Simple Summary
Classical swine fever is a highly contagious disease that severely affects the swine industry worldwide. The clinical symptoms of infected pigs are unusually complex, making it difficult to distinguish between wild-type and vaccinated pigs. In this study, a dual TaqMan-MGB RT-qPCR method for the identification of wild-type CSFV and attenuated vaccine strains was established. This newly developed method could specifically detect CSFV and HCLV with no cross-reactivity with other swine pathogens. The detection limit for the NS3 gene of CSFV and HCLV was 1.67 × 101 copies/μL, respectively. For precision testing, the repeatability and reproducibility of the relative standard deviation was less than 2%. This method was successfully used for rapid detection of 193 real samples. This dual TaqMan-MGB RT-qPCR technology meets the needs for early and rapid detection of CSFV, and can provide an efficient and rapid detection method for the investigation and analysis of CSFV field strain infections and vaccine immune status, thereby laying the foundation for better prevention and control of the occurrence and spread of CSF.
Abstract
To establish a rapid real-time RT-PCR method for differentiating wild-type classical swine fever virus (CSFV) strains from vaccine strains (HCLV), we designed a universal primer targeting the NS3 gene to detect wild-type CSFV strains and vaccine strains simultaneously, and two TaqMan-MGB probes were designed to differentiate between wild-type and vaccine strains. After optimizing the RT-qPCR conditions, a rapid dual TaqMan-MGB RT-qPCR method for the detection and identification of CSFV and HCLV was developed. The results showed that method could specifically detect CSFV and HCLV with no cross-reactivity with other swine pathogens. The analytic sensitivity for the NS3 gene of CSFV and HCLV were 1.67 × 101 copies/μL, respectively. For precision testing, the repeatability and reproducibility of the test was less than 2%. This method was successfully used for the rapid detection of 193 biological samples collected from CSFV-vaccinated pigs. This fast and accurate detection technology can be used for the detection of CSFV and is suitable for differentiating between wild-type CSFV strains and vaccine strains.
1. Introduction
Classical swine fever (CSF) is a highly contagious disease characterized by high fever and hemorrhage [1]. CSF has a severe impact on the swine industry and is listed as an A-class legal infectious disease by the World Organization for Animal Health (WOAH) [2]. CSF is caused by the classical swine fever virus (CSFV), which belongs to the Flaviviridae family and the Pestivirus genus [3]. Pigs of all ages can be infected with CSFV, which has strong infectivity, rapid spread, and causes a high mortality rate [4]. The clinical manifestations of CSF include fever, cough, ataxia, conjunctivitis, diarrhea, stillbirth, and abortion [5,6].
CSFV is an enveloped, positive-sense, single-stranded RNA virus with a genome size of 12.3 kb [7]. Based on the differences in the sequences of E2 gene, 5′ non-coding region (5′ NCR) and the polymerase gene (NS5B), CSFV can be divided into three genetic groups: I, II, and III [8]. Phylogenetic analysis indicated that strain C (vaccine strain) belongs to genetic group I and can elicit immune protection against highly virulent strains and various genetic groups. Epidemiological investigations of CSFV in China have shown that all wild-type CSFV strains belong to genetic group II [9], and the widespread use of the C strain vaccine makes it difficult to distinguish between the wild-type and C strains, even during laboratory testing [10]. Currently, there are reports on the use of real-time RT-PCR to differentiate wild-type CSFV strains from the C strain [11]. The TaqMan MGB probe is a molecular beacon based on the principle of fluorescence resonance energy transfer (FRET). It can detect single-nucleotide mismatches and achieve high-precision detection of gene mutations with a high sensitivity and specificity [12]. Additionally, MGB probes exhibit high stability, allowing for hybridization reactions under high-temperature conditions, thus avoiding the issue of easy degradation encountered by traditional molecular beacons at room temperature [13,14]. To facilitate the rapid identification of classical swine fever virus and to differentiate between wild-type CSFV infections and vaccination with attenuated vaccines, this study established a dual TaqMan-MGB RT-qPCR method for the identification of wild-type CSFV and attenuated vaccine strains.
In this study, a pair of universal primers was designed within the NS3 gene to detect the wild-type CSFV and the vaccine strain simultaneously, and two TaqMan MGB probes were designed to detect and differentiate between the wild-type and C strains. This dual TaqMan-MGB RT-qPCR provides an effective detection method for the rapid and accurate detection of wild-type CSFV and C strains in the swine industry.
2. Material and Methods
2.1. The Strains Used in This Study
Five reference strains of CSFV (Brescia, Shimen, RUCSFPLUM, Paderborn, SSFV39) and vaccines with a titer of 104.7 TCID50/μL–105.5 TCID50/μL, were obtained from Sairuisi Biotechnology (Jilin) Co., Ltd., Changchun, China. And other commerce vaccines include classical swine fever virus vaccine (CVCC AV1412), Chinese Swine Fever Rabbit Attenuated Vaccine Strain (HCLV), High-pathogenicity Porcine Reproduction and Respiratory Syndrome vaccine, live (Strain JXA1-R); Porcine Reproduction and Respiratory Syndrome vaccine, live (Strain R9); Porcine Reproduction and Respiratory Syndrome vaccine, live (Strain CH-1R); Porcine Reproduction and Respiratory Syndrome vaccine, inactivated (Strain CH-1a); Swine Epidemic Encephalitis Vaccine, live (Strain SA14-14-2); Porcine Parvovirus vaccine, inactivated (Strain WH-1); Swine influenza virus H1N1 subtype, inactivated vaccine (TJ strain); Porcine pseudorabies vaccine, live vaccine (HB-98 strain); Classical Swine Fever Live Vaccine (Cell-Derived) (CVCC AV1412 Strain); Inactivated porcine circovirus disease type 2 vaccine (SH strain); Porcine transmissible gastroenteritis, porcine epidemic diarrhea, porcine rotavirus (GP5 type) live vaccine (flower virus strain + CV777 strain + NX strain); Foot and mouth disease type O and A bivalent 3B protein epitope deletion vaccine, inactivated (O/rV-1 strain + A/rV-2 strain); Haemophilus parasuis quadrivalent propolis inactivated vaccine (type 4 SD02 strain + type 5 HN02 strain + type 12 GZ01 strain + type 13 JX03 strain); Staphylococcus suis propolis inactivated vaccine (Staphylococcus suis group C BHZZ-L1 strain + Staphylococcus suis type 2 BHZZ-L4 strain); Trivalent inactivated vaccine for virulent colibacillosis in piglets (containing K88, K99, and 987P flagellar antigens); Porcine Mycoplasmal Pneumonia Inactivated Vaccine (Strain J); Triple live vaccine for swine fever, swine erysipelas and pasteurellosis (cell source + G4T10 strain + EO630 strain); Inactivated vaccine against porcine atrophic rhinitis (Bordetella pertussis strain JB5); Porcine infectious pleuropneumonia trivalent inactivated vaccine (serotype 1 9901 strain, serotype 2 XT9904 strain, serotype 7 GZ9903 strain). All these commerce vaccines were preserved at −20 °C in our laboratory. The manufacturers’ information for commercial vaccine strains are in supplementary Table S1.
2.2. Main Reagents and Instruments
A viral magnetic bead-based DNA/RNA extraction kit and bacterial magnetic bead-based DNA extraction kit were purchased from Sairuisi Biotechnology (Jilin) Co., Ltd. (Changchun, China). The Hifair® V C58P2 Multiplex One Step RT-qPCR Probe Kit (UDG Plus) was purchased from Yeasen Biotechnology (Shanghai) Co., Ltd., Shanghai, China. The 4S GREEN dye, 2000 DNA Marker, 500 DNA Marker, M-MuLV First Strand cDNA Synthesis Kit, and gel extraction and purification kit were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). A classical porcine swine fever real-time fluorescence RT-PCR detection kit was purchased from Beijing Anheal Laboratories Co., Ltd., Beijing, China. A real-time PCR instrument (Gentier 96R) was purchased from Xi’an Tianlong Biotechnology Co., Ltd. (Xi’an, China).
2.3. Preparation of Nucleic Acids from Pathogenic Microorganisms
The viral DNA/RNA and the bacterial DNA were extracted using the viral and bacterial magnetic bead-based DNA extraction kits, respectively, and the RNAs of the RNA virus were reverse transcribed using the First-Strand cDNA Synthesis Kit and stored at −20 °C for later use.
2.4. Primer and Probe Design
The full genome sequences of CSFV (GenBank NO: AF333000) and HCLV (GenBank NO: AY805221) were downloaded from GenBank, and multi-sequence alignment analysis was performed using the alignment program in the Vector NTI v11.0 (Thermo Fisher Scientific, Waltham, MA, USA). Primers were designed based on the NS3 gene site, and two TaqMan-MGB probes were designed based on the SNP sites in the NS3 gene to differentiate CSFV wild-type strains from the C strain (Figure 1). The specificity of the primers and probes was validated using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 6 September 2023). The primer and probe sequences are listed in Table 1. The primers and probes were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China).
Figure 1.
Sequence alignment information for primers and probes. The yellow labeled area represents the conserved region of the genes; the green and white labeled area represents the variant site of the genes.

Table 1.
Primer and probe sequences.
2.5. Preparation of Standard Positive Plasmids
CSFV and HCLV RNA were amplified using conventional reverse RT-PCR. The target NS3 genes were recovered from the agarose gel, and ligated with the pUC57 and pUCm -T vector, respectively. The ligation mixtures were transformed into E. coli DH5α competent cells, and the positive clones were selected; these clones were identified by PCR and sequencing. The correct clones containing NS3 genes were named CSFV-NS3-Plasmid and HCLV-NS3-Plasmid. According to the formula: Y (copies/μL) = (6.02 × 1023 copies/mol) × (X ng/μL × 10−9)/(DNA length × 660). The concentrations of the plasmids of CSFV-NS3-Plasmid and HCLV-NS3-Plasmid were 3.34 × 1010 copies/µL and 5.8 × 1010 copies/µL, respectively.
The CSFV-NS3-Plasmid and HCLV-NS3-Plasmid were diluted by a 10-fold serial dilutions, respectively. The two plasmids at the same concentration level were mixed at a ratio of 1:1 (v/v), and the concentration of the plasmid mixtures (CSFV-NS3-Plasmid and HCLV-NS3-Plasmid) was 1.67 × 1010 copies/µL to 1.67 × 100 copies/µL.
2.6. Optimization of Dual TaqMan-MGB RT-qPCR
To obtain an optimal RT-qPCR reaction system, 107 copies/μL, 106 copies/μL, and 105 copies/μL of the plasmid mixtures were used as the template for dual TaqMan-MGB RT-qPCR. The primer concentration was set at 0.2, 0.4, and 0.6 μmol/L (the probes’ concentrations were 0.2 μmol/L), after the optimal primer concentration was determined, the probes’ concentrations (0.1, 0.2, 0.3, and 0.4 μmol/L) were also tested. After the primers’ and probes’ concentrations were determined, the annealing temperature (58 °C, 60 °C, and 62 °C) was optimized.
2.7. Establishment of the Standard Curve
To establish the standard curve, 103 copies/µL to 107 copies/µL of the plasmid mixtures were used as the DNA templates and then amplified by dual TaqMan-MGB RT-qPCR method. For the standard curve, each concentration was run in triplicate.
2.8. Analytic Specificity
To validate the specificity of the assay for CSFV detection, a dual TaqMan-MGB RT-qPCR was employed for CSFV detection, and HCLV and other pathogens (Section 2.1) were used for cross-reactivity testing. At the same time, to evaluate the specificity of the assay for HCLV detection, dual TaqMan-MGB RT-qPCR was employed for HCLV and CSFV detection, and other pathogens mentioned above were utilized for cross-reactivity testing.
2.9. Analytic Sensitivity
To determine the analytic sensitivity of dual TaqMan-MGB RT-qPCR, 100 copies/µL to 108 copies/µL of the plasmid mixture were used as the DNA templates, with ddH2O as the negative control. The analytic sensitivity of the dual TaqMan-MGB RT-qPCR was evaluated. Besides that, 10-fold gradient dilution of CSFV and HCLV attenuated vaccines, ranging from 105.5 TCID50/tube to 10−0.5 TCID50/tube. Dual TaqMan-MGB RT-qPCR was conducted to assess the sensitivity of the detection, using ddH2O as the negative control.
2.10. Inclusiveness of the Test
Five classic epidemic strains of CSFV (Brescia, Shimen, RUCSFPLUM, Paderborn, SSFV39) were chosen for inclusiveness assessment of the test. The strains were diluted to the lowest limit of detection of the test, and then each of these strains was tested 3 times using dual TaqMan-MGB RT-qPCR technology.
2.11. Precision of the Test
The reproducibility of the experiment was evaluated using a moderately positive sample consisting of a plasmid mixture at a concentration of 103 copies/µL, a weakly positive sample with a plasmid mixture at a concentration of 101 copies/µL, and a negative control composed of deionized water (ddH2O); the same batch of dual TaqMan-MGB RT-qPCR solution was tested three times.
For the reproducibility of the test: this test was performed at two laboratories [including Molecular Diagnostic Science and Technology Innovation Center of Jilin Agricultural Science and Technology University, and Research Laboratory of Sairuisi Biotechnology (Jilin) Co., Ltd.], with two brands of qPCR instruments (Gentier 96R, Tianlong Xian, China and StepOne Plus, Applied Biosystem, Foster City, CA, USA), and two operators and two batches of reagents (batch numbers 20240608 and 20240610) were used for qPCR amplification. Each dilution was repeated three times to calculate the standard deviation (SD) and relative standard deviation (RSD) of the Ct value, to assess the reproducibility of the experimental results.
2.12. Detection in Biological Samples Collected from CSFV-Vaccinated Pigs
During the sample collection process, samples were obtained from six pig farms, in Jilin City, China, where no swine fever epidemics had been reported. A total of six samples was collected, which including 35 lymph samples, 28 meat samples, 40 throat swabs, 40 whole blood samples, and 50 fecal samples from 70 pigs that had been vaccinated within the past two weeks. To verify the accuracy of the test, the 193 samples were selected and tested using a commercial real-time qPCR kit for CSFV (Beijing Shiji Yuanheng Animal Disease Control Technology Co., Ltd., Beijing, China).
3. Results
3.1. Optimization of Dual TaqMan-MGB qPCR for CSFV and HCLV
The reaction conditions were studied by optimizing the primer and probe concentrations and the RT-qPCR program. Table 2 indicates that when the concentrations of the primers were 0.2 μmol/L and 0.4 μmol/L, the Ct value was higher and the repeatability was poor, and when the primer concentration was 0.6 μmol/L, the Ct value was the lowest. Thus, 0.6 μmol/L of the primer was used in this test. Considering the amplification efficiency, Ct value, and repeatability, the optimal concentration of the probes was determined at 0.1 μmol/L. The difference in the detection results among different annealing temperatures were small, and the annealing temperature was selected as 60 °C. Therefore, the final detection reaction system was set as follows: primer concentration 0.6 μmol/L, probe concentration (CSFV-P and HCLV-P) 0.1 μmol/L, and optimal annealing temperature 60 °C.

Table 2.
Optimization results of dual real-time RT-PCR.
The optimized dual TaqMan-MGB qPCR reaction system was as follows: 15 µL of 2× Hifair®V C58P2 MP Buffer, 1.2 µL of Hifair®V C58P2 Enzyme Mix, 1.8 µL of the CSFV-F/CSFV-R (each at 10 µmol/L), 0.3 µL of the probe (CSFV-P and HCLV-P) (each at 10 µmol/L), 5 µL of template, and 4.6 µL of RNase Free H2O. The reaction program was as follows: 50 °C for 5 min; 95 °C for 10 s; 95 °C for 1 s, and 60 °C for 20 s, 45 cycles.
3.2. Preparation of the Standard Curve
To establish a standard curve for the test, 10-fold serial dilutions of the CSFV-NS3-Plasmid and HCLV-NS3-Plasmid were used as DNA templates and subjected to TaqMan-MGB RT-qPCR. The detection results showed that this real-time PCR technique has a good linear relationship for the detection of CSFV-NS3-Plasmid, with an amplification efficiency of 98.874% and a correlation coefficient R2 = 0.99. For HCLV-NS3-Plasmid, the detection also had a good linear relationship, with an amplification efficiency of 103.404% and a correlation coefficient R2 = 0.99 (Figure 2).
Figure 2.
Standard curve for the dual RT-qPCR.
3.3. Analytic Specificity
The DNA samples prepared as described in Section 2.3 were used to evaluate the specificity of the test. Three repeated detections of dual TaqMan-MGB RT-qPCR showed that there were only specific amplification curves for CSFV, and no amplification curves were observed for HCLV and the other DNA samples or the negative control. In addition, dual TaqMan-MGB RT-qPCR also showed that there were only specific amplification curves for HCLV, and no amplification curves were observed for CSFV and the other DNA samples or the negative control, indicating that the dual TaqMan-MGB RT-qPCR method established in this study had good specificity (Figure 3).
Figure 3.
Specificity of dual RT-qPCR. (A) Detection result for CSFV, (B) detection result for HCLV.
3.4. Analytic Sensitivity
To assess the analytic sensitivity of the test, 10-fold serial dilutions of the plasmid mixture were used for RT-qPCR (Figure 4). This showed that the analytic sensitivity of dual TaqMan-MGB RT-qPCR for NS3 genes was 1.67 × 101 copies/μL.
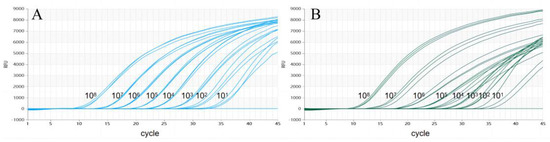
Figure 4.
The detection limit of dual RT-qPCR. (A) Detection result for CSFV-NS3-Plasmid. (B) Detection result for HCLV-NS3-Plasmid).
In addition, the analytic sensitivity of the dual TaqMan-MGB RT-qPCR method was also evaluated by testing the virus cultures. Figure 5 illustrates that a positive result (Ct value ≤ 35) was obtained for CSFV at a dilution degree of 100.5, with good repeatability and stability. However, at a dilution of 10−0.5, no typical amplification curve was observed. This method demonstrated a detection sensitivity of 3.16 TCID50/μL for CSFV virus. Meanwhile, for HCLV, a positive result (Ct value ≤ 35) was obtained at a dilution degree of 100.5, and no typical amplification curve was observed when the dilution degree was 10−0.5, indicating that the detection sensitivity of the test was 3.16 TCID50/μL for HCLV virus. Additionally, the detection limit of the commercial qPCR kit was also up to 3.16 TCID50/μL for HCLV virus.
Figure 5.
Detection limit testing. (A) The detection limit of dual RT-qPCR for CSFV and HCLV. The green amplification curves represent the detection result for CSFV, the orange amplification curves represent the detection result for HCLV (B) The detection limit of commercial qPCR kit for HCLV.
3.5. Inclusiveness of the Test
In this test, three repeated tests demonstrated that dual TaqMan-MGB RT-qPCR was able to identify five genotypes of strains, achieving a detection rate of 100%. The detection results are shown in Figure 6. This result showed that the newly developed technology has a good inclusiveness for the five classic epidemic strains.
Figure 6.
Inclusiveness of the test. The green amplification curves represent the amplification results of the pathogens, the horizontal straight lines represent the negative controls (ddH2O).
3.6. Precision of the Test
The repeatability of the test was evaluated as described in Section 2.11. The RSD values were ≤2% (Table 3). Indicating that, the method developed in this study has a high detection precision.

Table 3.
Detection precision for dual real-time RT-PCR.
For the reproducibility of the test, different instruments, reagent batches, laboratories, and operators were chosen for method reproducibility experiments. The results indicated that the SD values of the detection on three different concentrations of plasmid mixtures were less than 1.0, and the RSD values were less than 2.0%, which demonstrates that there was a good reproducibility of the method.
3.7. Detection in Biological Samples Collected from CSFV-Vaccinated Pigs
A total of 193 samples were collected and tested using this method. The results showed that the positive rate for CSFV detection was 16.1% (lymph samples, 2.0%; meat, 1.0%; pharyngeal swabs, 3.6%; whole blood, 3.6%; feces, 5.6%), and the positive rate for HCLV detection was 2.9% (lymph samples, 0%; meat, 1%; pharyngeal swabs, 0%; whole blood, 1.5%; feces, 0%). To verify the accuracy of the assay, 193 samples were tested using a commercial qPCR kit. The results showed that the positive rate for CSFV detection was 18.1% (lymph samples 2%, meat 1.5%, pharyngeal swabs 3.6%, whole blood 5.6%, feces 5.2%). The results showed that TaqMan-MGB dual RT-qPCR is comparable to the commercial qPCR kit (Table 4). The samples detected as positive for HCLV in the dual TaqMan-MGB RT-qPCR assay that could not be detected by the commercial qPCR kit were confirmed as positive using DNA sequencing. These results indicated that dual TaqMan-MGB RT-qPCR could be used for the differential diagnosis of CSFV and HCLV in field samples.

Table 4.
Comparison of dual real-time PCR and commercial kits.
4. Discussion
Establishing a rapid, accurate, and specific method for differentiating between wild-type and vaccine strains of CSF is crucial for the subsequent prevention and control of CSF. Currently, there have been numerous studies on CSFV detection methods utilizing qPCR, with a primary focus on developing qPCR assays for CSFV. Limited research exists on distinguishing between wild-type CSFV and vaccine strains [15,16]. In this study, based on the alignment results of representative gene sequences of CSFV and HCLV, the highly conserved gene NS3 was selected as the target for gene primer and probe design. Within the primers, two TaqMan-MGB probes were designed based on the SNP sites of CSFV and HCLV, with FAM and VIC fluorescent dyes labeled at the 5′ ends and MGB labeled at the 3′ ends of the probes. After systematic optimization of the RT-qPCR conditions, a one-step dual TaqMan-MGB fluorescent quantitative RT-PCR method for CSFV and HCLV was established.
In this test, the specificity, sensitivity, and detection ability of the biological samples collected from CSFV-vaccinated pigs were evaluated. This study showed that the method is specific for the detection of CSFV and HCLV, and it yielded negative results for 19 other common pig pathogens. The detection limits for CSFV and HCLV were 1.67 × 101 copies/μL, respectively. The detection of CSFV and HCLV in 193 biological samples collected from CSFV-vaccinated pigs showed that the positive detection rates of CSFV and HCLV in the samples were 16.1% and 2.9%, respectively. The samples positive for HCLV in the dual TaqMan-MGB RT-qPCR assay were also confirmed to be positive by DNA sequencing. Indicating that dual TaqMan-MGB RT-qPCR can be used for the differential diagnosis of CSFV and HCLV in field samples.
Despite recent advances, the newly developed assay is also dependent on traditional multi-step DNA extraction before PCR amplification processes, which restricts its usefulness for on-site pathogen detection. Future research will investigate direct amplification qPCR as a possible solution for on-site CSFV and HCLV detection. In summary, the one-step dual TaqMan-MGB RT-qPCR technology established in this study meets the needs for early and rapid detection of CSFV, and can provide an efficient and rapid detection method for the investigation and analysis of CSFV field strain infections and vaccine immune status, thereby laying the foundation for better prevention and control of the occurrence and spread of CSF.
5. Conclusions
This study established a new one-step dual TaqMan-MGB RT-qPCR detection technology for the simultaneous differentiation of wild-type CSFV and vaccine strains. This technology has high specificity, high sensitivity, and high precision and has been successfully used for the detection of CSFV on pig farms. This technology is a useful tool for distinguishing natural infections from wild-type CSFV in vaccinated pig herds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci11070289/s1, Table S1. Vaccine details.
Author Contributions
Data curation, Y.Z., M.W. and H.Z.; investigation, Y.Z. and X.X.; methodology, Y.Z., M.W. and Y.S.; writing—original draft preparation, S.W.; Resources, P.L.; validation, Y.L. and Y.M.; conceptualization, R.Y.; writing—Review & Editing, R.Y.; funding acquisition, Y.S. and R.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by Jilin Province Science and Technology Development Plan Project (20210204115YY, 20210203180SF). Meanwhile, the present study was supported by the innovation and entrepreneurship project for college students of Jilin province (GJ202211439020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are included within this article and Supplementary Materials.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Blome, S.; Staubach, C.; Henke, J.; Carlson, J.; Beer, M. Classical Swine Fever-An Updated Review. Viruses 2017, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Guo, Z.; Ding, N.Z.; He, C.Q. Studying classical swine fever virus: Making the best of a bad virus. Virus Res. 2015, 197, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeier, H.; Strebelow, G.; Depner, K.; Hoffmann, B.; Beer, M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J. Gen. Virol. 2004, 85, 3647–3652. [Google Scholar] [CrossRef] [PubMed]
- Moennig, V. The control of classical swine fever in wild boar. Front. Microbiol. 2015, 6, 160779. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Avalos Ramirez, R.; Orlich, M.; Cedillo Rosales, S.; Konig, M.; Schweizer, M.; Stalder, H.; Schirrmeier, H.; Thiel, H.J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Brown, V.R.; Bevins, S.N. A Review of Classical Swine Fever Virus and Routes of Introduction into the United States and the Potential for Virus Establishment. Front. Vet. Sci. 2018, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Ganges, L.; Crooke, H.R.; Bohórquez, J.A.; Postel, A.; Sakoda, Y.; Becher, P.; Ruggli, N. Classical swine fever virus: The past, present and future. Virus Res. 2020, 289, 198151. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Schmeiser, S.; Bernau, J.; Meindl-Boehmer, A.; Pridotkas, G.; Dirbakova, Z.; Mojzis, M.; Becher, P. Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet. Res. 2012, 43, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B. Classical Swine Fever in China-An Update Minireview. Front. Vet. Sci. 2019, 6, 187. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Liao, Y.; Zhang, M.; Liu, C.; Li, Z.; Li, Y.; Li, X.; Wu, K.; Yi, L.; Ding, H.; et al. Anti-Classical Swine Fever Virus Strategies. Microorganisms 2021, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, H.; Everett, H.; Sosan, O.; Crooke, H.; Meindl-Bohmer, A.; Qiu, H.J.; Moennig, V.; Belak, S.; Widen, F. A generic real-time TaqMan assay for specific detection of lapinized Chinese vaccines against classical swine fever. J. Virol. Methods 2011, 175, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hoffmann, B.; Baule, C.; Beer, M.; Belak, S.; Widen, F. Two real-time RT-PCR assays of classical swine fever virus, developed for the genetic differentiation of naturally infected from vaccinated wild boars. J. Virol. Methods 2009, 159, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Jamnikar Ciglenecki, U.; Grom, J.; Toplak, I.; Jemersic, L.; Barlic-Maganja, D. Real-time RT-PCR assay for rapid and specific detection of classical swine fever virus: Comparison of SYBR Green and TaqMan MGB detection methods using novel MGB probes. J. Virol. Methods 2008, 147, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, H.; Belak, S.; Widen, F. Development of a primer-probe energy transfer real-time PCR assay for improved detection of classical swine fever virus. J. Virol. Methods 2009, 160, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Dias, N.L.; Fonseca Júnior, A.A.; Oliveira, A.M.; Sales, E.B.; Alves, B.R.; Dorella, F.A.; Camargos, M.F. Validation of a real time PCR for classical Swine Fever diagnosis. Vet. Med. Int. 2014, 2014, 171235. [Google Scholar] [CrossRef] [PubMed]
- Haines, F.J.; Hofmann, M.A.; King, D.P.; Drew, T.W.; Crooke, H.R. Development and validation of a multiplex, real-time RT PCR assay for the simultaneous detection of classical and African swine fever viruses. PLoS ONE 2013, 8, e71019. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).