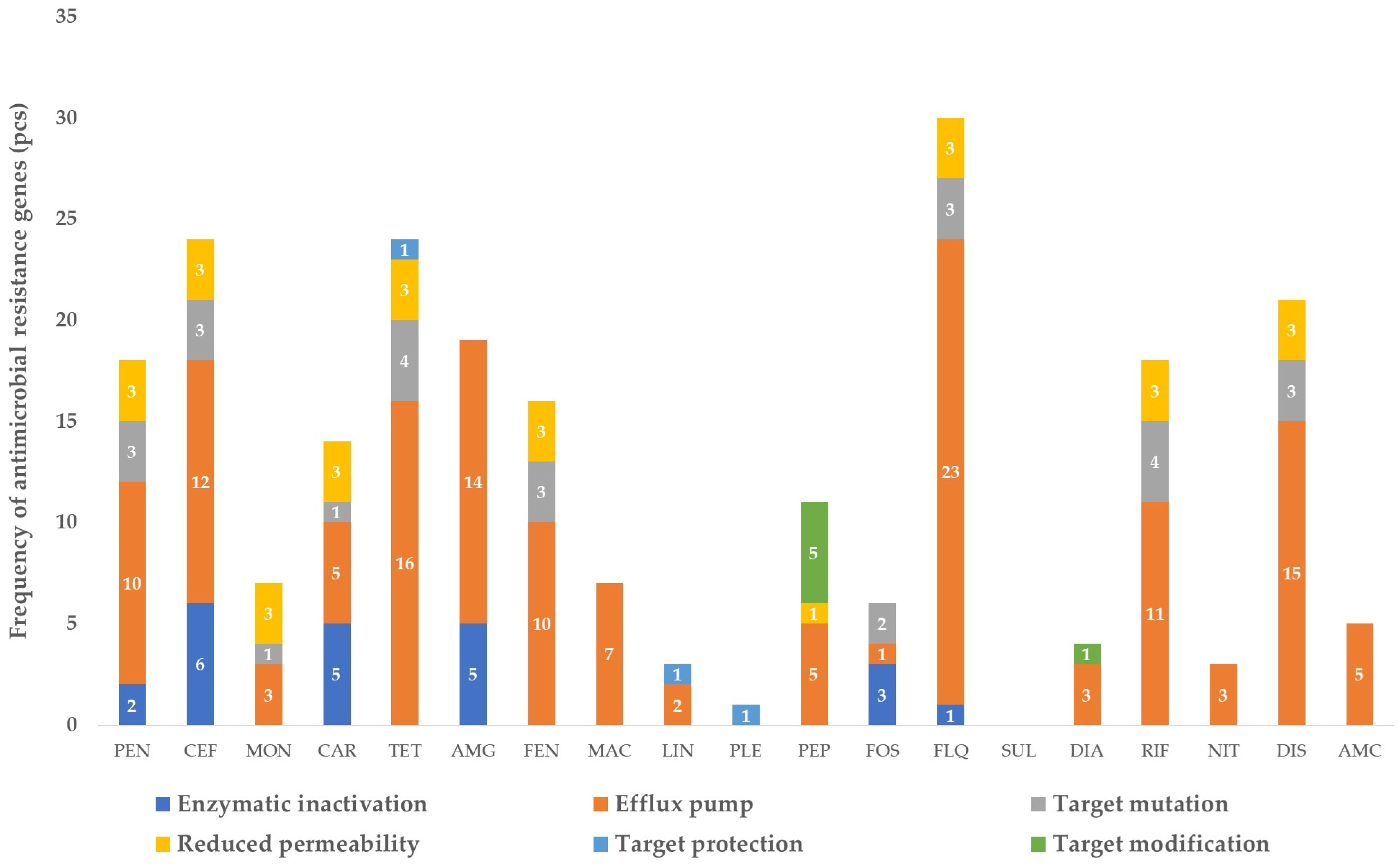
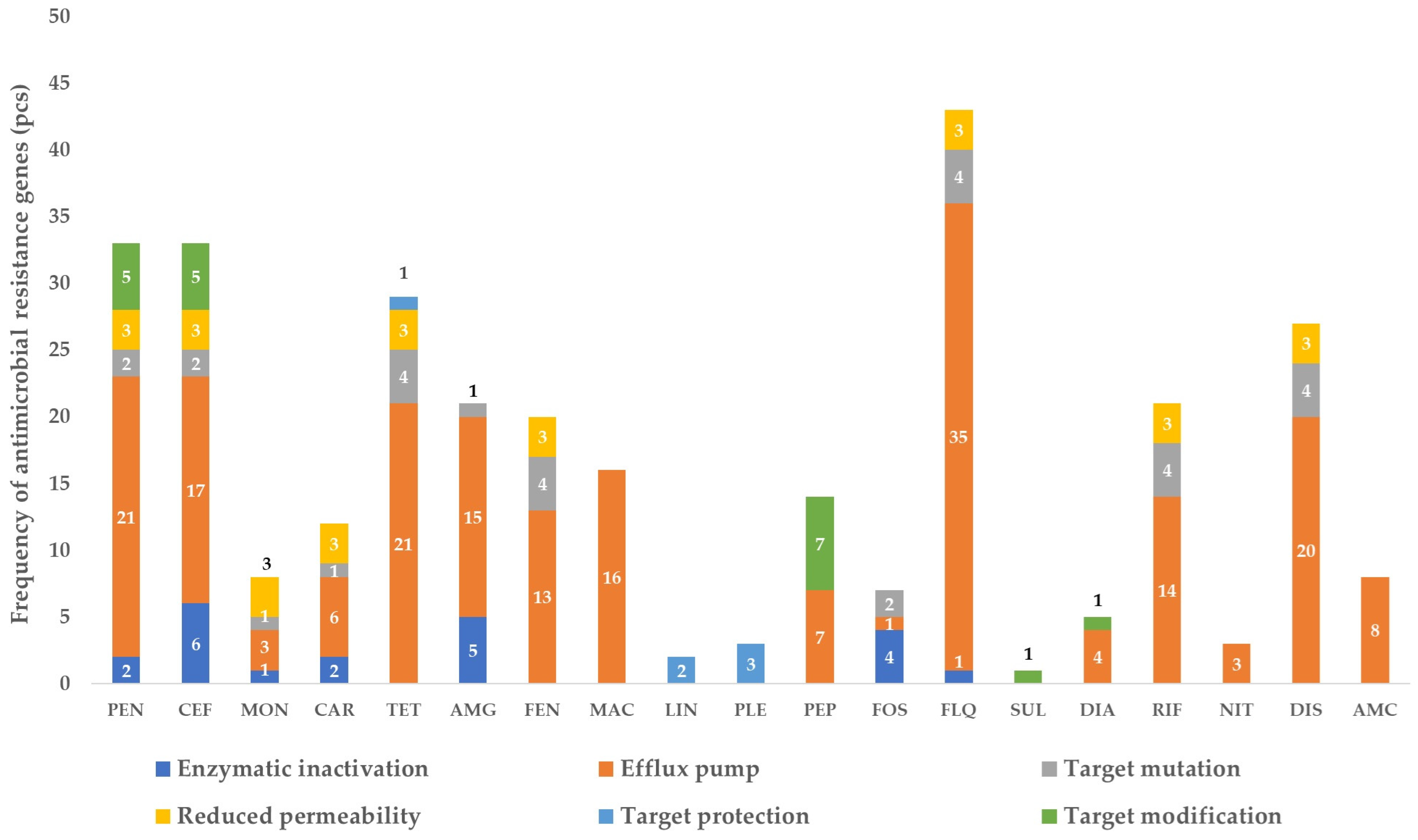
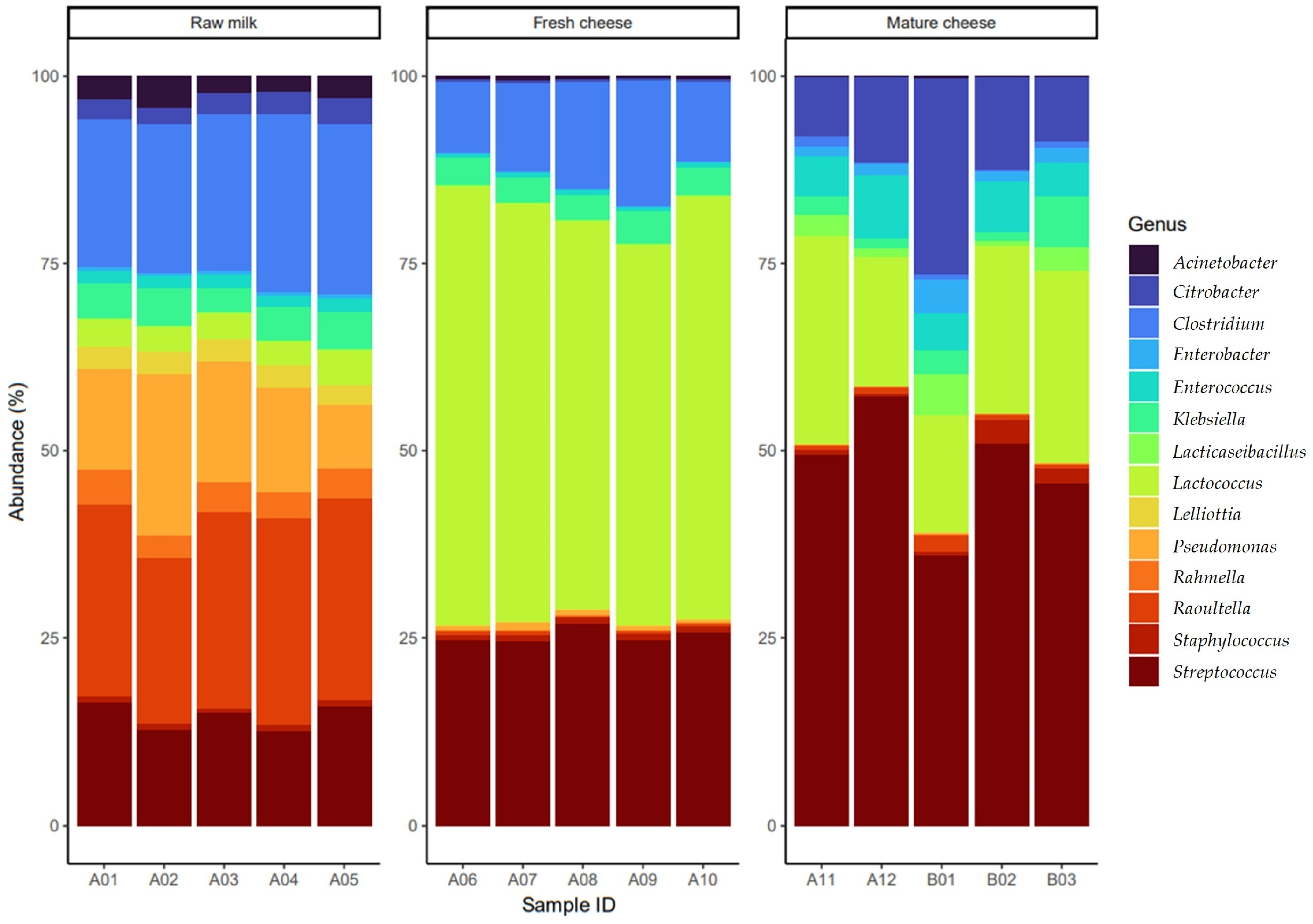
3. Results
3.1. The Gene Pool Identified from the Samples
3.2. Frequency of the Identified Genes per Sample
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Veeraraghavan, B.; Elangovan, R.; Vivekanandan, P. Antibiotic Resistance and Epigenetics: More to It than Meets the Eye. Antimicrob. Agents Chemother. 2020, 64, e02225-19. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F. Threats of Antibiotic Resistance: An Obliged Reappraisal. Int. Microbiol. 2021, 24, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic Resistance and Persistence—Implications for Human Health and Treatment Perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef] [PubMed]
- Benmazouz, I.; Kövér, L.; Kardos, G. The Rise of Antimicrobial Resistance in Wild Birds: Potential AMR Sources and Wild Birds as AMR Reservoirs and Disseminators: Literature Review. Hung. Vet. J. 2024, 146, 91–105. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Use on Australian Dairy Cattle Farms—A Survey of Veterinarians—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/35325612/ (accessed on 23 July 2023).
- Masebo, N.T.; Marliani, G.; Shannon Del Re, F.; Abram, L.; Cavallini, D.; Di Pietro, M.; Beltrame, A.; Schiavon, E.; Bolcato, M.; Hernandez Bermudez, J.; et al. Evaluation of Antimicrobial and Non-Steroidal Anti-Inflammatory Treatments for BRD on Health and Welfare in Fattening Bulls: A Cross-Sectional Study. Vet. Q. 2024, 44, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Masebo, N.T.; Marliani, G.; Cavallini, D.; Accorsi, P.A.; Di Pietro, M.; Beltrame, A.; Gentile, A.; Jacinto, J.G.P. Health and Welfare Assessment of Beef Cattle during the Adaptation Period in a Specialized Commercial Fattening Unit. Res. Vet. Sci. 2023, 158, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Activity of Selected Antimicrobial Agents against Strains of Staphylococcus Aureus Isolated from Bovine Intramammary Infections That Produce Beta-Lactamase—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/9149974/ (accessed on 10 August 2023).
- Güler, L.; Ok, Ü.; Gündüz, K.; Gülcü, Y.; Hadimli, H.H. Antimicrobial Susceptibility and Coagulase Gene Typing of Staphylococcus Aureus Isolated from Bovine Clinical Mastitis Cases in Turkey. J. Dairy Sci. 2005, 88, 3149–3154. [Google Scholar] [CrossRef]
- Myllyniemi, A.-L.; Evira, E.; Lääkelaitos (Eds.) FINRES-Vet. 2005–2006: Finnish Veterinary Antimicrobial Resistance Monitoring and Consumption of Antimicrobial Agents; Evira Publications; Finnish Food Safety Authority Evira: National Agency for Medicines: Helsinki, Finland, 2007; ISBN 978-952-5662-99-3. [Google Scholar]
- Saini, V.; McClure, J.T.; Scholl, D.T.; DeVries, T.J.; Barkema, H.W. Herd-Level Association between Antimicrobial Use and Antimicrobial Resistance in Bovine Mastitis Staphylococcus Aureus Isolates on Canadian Dairy Farms. J. Dairy Sci. 2012, 95, 1921–1929. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.-M.; Lu, L.-M.; Ren, G.-W.N.; Cao, X.-Y.; Shen, J.-Z. Macrolide-Lincosamide-Resistant Phenotypes and Genotypes of Staphylococcus Aureus Isolated from Bovine Clinical Mastitis. Vet. Microbiol. 2008, 130, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Makovec, J.A.; Ruegg, P.L. Results of Milk Samples Submitted for Microbiological Examination in Wisconsin from 1994 to 2001. J. Dairy Sci. 2003, 86, 3466–3472. [Google Scholar] [CrossRef] [PubMed]
- Sabour, P.M.; Gill, J.J.; Lepp, D.; Pacan, J.C.; Ahmed, R.; Dingwell, R.; Leslie, K. Molecular Typing and Distribution of Staphylococcus Aureus Isolates in Eastern Canadian Dairy Herds. J. Clin. Microbiol. 2004, 42, 3449–3455. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, Á.V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial Resistance Genes in Raw Milk for Human Consumption. Sci. Rep. 2020, 10, 7464. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of Antimicrobial Resistance Genes in Retail Raw Milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Scientific Opinion on the Public Health Risks Related to the Consumption of Raw Drinking Milk | EFSA. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/3940 (accessed on 6 August 2023).
- The European Union One Health 2020 Zoonoses Report—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/36329690/ (accessed on 6 August 2023).
- Nikoloudaki, O.; Lemos Junior, W.J.F.; Campanaro, S.; Di Cagno, R.; Gobbetti, M. Role Prediction of Gram-Negative Species in the Resistome of Raw Cow’s Milk. Int. J. Food Microbiol. 2021, 340, 109045. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing Technologies—The next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 April 2022).
- Krueger, F.; James, F.; Ewels, P.; Afyounian, E.; Schuster-Boeckler, B. FelixKrueger/TrimGalore: v0.6.7—DOI via Zenodo. 2021. Available online: https://zenodo.org/records/5127899 (accessed on 6 August 2023).
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Czajkowski, M.D.; Vance, D.P.; Frese, S.A.; Casaburi, G. GenCoF: A Graphical User Interface to Rapidly Remove Human Genome Contaminants from Metagenomic Datasets. Bioinformatics 2019, 35, 2318–2319. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-Node SOLUTION for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 15, 1674–1676. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of Mobile Genetic Elements Associated with Antibiotic Resistance in Salmonella Enterica Using a Newly Developed Web Tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, K.; Ben Abdelaziz, A. Principal Component Analysis (PCA). Tunis. Med. 2021, 99, 383–389. [Google Scholar] [PubMed]
- Wang, Q.Q.; Yu, S.C.; Qi, X.; Hu, Y.H.; Zheng, W.J.; Shi, J.X.; Yao, H.Y. Overview of logistic regression model analysis and application. Zhonghua Yu Fang Yi Xue Za Zhi 2019, 53, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Dexter, F. Wilcoxon-Mann-Whitney Test Used for Data That Are Not Normally Distributed. Anesth. Analg. 2013, 117, 537–538. [Google Scholar] [CrossRef]
- De’ath, G. The Multinomial Diversity Model: Linking Shannon Diversity to Multiple Predictors. Ecology 2012, 93, 2286–2296. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Chen, G.; Alekseyenko, A.V. PERMANOVA-S: Association Test for Microbial Community Composition That Accommodates Confounders and Multiple Distances. Bioinformatics 2016, 32, 2618–2625. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Milk and Milk Product Statistics. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Milk_and_milk_product_statistics (accessed on 6 August 2023).
- Silveira, A.; Carvalho, J.P.; Loh, L.; Benusic, M. Public Health Risks of Raw Milk Consumption: Lessons from a Case of Paediatric Hemolytic Uremic Syndrome. Can. Commun. Dis. Rep. 2023, 49, 375–379. [Google Scholar] [CrossRef]
- Costard, S.; Espejo, L.; Groenendaal, H.; Zagmutt, F.J. Outbreak-Related Disease Burden Associated with Consumption of Unpasteurized Cow’s Milk and Cheese, United States, 2009–2014. Emerg. Infect. Dis. J. 2017, 23, 6. [Google Scholar] [CrossRef] [PubMed]
- Alegbeleye, O.O.; Guimarães, J.T.; Cruz, A.G.; Sant’Ana, A.S. Hazards of a ‘Healthy’ Trend? An Appraisal of the Risks of Raw Milk Consumption and the Potential of Novel Treatment Technologies to Serve as Alternatives to Pasteurization. Trends Food Sci. Technol. 2018, 82, 148–166. [Google Scholar] [CrossRef]
- Andriyanov, P.A.; Zhurilov, P.A.; Kashina, D.D.; Tutrina, A.I.; Liskova, E.A.; Razheva, I.V.; Kolbasov, D.V.; Ermolaeva, S.A. Antimicrobial Resistance and Comparative Genomic Analysis of Elizabethkingia Anophelis Subsp. Endophytica Isolated from Raw Milk. Antibiotics 2022, 11, 648. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Maróti, G.; Jerzsele, Á.; Dubecz, A.; Patai, Á.V.; Judge, M.F.; Nagy, S.Á.; Makrai, L.; Bányai, K.; et al. A Glimpse of Antimicrobial Resistance Gene Diversity in Kefir and Yoghurt. Sci. Rep. 2020, 10, 22458. [Google Scholar] [CrossRef] [PubMed]
- Aragão, B.B.; Trajano, S.C.; Silva, J.G.; Silva, B.P.; Oliveira, R.P.; Junior, J.W.P.; Peixoto, R.M.; Mota, R.A. Short Communication: High Frequency of β-Lactam-Resistant Staphylococcus Aureus in Artisanal Coalho Cheese Made from Goat Milk Produced in Northeastern Brazil. J. Dairy. Sci. 2019, 102, 6923–6927. [Google Scholar] [CrossRef] [PubMed]
- Elafify, M.; Khalifa, H.O.; Al-Ashmawy, M.; Elsherbini, M.; El Latif, A.A.; Okanda, T.; Matsumoto, T.; Koseki, S.; Abdelkhalek, A. Prevalence and Antimicrobial Resistance of Shiga Toxin-Producing Escherichia Coli in Milk and Dairy Products in Egypt. J. Environ. Sci. Health B 2020, 55, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Alexa Oniciuc, E.A.; Walsh, C.J.; Coughlan, L.M.; Awad, A.; Simon, C.A.; Ruiz, L.; Crispie, F.; Cotter, P.D.; Alvarez-Ordóñez, A. Dairy Products and Dairy-Processing Environments as a Reservoir of Antibiotic Resistance and Quorum-Quenching Determinants as Revealed through Functional Metagenomics. mSystems 2020, 5, e00723-19. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Zhou, R.; Li, M. Antimicrobial Resistance and Molecular Typing of Staphylococcus Aureus Isolates from Raw Milk in Hunan Province. PeerJ 2023, 11, e15847. [Google Scholar] [CrossRef]
- Rodrigues, M.X.; Silva, N.C.C.; Trevilin, J.H.; Cruzado, M.M.B.; Mui, T.S.; Duarte, F.R.S.; Castillo, C.J.C.; Canniatti-Brazaca, S.G.; Porto, E. Molecular Characterization and Antibiotic Resistance of Staphylococcus spp. isolated from cheese processing plants. J. Dairy Sci. 2017, 100, 5167–5175. [Google Scholar] [CrossRef]
- Ashraf, D.; Ombarak, R.A.; Samir, A.; Abdel-Salam, A.B. Characterization of Multidrug-Resistant Potential Pathogens Isolated from Milk and Some Dairy Products in Egypt. J. Adv. Vet. Anim. Res. 2023, 10, 275–283. [Google Scholar] [CrossRef]
- Liu, H.; Dong, L.; Zhao, Y.; Meng, L.; Wang, J.; Wang, C.; Zheng, N. Antimicrobial Susceptibility, and Molecular Characterization of Staphylococcus Aureus Isolated from Different Raw Milk Samples in China. Front. Microbiol. 2022, 13, 840670. [Google Scholar] [CrossRef] [PubMed]
- Endres, C.M.; Moreira, E.; de Freitas, A.B.; Castel, A.P.D.; Graciano, F.; Mann, M.B.; Frazzon, A.P.G.; Mayer, F.Q.; Frazzon, J. Evaluation of Enterotoxins and Antimicrobial Resistance in Microorganisms Isolated from Raw Sheep Milk and Cheese: Ensuring the Microbiological Safety of These Products in Southern Brazil. Microorganisms 2023, 11, 1618. [Google Scholar] [CrossRef] [PubMed]
- Parry-Hanson Kunadu, A.; Holmes, M.; Miller, E.L.; Grant, A.J. Microbiological Quality and Antimicrobial Resistance Characterization of Salmonella spp. in Fresh Milk Value Chains in Ghana. Int. J. Food Microbiol. 2018, 277, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (U.S.). Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GA, USA, 2019. [Google Scholar]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-Generation Approaches to Understand and Combat the Antibiotic Resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic Antibiotic Resistance: Mechanisms, Origins, Challenges and Solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.-C. Monitoring Bacterial Communities in Raw Milk and Cheese by Culture-Dependent and -Independent 16S rRNA Gene-Based Analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.K.; Carstens, C.K.; Ramachandran, P.; Shazer, A.G.; Narula, S.S.; Reed, E.; Ottesen, A.; Schill, K.M. Metagenomics of Pasteurized and Unpasteurized Gouda Cheese Using Targeted 16S rDNA Sequencing. BMC Microbiol. 2018, 18, 189. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for Detection of Subpopulations of Bacteria Not Previously Associated with Artisanal Cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef]
- Lusk, T.S.; Ottesen, A.R.; White, J.R.; Allard, M.W.; Brown, E.W.; Kase, J.A. Characterization of Microflora in Latin-Style Cheeses by Next-Generation Sequencing Technology. BMC Microbiol. 2012, 12, 254. [Google Scholar] [CrossRef]
- Place, R.B.; Hiestand, D.; Gallmann, H.R.; Teuber, M. Staphylococcus equorum Subsp. Linens, Subsp. Nov., a Starter Culture Component for Surface Ripened Semi-Hard Cheeses. Syst. Appl. Microbiol. 2003, 26, 30–37. [Google Scholar] [CrossRef]
- Massa, S.; Gardini, F.; Sinigaglia, M.; Guerzoni, M.E. Klebsiella Pneumoniae as a Spoilage Organism in Mozzarella Cheese. J. Dairy Sci. 1992, 75, 1411–1414. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.; Kiviniemi, K.; Korkeala, H. Hazard and Control of Group II (Non-Proteolytic) Clostridium Botulinum in Modern Food Processing. Int. J. Food Microbiol. 2006, 108, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, H.R.; Meshgi, M.A.; Jafari, N.J.; Izadi, M.; Ranjbar, R.; Fooladi, A.A.I. A Survey of Traditional Iranian Food Products for Contamination with Toxigenic Clostridium Botulinum. J. Infect. Public Health 2009, 2, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Kovács, D.; Palkovicsné Pézsa, N.; Farkas, O.; Jerzsele, Á. Usage of Antibiotic Alternatives in Pig Farming: Literature Review. Hung. Vet. J. 2021, 143, 281–282. [Google Scholar]
- Sebők, C.; Márton, R.A.; Meckei, M.; Neogrády, Z.; Mátis, G. Antimicrobial Peptides as New Tools to Combat Infectious Diseases. Hung. Vet. J. 2024, 146, 181–191. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antibacterial Efficiency of Propolis—Part 1. Hung. Vet. J. 2022, 144, 285–298. [Google Scholar]
- Olasz, Á.; Jerzsele, Á.; Balta, L.; Dobra, P.F.; Kerek, Á. In Vivo Efficacy of Different Extracts of Propolis in Broiler Salmonellosis. Hung. Vet. J. 2023, 145, 461–475. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antiprotozoal and Antifungal Efficiency of Propolis—Part 2. Hung. Vet. J. 2022, 144, 691–704. [Google Scholar]
- Kerek, Á.; Csanády, P.; Tuska-Szalay, B.; Kovács, L.; Jerzsele, Á. In Vitro Efficacy of Hungarian Propolis against Bacteria, Yeast, and Trichomonas Gallinae Isolated from Pigeons—A Possible Antibiotic Alternative? Resources 2023, 12, 101. [Google Scholar] [CrossRef]
- Kovács, L.; Nagy, D.; Könyves, L.; Jerzsele, Á.; Kerek, Á. Antimicrobial Properties of Essential Oils—Animal Health Aspects. Hung. Vet. J. 2023, 145, 497–510. [Google Scholar]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What Is a Resistance Gene? Ranking Risk in Resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Hankin, L.; Lacy, G.H.; Stephens, G.R.; Dillman, W.F. Antibiotic-Resistant Bacteria in Raw Milk and Ability of Some to Transfer Antibiotic Resistance to Escherichia Coli. J. Food Prot. 1979, 42, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Kerek, Á.; Török, B.; Jerzsele, Á. MEGA-Plate—New Evolutionary and Coselection Microbiological Method. Hung. Vet. J. 2022, 144, 429–439. [Google Scholar]
- Kerek, Á.; Török, B.; Laczkó, L.; Kardos, G.; Bányai, K.; Somogyi, Z.; Kaszab, E.; Bali, K.; Jerzsele, Á. In Vitro Microevolution and Co-Selection Assessment of Florfenicol Impact on Escherichia Coli Resistance Development. Antibiotics 2023, 12, 1728. [Google Scholar] [CrossRef] [PubMed]
- Kerek, Á.; Török, B.; Laczkó, L.; Somogyi, Z.; Kardos, G.; Bányai, K.; Kaszab, E.; Bali, K.; Jerzsele, Á. In Vitro Microevolution and Co-Selection Assessment of Amoxicillin and Cefotaxime Impact on Escherichia Coli Resistance Development. Antibiotics 2024, 13, 247. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, R.; Gopal, D.R.; Dhandapani, R.; Subbarayalu, R.; Elangovan, M.P.; Prabhu, B.; Veerappan, V.; Nandheeswaran, A.; Paramasivam, S.; Muthupandian, S. Is AMR in Dairy Products a Threat to Human Health? An Updated Review on the Origin, Prevention, Treatment, and Economic Impacts of Subclinical Mastitis. Infect. Drug Resist. 2023, 16, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Muca, E.; Buonaiuto, G.; Lamanna, M.; Silvestrelli, S.; Ghiaccio, F.; Federiconi, A.; De Matos Vettori, J.; Colleluori, R.; Fusaro, I.; Raspa, F.; et al. Reaching a Wider Audience: Instagram’s Role in Dairy Cow Nutrition Education and Engagement. Animals 2023, 13, 3503. [Google Scholar] [CrossRef]
- Muca, E.; Cavallini, D.; Raspa, F.; Bordin, C.; Bergero, D.; Valle, E. Integrating New Learning Methods into Equine Nutrition Classrooms: The Importance of Students’ Perceptions. J. Equine Vet. Sci. 2023, 126, 104537. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerek, Á.; Németh, V.; Szabó, Á.; Papp, M.; Bányai, K.; Kardos, G.; Kaszab, E.; Bali, K.; Nagy, Z.; Süth, M.; et al. Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption. Vet. Sci. 2024, 11, 265. https://doi.org/10.3390/vetsci11060265
Kerek Á, Németh V, Szabó Á, Papp M, Bányai K, Kardos G, Kaszab E, Bali K, Nagy Z, Süth M, et al. Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption. Veterinary Sciences. 2024; 11(6):265. https://doi.org/10.3390/vetsci11060265
Chicago/Turabian StyleKerek, Ádám, Virág Németh, Ábel Szabó, Márton Papp, Krisztián Bányai, Gábor Kardos, Eszter Kaszab, Krisztina Bali, Zoltán Nagy, Miklós Süth, and et al. 2024. "Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption" Veterinary Sciences 11, no. 6: 265. https://doi.org/10.3390/vetsci11060265
APA StyleKerek, Á., Németh, V., Szabó, Á., Papp, M., Bányai, K., Kardos, G., Kaszab, E., Bali, K., Nagy, Z., Süth, M., & Jerzsele, Á. (2024). Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption. Veterinary Sciences, 11(6), 265. https://doi.org/10.3390/vetsci11060265







