Simple Summary
The infection of animals and humans with Entamoeba spp. causes significant economic losses to the livestock industry and threatens public health. Animals are potential reservoirs for human infection with Entamoeba spp. Hence, investigations of Entamoeba infections in livestock are important for better control of human infection with Entamoeba spp. However, to date, infection of Entamoeba spp. in sheep and cattle in Shanxi Province remains unknown. In the present study, fecal samples were collected from 311 sheep, 392 dairy cattle, and 393 beef cattle from three representative counties in the northern, central, and southern regions of Shanxi Province and were investigated for Entamoeba spp. by PCR amplification and sequencing. The results showed that the overall infection rates of Entamoeba were 51.5% (160/311), 82.9% (325/392), and 79.1% (311/393) in sheep, dairy cattle, and beef cattle, respectively; and several Entamoeba species were identified, including Entamoeba bovis, Entamoeba Ribosomal Lineage (RL) 2, Entamoeba RL4, and Entamoeba RL8. The present study reports the occurrence and prevalence of Entamoeba spp. for the first time in sheep and cattle in Shanxi Province, which extended the geographical distribution of Entamoeba spp.
Abstract
Entamoeba spp. are common zoonotic intestinal protozoa, which can lead to serious intestinal diseases in both humans and animals through fecal–oral transmission, leading to significant economic losses and public health challenges. To reveal the prevalence of Entamoeba in sheep and cattle in Shanxi Province, North China, fecal samples were collected from 311 sheep, 392 dairy cattle, and 393 beef cattle from three representative counties in the northern, central, and southern regions of Shanxi Province. DNA was extracted from the fecal samples and amplified by PCR with primers targeting the nuclear small subunit ribosomal RNA (SSU rRNA) gene of Entamoeba spp., followed by the sequencing of the positive products. The overall infection rates of Entamoeba were 51.5% (160/311), 82.9% (325/392), and 79.1% (311/393) in sheep, dairy cattle, and beef cattle, respectively. Statistical analysis showed a significant correlation between the infection rate of Entamoeba and the location factor in sheep, dairy cattle, and beef cattle (p < 0.001). According to the obtained SSU rRNA sequences, several Entamoeba species, namely Entamoeba bovis, Entamoeba Ribosomal Lineage (RL) 2, Entamoeba RL4, and Entamoeba RL8, were identified. This study represents the first molecular survey of Entamoeba prevalence in sheep, beef cattle, and dairy cattle in Shanxi Province. The findings extend the geographical distribution of Entamoeba spp. and provide valuable scientific data for the prevention and control of amoebiasis in Shanxi Province.
1. Introduction
Entamoeba spp. are zoonotic protozoan parasites that inhabit the intestine and other organs of their hosts and are widely distributed in the natural environment [1]. Humans and animals are primarily infected with Entamoeba through the ingestion of mature cysts via the fecal–oral transmission route [1,2]. The genus Entamoeba includes several species capable of infecting humans, including Entamoeba histolytica, E. dispar, E. moshkovskii, E. coli, E. polecki, and E. hartmanni [3,4]. Of these, E. histolytica is recognized as the only pathogenic species of Entamoeba that causes amebic dysentery and other far-ranging diseases, including self-limiting colitis, invasive colitis, extra-intestinal infection, and invasive organ abscesses [5,6,7].
Amebiasis ranks as the third leading cause of parasitic mortality worldwide, following malaria and schistosomiasis [8,9]. It is estimated that approximately 500 million of the global population is infected with Entamoeba, with around 40,000–100,000 deaths annually due to amebiasis [1]. An epidemiological survey conducted in a small town in Ethiopia showed that 13.17% of children were infected with E. histolytica [10,11], highlighting its significant impact on child health in developing countries [1,12]. In addition to being identified in humans, Entamoeba infection has been reported in various animals, including pigs, alpaca, deer, and yaks, across multiple regions such as Australia, China, Costa Rica, Iceland, Japan, Libya, Sweden, Uganda, and the United Kingdom [13,14,15,16,17,18].
So far, several Entamoeba species/genotypes have been reported in ruminants, such as E. bovis and Entamoeba Ribosomal Lineage (RL) 1, Entamoeba RL2, and Entamoeba RL4 [19]. However, the prevalence and genetic characteristics of Entamoeba infection in sheep, dairy cattle, and beef cattle remain limited in China. Thus, the present study aimed to investigate the prevalence of Entamoeba in sheep, dairy cattle, and beef cattle in Shanxi Province, North China, and to identify the species/genotypes through PCR amplification and sequencing of the nuclear small subunit ribosomal RNA (SSU rRNA) sequences [20]. The findings are intended to provide essential data for the prevention and control of amoebiasis in Shanxi province.
2. Materials and Methods
2.1. Sample Collection
From May to November 2020, 311 fresh fecal samples from sheep (97 from Qi County, 108 from Jishan County, and 106 from Shanyin County), 392 fecal samples from dairy cattle (137 from Qi County, 49 from Jishan County, and 206 from Shanyin County), and 393 fecal samples from beef cattle (174 from Qi County and 219 from Jishan County) were collected from three representative counties of Shanxi province (Figure 1), respectively. The fecal samples were carefully separated using sterile polyethylene gloves to prevent contamination, and relevant sample information (e.g., region, sex, age, and host species/breed) was recorded. Each sample weighed between 10 g and 20 g. The collected samples were transported to the Laboratory of Parasitic Diseases, College of Veterinary Medicine, Shanxi Agricultural University, and stored at −20 °C for further analysis [21,22].
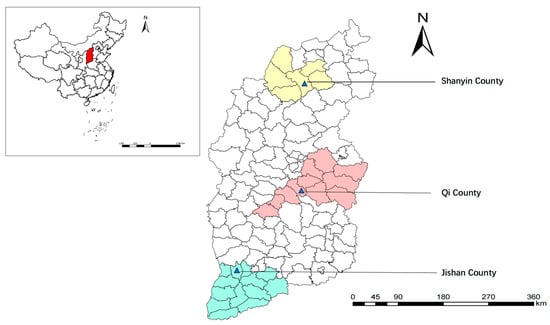
Figure 1.
Sampling sites of sheep and cattle feces in Shanxi province, North China. The map was generated by ArcGIS10.8 using data from the Resource and Environmental Science and Data Center of the Chinese Academy of Sciences.
2.2. DNA Extraction and PCR Amplification
The genomic DNA was extracted from each of the mixed fecal samples (approximately 200 mg) using the E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA) following the manufacturer’s instructions and was stored at −20 °C. The SSU rRNA gene of Entamoeba spp. was amplified by specific primers, EntboF2 (5′-TAAGAGGAACAATTGGGGTGAT-3′) and R3 (5′-AGGAATTCCTCGTTCAAAACAAA-3′), as described in a previous study [16]. The PCR reaction mixture consisted of 17.8 µL of ddH2O, 2.5 µL of 10× buffer (Mg2+ plus), 2 µL of dNTP (2.5 mM), 0.2 µL of Ex Taq, 1.5 µL of template DNA, and 0.5 µL of upstream and downstream primers. The PCR reaction conditions involved an initial pre-denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min, concluding with a final extension at 72 °C for 5 min, and cooling to 16 °C. Subsequently, the PCR products were examined through electrophoresis on a 1.5% agarose gel containing ethidium bromide and were visualized under ultraviolet light.
2.3. Sequence and Phylogenetic Analyses
Positive PCR amplicons (approximately 800 bp) were sent to Sangon Biotech Company (Shanghai, China) for sequencing. The species/genotypes of Entamoeba were determined by aligning and comparing the obtained sequences with known sequences available in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/, accessed on 5 July 2024). The phylogenetic relationship of species/genotypes was established by the neighbor-joining (NJ) method using the Kimura-2-parameter model in MEGA 7 software. The bootstrap value was set to 1000 times to evaluate the robustness of clusters.
2.4. Statistical Analysis
In this study, statistical analysis was performed using SPSS 26.0 (IBM, Chicago, IL, USA). The relationships between factors (e.g., region, age, and sex) and the infection rate of Entamoeba was assessed using the Chi-square test (χ2). The odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were calculated, with a p-value < 0.05 being considered statistically significant.
3. Results
3.1. Prevalence of Entamoeba spp. in Sheep
In this study, 160 of 311 sheep fecal samples tested positive for Entamoeba, resulting in an overall infection rate of 51.5% (160/311, 95% CI: 45.9–57.0) (Table 1). The highest infection rate was observed in sheep in Qi County (72.2%, 70/97, 95% CI: 63.2–81.1), followed by Jishan County (44.3%, 47/106, 95% CI: 34.9–53.8) and Shanyin County (39.8%, 43/108, 95% CI: 30.6–49.0), with significant regional differences (p < 0.001). In addition, lamb (≤6 months) had a significantly higher infection rate (75.7%, 53/70, 95% CI: 65.7–85.8) than older sheep (>6 months) (44.4%, 107/241, 95% CI: 38.1–50.7), showing a strong correlation between Entamoeba infection and age in sheep (p < 0.001).

Table 1.
Factors associated with prevalence of Entamoeba spp. in sheep.
3.2. Prevalence of Entamoeba spp. in Dairy Cattle
In dairy cattle, the prevalence of Entamoeba spp. was 82.9% (325/392, 95% CI: 79.2–86.6) in this study (Table 2). The highest prevalence was observed in dairy cattle in Jishan County (95.9%, 47/49, 95% CI: 90.4–100.0), followed by Qi County (89.1%, 122/137, 95% CI: 83.8–94.3) and Shanyin County (75.7%, 156/206, 95% CI: 69.9–81.6). Furthermore, the infection rates of Entamoeba were 82.7% (268/324, 95% CI: 78.6–86.8) in female cattle and 83.8% (57/68, 95% CI: 75.1–92.6) in male cattle, showing no significant gender difference (p > 0.05). Among the age groups, the infection rates of Entamoeba were 83.3% (180/216, 95% CI: 78.4–88.3) in adult dairy cattle (>18 months) and 82.4% (145/176, 95% CI: 76.8–88.0) in young dairy cattle (≤18 months), with no significant difference (p > 0.05).

Table 2.
Factors associated with prevalence of Entamoeba spp. in dairy cattle.
3.3. Prevalence of Entamoeba spp. in Beef Cattle
The overall prevalence of Entamoeba spp. in beef cattle from Qi County and Jishan County was 79.1% (311/393, 95% CI: 75.1–83.2) (Table 3). Notably, beef cattle in Jishan County had a significantly higher Entamoeba prevalence (85.8%, 188/219, 95% CI: 81.2–90.5) than those in Qi County (70.7%, 123/174, 95% CI: 63.9–77.5) (p < 0.001). No significant difference was found in the prevalence of Entamoeba between male beef cattle (81.6%, 151/185, 95% CI: 76.0–87.2) and female beef cattle (76.9%, 160/208, 95% CI: 71.2–82.6) (p > 0.05). However, the prevalence of Entamoeba in young beef cattle (≤12 months) (81.8%, 243/297, 95% CI: 77.4–86.2) was higher than that in adult beef cattle (>12 months) (70.8%, 68/96, 95% CI: 61.7–79.9), with a statistically significant difference (p < 0.05).

Table 3.
Factors associated with prevalence of Entamoeba spp. in beef cattle.
3.4. Sequence Alignment and Phylogenetic Analysis of Entamoeba
The samples with successful PCR amplification (approximately 800 bp) were sequenced, and phylogenetic analysis was performed. In the present study, 15 distinct sequences were identified (GenBank accession numbers PQ579357, PQ579823, and PQ604607–PQ604619) through sequencing analysis. The genetic distance between different species/genotypes was analyzed to determine their relationship. The results showed that the Entamoeba species/genotype infecting sheep, dairy cattle, and beef cattle in Shanxi Province were E. bovis, Entamoeba RL2, Entamoeba RL4, and Entamoeba RL8 (Figure 2).
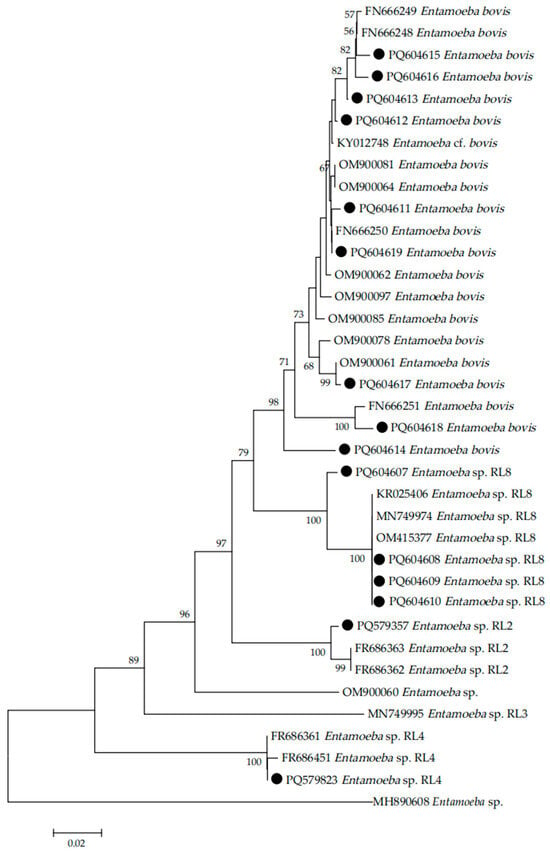
Figure 2.
Phylogenetic relationships of Entamoeba spp. based on the sequences of the small subunit ribosomal RNA (SSU rRNA) using the neighbor-joining (NJ) method. Representative Entamoeba SSU rRNA sequences obtained in this study are labeled with black circles (●). The bootstrap value is shown when >50%.
4. Discussion
Entamoeba spp. are among the most prevalent intestinal parasites in humans and large mammals, and they often parasitize ruminant animals such as cattle and sheep [8]. The prevalence of Entamoeba infection in animals has been documented domestically and internationally [23,24]. However, at present, research on enteric Entamoeba infection in even-toed ungulates in China is limited, particularly in Shanxi Province. To the best of our knowledge, the present study is the first to investigate Entamoeba spp. infection in sheep, dairy cattle, and beef cattle in Shanxi Province, North China using a molecular approach.
Previous studies have reported the infection of Entamoeba in ruminants in China by PCR technology. The infection rate of Entamoeba in sika deer in Anhui Province, China, was 31.5% (106/336); in Qinghai Province, China, the overall positive rate of Entamoeba among yaks was 36.32% (373/1027); in Shanxi Province, China, the overall positive rate of Entamoeba among alpacas was 18.03% (66/366); and it was 100% (57/57) in farm animals on the Qinghai-Tibetan Plateau of China [14,23,25,26]. The differences in the prevalence of Entamoeba may be due to several factors, such as geographical locations, detection methods, management patterns, and sampling sizes. Our study revealed a high Entamoeba prevalence of 82.9% (325/392) in dairy cattle and 79.1% (311/393) in beef cattle.
In the present study, the prevalence of Entamoeba infection in sheep, dairy cattle, and beef cattle was significantly different (p < 0.001) in different regions. In sheep, the highest prevalence of Entamoeba was detected in Qi County (72.2%), followed by Jishan County (44.3%) and Shanyin County (39.8%). The highest Entamoeba prevalence in cattle was observed in Jishan County, where the prevalence in dairy cattle (95.9%) and beef cattle (85.8%) was notably higher than that in Qi County (89.1% in dairy cattle and 70.7% in beef cattle) and Shanyin County (75.7% in dairy cattle). According to these findings, the higher prevalence of Entamoeba in sheep in Qi County might be related to local climate, farm hygiene conditions, feeding management patterns, and sample sizes [13]. However, the elevated positivity rate in dairy cattle and beef cattle in Jishan County could be associated with the regional latitude [21,27].
In terms of sex groups, the prevalence of Entamoeba did not exhibit significant differences between male dairy cattle (83.8%) and female dairy cattle (82.7%) (p > 0.05) or between male beef cattle (81.6%) and female beef cattle (76.9%) (p > 0.05). These findings suggest that sex does not play a significant role in the prevalence of Entamoeba in either dairy cattle or beef cattle. Our study revealed significant differences in the prevalence of Entamoeba in different age groups in sheep (p < 0.001), with lambs exhibiting a higher prevalence than older sheep. This suggests that age may influence the susceptibility to Entamoeba infection in sheep, potentially due to factors such as immune system development [25]. Additionally, a significant difference was observed in the prevalence of Entamoeba in beef cattle (p = 0.021), whereas no significant difference was detected in the prevalence of Entamoeba in dairy cattle (p > 0.05). These findings suggest that while sex does not appear to influence Entamoeba prevalence in either dairy or beef cattle, age-related differences in sheep and certain variations between beef cattle and dairy cattle indicate that factors other than sex, such as age and possibly breed, may play a more significant role in Entamoeba infection dynamics [28]. The high prevalence of Entamoeba observed in this study could be attributed to insufficient awareness and lack of preventive measures among managers of these farms, which may exacerbate the spread of these parasites.
In this study, phylogenetic analysis revealed the presence of E. bovis, Entamoeba RL2, Entamoeba RL4, and Entamoeba RL8 in the sampled ruminant populations, which extends our understanding of the genetic diversity and host range of Entamoeba spp. in Shanxi Province. The term “ribosomal lineage” (RL) refers to newly discovered Entamoeba 18S rRNA sequences that diverge by more than 5% from known species [29]. RLs are used to classify organisms with phylogenetic branches distinct from previously described Entamoeba species, and 11 RLs have been identified [19,30]. The phylogenetic evolutionary analysis suggests that E. bovis can be isolated from ruminant hosts other than cattle [31,32]. To our knowledge, this study represents the first species-specific identification of Entamoeba in sheep, dairy cattle, and beef cattle in Shanxi Province, and further studies are warranted to enhance the better understanding of Entamoeba infection in ruminants in Shanxi province by sampling larger numbers of animals and more geographical locations.
5. Conclusions
This study represents the first on the occurrence and prevalence of Entamoeba spp. in sheep, dairy cattle, and beef cattle in Shanxi Province. Entamoeba prevalence was 51.5%, 82.9%, and 79.1%, respectively, in these ruminants. According to the obtained SSU rRNA sequences, the identified Entamoeba included E. bovis, Entamoeba RL2, Entamoeba RL4, and Entamoeba RL8, extending our understanding of the geographical distribution of Entamoeba in ruminants. Furthermore, these findings underscore the necessity for ongoing research to fully comprehend the prevalence and impact of Entamoeba spp. in different regions and host species.
Author Contributions
X.-Q.Z. and W.-B.Z. conceived and designed the study. Z.-X.W., H.-D.X., Y.-H.H., and S.-B.H. performed the experiments and analyzed the data. Z.-X.W. wrote the manuscript. J.L. and Y.K. participated in the implementation of the study. X.-Q.Z. and W.-B.Z. critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Research Fund of Shanxi Province for Introduced High-level Leading Talents (Grant No. RFSXIHLT202101), the Special Research Fund of Shanxi Agricultural University for High-level Talents (Grant No. 2021XG001), and the Yunnan Key Laboratory of Veterinary Etiological Biology (Grant No. 202449CE340019).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the results of this article have been submitted to GenBank, and the accession numbers are shown in the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Carrero, J.C.; Reyes-López, M.; Serrano-Luna, J.; Shibayama, M.; Unzueta, J.; León-Sicairos, N.; Garza, M. Intestinal amoebiasis: 160 years of its first detection and still remains as a health problem in developing countries. Int. J. Med. Microbiol. 2020, 310, 151358. [Google Scholar] [CrossRef] [PubMed]
- Morf, L.; Singh, U. Entamoeba histolytica: A snapshot of current research and methods for genetic analysis. Curr. Opin. Microbiol. 2012, 15, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Fotedar, R.; Stark, D.; Beebe, N.; Marriott, D.; Ellis, J.; Harkness, J. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 2007, 20, 511–532. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Ehrenkaufer, G.M.; Lozano-Amado, D.; Singh, U. Entamoeba stage conversion: Progress and new insights. Curr. Opin. Microbiol. 2020, 58, 62–68. [Google Scholar] [CrossRef]
- Guillén, N. Pathogenicity and virulence of Entamoeba histolytica, the agent of amoebiasis. Virulence 2023, 14, 2158656. [Google Scholar] [CrossRef]
- Showler, A.J.; Boggild, A.K. Entamoeba histolytica . CMAJ 2013, 185, 1064. [Google Scholar] [CrossRef]
- Morán, P.; Serrano-Vázquez, A.; Rojas-Velázquez, L.; González, E.; Pérez-Juárez, H.; Hernández, E.G.; Padilla, M.L.A.; Zaragoza, M.E.; Portillo-Bobadilla, T.; Ramiro, M.; et al. Amoebiasis: Advances in diagnosis, treatment, immunology features and the interaction with the intestinal ecosystem. Int. J. Mol. Sci. 2023, 24, 11755. [Google Scholar] [CrossRef]
- Stanley, S.L. Amoebiasis. Lancet 2003, 361, 1025–1034. [Google Scholar] [CrossRef]
- Marie, C.; Petri, W.A. Regulation of virulence of Entamoeba histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef]
- Rojas, L.; Morán, P.; Valadez, A.; Gómez, A.; González, E.; Hernández, E.; Partida, O.; Nieves, M.; Gudiño, M.; Magaña, U.; et al. Entamoeba histolytica and Entamoeba dispar infection in Mexican school children: Genotyping and phylogenetic relationship. BMC Infect. Dis. 2016, 16, 485. [Google Scholar] [CrossRef]
- Nakada-Tsukui, K.; Nozaki, T. Immune response of Amebiasis and immune evasion by Entamoeba histolytica. Front. Immunol. 2016, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.K.; Clark, C.G.; Petri, W.A. Molecular epidemiology of amebiasis. Infect. Genet. Evol. 2008, 8, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, S.; Zou, Y.; Han, R.Y.; Wang, P.; Song, D.P.; Wang, C.B.; Chen, X.Q. Molecular characterization of Entamoeba spp. in pigs with diarrhea in southern China. Animals 2022, 12, 1476. [Google Scholar] [CrossRef]
- Gao, W.W.; Ma, Y.T.; Ma, Y.Y.; Li, R.L.; Li, J.; Zheng, F.G.; Zheng, W.B.; Liu, Q.; Zhu, X.Q. First report of Eimeria and Entamoeba infection in alpacas (Vicugna pacos) in Shanxi Province, northern China. Parasitol. Res. 2021, 120, 2031–2035. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Berg, R.; Maloney, J.G.; Molokin, A.; Santin, M. Molecular characterization of Blastocystis and Entamoeba of muskoxen and sheep in Greenland. Int. J. Parasitol. 2023, 53, 673–685. [Google Scholar] [CrossRef]
- Matsubayashi, M.; Matsuura, Y.; Nukata, S.; Daizi, Y.; Shibahara, T.; Teramoto, I.; Matsuo, T.; Uni, S.; Hatta, T.; Kaneko, A.; et al. First detection and molecular identification of Entamoeba bovis from Japanese cattle. Parasitol. Res. 2018, 117, 339–432. [Google Scholar] [CrossRef]
- Aryal, M.; Adhikari, R.B.; Kandel, P.; Ghimire, T.R.; Khadka, D.; Maharjan, J.; Gaire, K.P.; Shrestha, S.; Manandhar, K.D.; Kandel, R.C.; et al. First report on the molecular detection of Entamoeba bovis from the endangered wild water buffalo (Bubalus arnee) in Nepal. Vet. Med. Sci. 2022, 8, 799–807. [Google Scholar] [CrossRef]
- Huaman, J.L.; Pacioni, C.; Kenchington-Evans, L.; Doyle, M.; Helbig, K.J.; Carvalho, T.G. First evidence of Entamoeba parasites in Australian wild deer and assessment of transmission to cattle. Front. Cell Infect. Microbiol. 2022, 12, 883031. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Lebbad, M.; Victory, E.L.; Verweij, J.J.; Tannich, E.; Alfellani, M.; Legarraga, P.; Clark, C.G. Increased sampling reveals novel lineages of Entamoeba: Consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 2011, 162, 525–541. [Google Scholar] [CrossRef]
- Verweij, J.J.; Laeijendecker, D.; Brienen, E.A.; Lieshout, L.; Polderman, A.M. Detection and identification of Entamoeba species in stool samples by a reverse line hybridization assay. J. Clin. Microbiol. 2003, 41, 5041–5045. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, Y.Y.; Mei, J.J.; Zheng, W.B.; Liu, Q.; Gao, W.W.; Zhu, X.Q.; Xie, S.C. Molecular identification and genotyping of Cryptosporidium spp. and Blastocystis sp. in cattle in representative areas of Shanxi province, north China. Animals 2023, 13, 2929. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.L.; Liu, Y.Y.; Mei, J.J.; Zou, Y.; Zhang, Z.H.; Zheng, W.B.; Liu, Q.; Gao, W.W.; Xie, S.C.; Zhu, X.Q. Molecular identification and genotyping of Enterocytozoon bieneusi in sheep in Shanxi province, north China. Animals 2022, 12, 993. [Google Scholar] [CrossRef] [PubMed]
- Ai, S.; Zhang, Z.; Wang, X.; Zhang, Q.; Yin, W.; Duan, Z. The first survey and molecular identification of Entamoeba spp. in farm animals on Qinghai-Tibetan Plateau of China. Comp. Immunol. Microbiol. Infect. Dis. 2021, 75, 101607. [Google Scholar] [CrossRef]
- Al-Habsi, K.; Yang, R.; Ryan, U.; Jacobson, C.; Miller, D.W. Morphological and molecular characterization of an uninucleated cyst-producing Entamoeba spp. in captured Rangeland goats in Western Australia. Vet. Parasitol. 2017, 235, 41–46. [Google Scholar] [CrossRef]
- Liu, X.C.; Ren, Q.; Guo, J.; Chen, D.Q.; Li, Q.Q.; Luo, X.Y.; Gu, Y.F.; Li, W.C. First detection and molecular identification of Entamoeba bovis in farm-raised Sika deer from Anhui province, China. Acta Parasitol. 2022, 67, 1782–1787. [Google Scholar] [CrossRef]
- Ren, M.; Yang, F.; Gou, J.M.; Wang, P.X.; Zou, M.; Zhong, X.H.; Lin, Q. First detection and molecular identification of Entamoeba in yaks from China. Acta Parasitol. 2021, 66, 264–270. [Google Scholar] [CrossRef]
- Kang, Y.; Lu, X.S.; He, Y.H.; Wang, C.; Wu, Z.X.; Wang, L.; Wu, X.J.; Hu, J.J.; Zhu, X.Q. First molecular identification and prevalence of Sarcocystis spp. in sheep intended for human consumption in Shanxi province, China. Vet. Sci. 2024, 11, 504. [Google Scholar] [CrossRef]
- Souza, J.B.; Silva, Z.A.; Alves-Ribeiro, B.S.; Moraes, I.S.; Alves-Sobrinho, A.V.; Saturnino, K.C.; Ferraz, H.T.; Machado, M.F.; Braga, Í.A.; Ramos, D.S. Prevalence of intestinal parasites, risk factors and zoonotic aspects in dog and cat populations from Goiás, Brazil. Vet. Sci. 2023, 10, 492. [Google Scholar] [CrossRef]
- Jacob, A.S.; Busby, E.J.; Levy, A.D.; Komm, N.; Clark, C.G. Expanding the Entamoeba universe: New hosts yield novel ribosomal lineages. J. Eukaryot. Microbiol. 2016, 63, 69–78. [Google Scholar] [CrossRef]
- Clark, C.G.; Kaffashian, F.; Tawari, B.; Windsor, J.J.; Twigg-Flesner, A.; Davies-Morel, M.G.; Blessmann, J.; Ebert, F.; Peschel, B.; Van, A.L.; et al. New insights into the phylogeny of Entamoeba species provided by analysis of four new small-subunit rRNA genes. Int. J. Syst. Evol. Microbiol. 2006, 56, 2235–2239. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Lebbad, M.; Clark, C.G. Genetic characterisation of uninucleated cyst-producing Entamoeba spp. from ruminants. Int. J. Parasitol. 2010, 40, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Al-Dabbagh, S.K.; Alseady, H.H.; Alhadad, E.J. Molecular identification of Entamoeba spp. in humans and cattle in Baghdad, Iraq. Vet. World 2024, 17, 1348–1355. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).