Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum and Related Strains in Cattle from Henan, China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
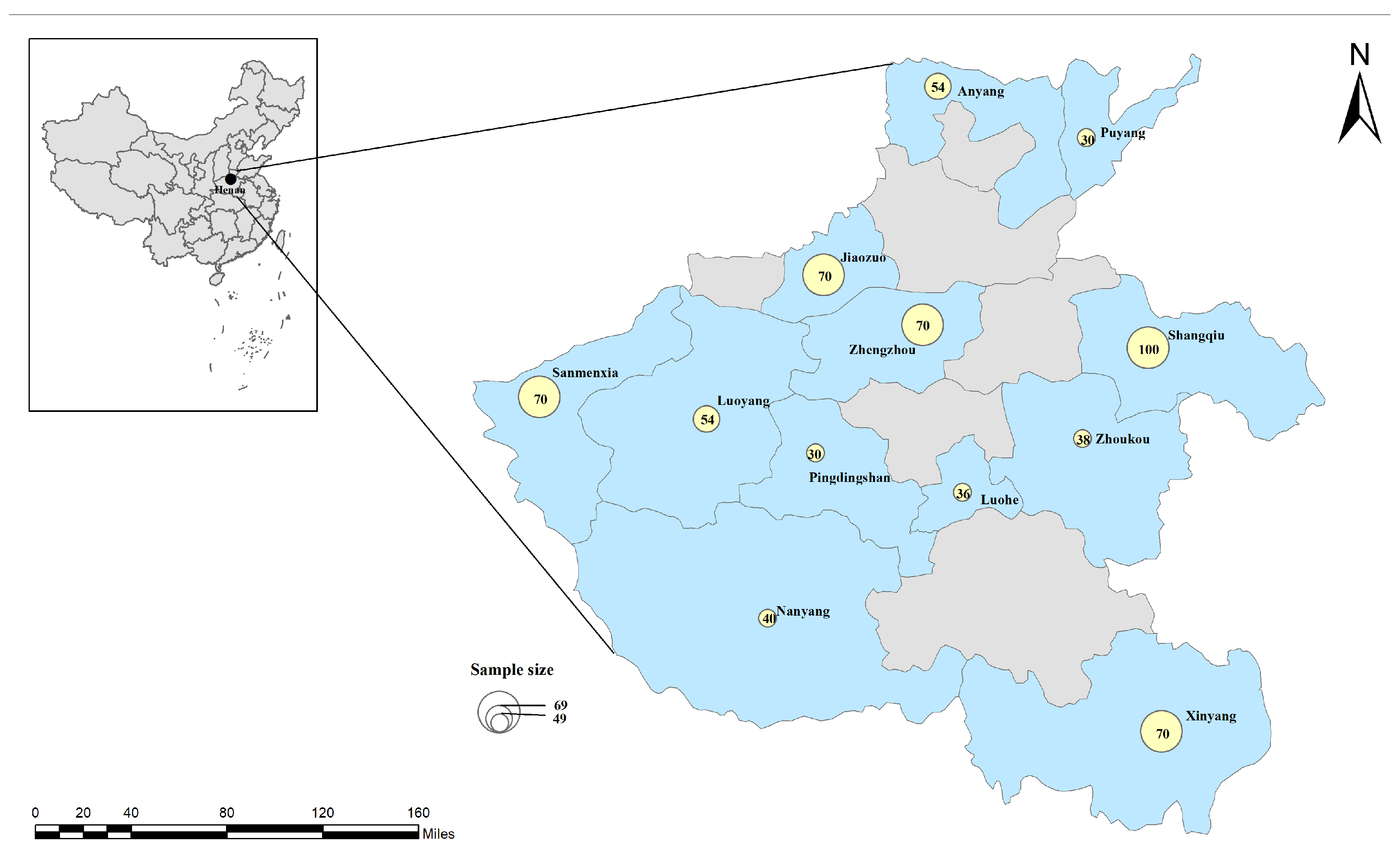
2.1. Sample Collection and DNA Extraction
2.2. Detection of Strains by PCR and Restriction Fragment Length Polymorphism (RFLP)
2.3. DNA Cloning
2.4. Analysis of Sequencing and Phylogenetics
2.5. Statistical Analysis
2.6. Accession Numbers for Nucleotide Sequences
3. Results
3.1. Anaplasma spp. Frequency
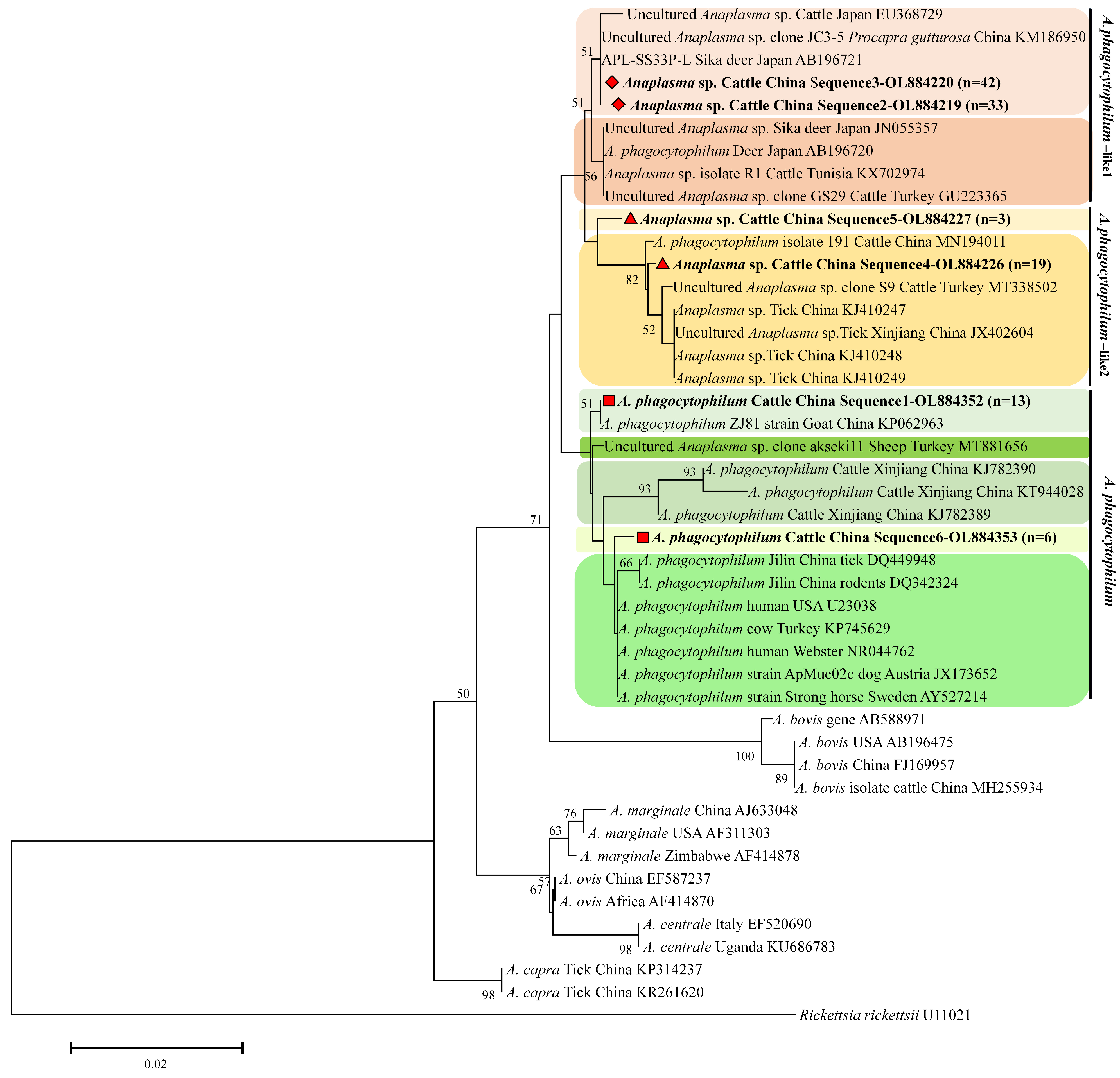
3.2. Molecular Characterization of Anaplasma spp. 16S rRNA Sequence Types
3.3. Molecular Characterization of Anaplasma spp. GroEL Sequence Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dumler, J.; Barbet, A.; Bekker, C.; Dasch, G.; Palmer, G.; Ray, S.; Rikihisa, Y.; Rurangirwa, F. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [CrossRef] [PubMed]
- Rar, V.; Tkachev, S.; Tikunova, N. Genetic diversity of Anaplasma bacteria: Twenty years later. Infect. Genet. Evol. 2021, 91, 104833. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Lu, C.; Gong, P.; Pei, Z.; Peng, Y.; Jian, F.; Wang, R.; Zhang, L.; Qi, M.; Ning, C. Molecular detection and phylogeny of Anaplasma spp. closely related to Anaplasma phagocytophilum in small ruminants from China. Ticks Tick Borne Dis. 2022, 13, 101992. [Google Scholar] [CrossRef] [PubMed]
- Stuen, S.; Granquist, E.; Silaghi, C. Anaplasma phagocytophilum—A widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 2013, 3, 31–64. [Google Scholar] [CrossRef]
- Ismail, N.; McBride, J. Tick-Borne Emerging Infections: Ehrlichiosis and Anaplasmosis. Clin. Lab. Med. 2017, 37, 317–340. [Google Scholar] [CrossRef]
- Woldehiwet, Z. The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 2010, 167, 108–122. [Google Scholar] [CrossRef]
- Woldehiwet, Z. Immune evasion and immunosuppression by Anaplasma phagocytophilum, the causative agent of tick-borne fever of ruminants and human granulocytic anaplasmosis. Vet. J. 2008, 175, 37–44. [Google Scholar] [CrossRef]
- Pusterla, N.; Huder, J.; Wolfensberger, C.; Braun, U.; Lutz, H. Laboratory findings in cows after experimental infection with Ehrlichia phagocytophila. Clin. Diagn. Lab. Immunol. 1997, 4, 643–647. [Google Scholar] [CrossRef]
- Katherine, K.; de la Fuente, J.; Blouin, E.; Coetzee, J.; Ewing, S. The natural history of Anaplasma marginale. Vet. Parasitol. 2010, 167, 95–107. [Google Scholar] [CrossRef]
- Aubry, P.; Geale, D.W. A review of bovine anaplasmosis. Transbound. Emerg. Dis. 2011, 58, 1–30. [Google Scholar] [CrossRef]
- Aktas, M.; Özübek, S. Bovine anaplasmosis in Turkey: First laboratory confirmed clinical cases caused by Anaplasma phagocytophilum. Vet. Microbiol. 2015, 178, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Ybañez, P.; Kishimoto, T. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet. Microbiol. 2012, 157, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Diao, X.; Zhao, G.; Chen, M.; Xiong, Y.; Shi, M.; Fu, W.; Guo, Y.; Pan, B.; Chen, X.; et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol. Biol. 2014, 30, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Said, M.; Belkahia, H.; Alberti, A.; Zobba, R.; Bousrih, M.; Yahiaoui, M.; Daaloul-Jedidi, M.; Mamlouk, A.; Gharbi, M.; Messadi, L. Molecular Survey of Anaplasma Species in Small Ruminants Reveals the Presence of Novel Strains Closely Related to A. phagocytophilum in Tunisia. Vector Borne Zoonotic Dis. 2015, 15, 580–590. [Google Scholar] [CrossRef]
- Said, M.; Belkahia, H.; Mabrouk, N.; Saidani, M.; Ben, M.; Alberti, A.; Zobba, R.; Bouattour, S.; Bouattour, A.; Messadi, L. Molecular typing and diagnosis of Anaplasma spp. closely related to Anaplasma phagocytophilum in ruminants from Tunisia. Ticks Tick Borne Dis. 2017, 8, 412–422. [Google Scholar] [CrossRef]
- Zobba, R.; Said, M.; Belkahia, H.; Pittau, M.; Cacciotto, C.; Parpaglia, M.; Messadi, L.; Alberti, A. Molecular epidemiology of Anaplasma spp. related to A. phagocytophilum in Mediterranean small ruminants. Acta Trop. 2020, 202, 105286. [Google Scholar] [CrossRef]
- Aktas, M.; Çolak, S. Molecular detection and phylogeny of Anaplasma spp. in cattle reveals the presence of novel strains closely related to A. phagocytophilum in Turkey. Ticks Tick Borne Dis. 2021, 12, 101604. [Google Scholar] [CrossRef]
- Altay, K.; Erol, U.; Sahin, O.; Aytmirzakizi, A. First molecular detection of Anaplasma species in cattle from Kyrgyzstan; molecular identification of human pathogenic novel genotype Anaplasma capra and Anaplasma phagocytophilum related strain. Ticks Tick Borne Dis. 2021, 13, 101861. [Google Scholar] [CrossRef]
- Erol, U.; Şahin, Ö.; Altay, K. Molecular Survey of Anaplasma phagocytophilum and Related Strains in Sheep and Goats from Sivas; with a High Prevalence of Anaplasma phagocytophilum-like 1. Turk. Parazitol. Derg. 2022, 46, 293–300. [Google Scholar] [CrossRef]
- Aktaş, M.; Özübek, S.; Uluçeşme, M. Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey. Animals 2021, 11, 814. [Google Scholar] [CrossRef]
- Jilintai, S.N.; Hayakawa, D.; Suzuki, M.; Hata, H.; Kondo, S.; Matsumoto, K.; Yokoyama, N.; Inokuma, H. Molecular survey for Anaplasma bovis and Anaplasma phagocytophilum infection in cattle in a pastureland where sika deer appear in Hokkaido, Japan. Jpn J. Infect. Dis. 2009, 62, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Inayoshi, M.; Kitamura, K.; Kawamori, F.; Kawaguchi, D.; Nishimura, Y.; Naitou, H.; Hiroi, M.; Masuzawa, T. Anaplasma phagocytophilum-infected ticks, Japan. Emerg. Infect. Dis. 2005, 11, 1780–1783. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Rikihisa, Y.; Lin, Q.; Isogai, E.; Tahara, K.; Itagaki, A.; Hiramitsu, Y.; Tajima, T. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 2006, 72, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Ouh, I.; Kwon, O.; Kwak, D. Molecular detection of Anaplasma phagocytophilum-like Anaplasma spp. and pathogenic A. Phagocytophilum in cattle from South Korea. Mol. Phylogenet. Evol. 2018, 126, 23–30. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, Y.; Tao, D.; Zhao, A.; Qi, M.; Ning, C. Molecular detection of Anaplasma spp. in dairy cattle in southern Xinjiang, China. Vet. Parasitol. Reg. Stud. Rep. 2020, 20, 100406. [Google Scholar] [CrossRef]
- Selmi, R.; Belkahia, H.; Dhibi, M.; Abdelaali, H.; Lahmar, S.; Said, M.; Messadi, L. Zoonotic vector-borne bacteria in wild rodents and associated ectoparasites from Tunisia. Infect. Genet. Evol. 2021, 95, 105039. [Google Scholar] [CrossRef]
- Erol, U.; Sahin, O.; Urhan, O.; Atas, A.; Altay, K. Molecular investigation of Anaplasma phagocytophilum and related strains among sheep flocks from different parts of Türkiye; with a note of phylogenetic analyses of Anaplasma phagocytophilum-like 1. Comp. Immunol. Microbiol. Infect. Dis. 2024, 107, 102154. [Google Scholar] [CrossRef]
- Sahin, O.; Erol, U.; Duzlu, O.; Altay, K. Molecular survey of Anaplasma phagocytophilum and related variants in water buffaloes: The first detection of Anaplasma phagocytophilum-like 1. Comp. Immunol. Microbiol. Infect. Dis. 2023, 98, 102004. [Google Scholar] [CrossRef]
- Aung, A.; Narapakdeesakul, D.; Arnuphapprasert, A.; Nugraheni, Y.; Wattanachant, C.; Kaewlamun, W.; Kaewthamasorn, M. Multi-locus sequence analysis of Anaplasma bovis in goats and ticks from Thailand, with the initial identification of an uncultured Anaplasma species closely related to Anaplasma phagocytophilum-like 1. Comp. Immunol. Microbiol. Infect. Dis. 2024, 109, 102181. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Liu, Z.; Liu, J.; Niu, Q.; Ren, Q.; Chen, Z.; Guan, G.; Luo, J.; Yin, H. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, northwest China. Parasites Vectors 2015, 8, 108. [Google Scholar] [CrossRef]
- Dugat, T.; Lagree, A.C.; Maillard, R.; Boulouis, H.J.; Haddad, N. Opening the black box of Anaplasma phagocytophilum diversity: Current situation and future perspectives. Front. Cell. Infect. Microbiol. 2015, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Z.; Guan, G.; Liu, Q.; Li, Y.; Chen, Z.; Ma, M.; Liu, A.; Ren, Q.; Luo, J.; et al. Prevalence of Anaplasma phagocytophilum in ruminants, rodents and ticks in Gansu, north-western China. J. Med. Microbiol. 2013, 62, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, K.; Sun, Y.; Shi, J.; Li, H.; Chen, Y.; Yang, H.; Li, X.; Wu, B.; Li, X.; et al. Molecular epidemiology and risk factors of Anaplasma spp., Babesia spp. and Theileria spp. infection in cattle in Chongqing, China. PLoS ONE 2019, 14, e0215585. [Google Scholar] [CrossRef] [PubMed]
- Department of Agriculture and Rural Affairs of Hainan Province. Henan Animal Husbandry Development Achieves a Good Start in 2021. Available online: https://www.henan.gov.cn/2021/05-25/2151291.html (accessed on 25 May 2021).
- Barlough, J.; Madigan, J.; DeRock, E.; Bigornia, L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet. Parasitol. 1996, 63, 319–329. [Google Scholar] [CrossRef]
- Silaghi, C.; Liebisch, G.; Pfister, K. Genetic variants of Anaplasma phagocytophilum from 14 equine granulocytic anaplasmosis cases. Parasites Vectors 2011, 4, 161–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Y.; Zhang, F.; Zhang, W.; Wang, J.; Cui, Y.; Wang, R.; Jian, F.; Zhang, L.; Ning, C. Molecular and phylogenetic analysis of Anaplasma spp. in sheep and goats from six provinces of China. J. Vet. Sci. 2016, 17, 523–529. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2001. [Google Scholar]
- Zhang, L.; Liu, H.; Xu, B.; Lu, Q.; Li, L.; Chang, L.; Zhang, X.; Fan, D.; Li, G.; Jin, Y.; et al. Anaplasma phagocytophilum infection in domestic animals in ten provinces/cities of China. Am. J. Trop. Med. Hyg. 2012, 87, 185–189. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Z.; Niu, Q.; Liu, J.; Xie, J.; Chen, Q.; Chen, Z.; Guan, G.; Liu, G.; Luo, J.; et al. Evaluation of different nested PCRs for detection of Anaplasma phagocytophilum in ruminants and ticks. BMC Vet. Res. 2016, 12, 35–41. [Google Scholar] [CrossRef]
- Noaman, V. Epidemiological study on Anaplasma phagocytophilum in cattle: Molecular prevalence and risk factors assessment in different ecological zones in Iran. Prev. Vet. Med. 2020, 183, 105118. [Google Scholar] [CrossRef]
- Wang, K.; Yan, Y.; Zhou, Y.; Zhao, S.; Jian, F.; Wang, R.; Zhang, L.; Ning, C. Seasonal dynamics of Anaplasma spp. in goats in warm-temperate zone of China. Ticks Tick Borne Dis. 2021, 12, 101673. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, K.; Cui, Y.; Zhou, Y.; Zhao, S.; Zhang, Y.; Jian, F.; Wang, R.; Zhang, L.; Qi, M.; et al. Molecular detection and phylogenetic analyses of Anaplasma spp. in Haemaphysalis longicornis from goats in four provinces of China. Sci. Rep. 2021, 11, 14155. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Abdugani, A.; Sahin, O.; Muratova, R.; Ero, L.; Attokurov, K.; Abdurasulov, I.; Sakar, H.; Risvanli, A. A comprehensive molecular survey of vector-borne blood parasites in cattle in Kyrgyzstan with a note of the first molecular detection of Anaplasma bovis and Candidatus Anaplasma Camelii. Trop. Anim. Health Prod. 2024, 56, 266–289. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Huang, L.; Lin, Y.; Bhowmick, B.; Zhao, J.; Liao, C.; Guan, Q.; Wang, J.; Han, Q. Molecular surveillance and genetic diversity of Anaplasma spp. in cattle (Bos taurus) and goat (Capra aegagrus hircus) from Hainan island/province, China. BMC Vet. Res. 2023, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Shafy, S.; Abdullah, H.; Elbayoumy, M.K.; Elsawy, B.S.M.; Hassan, M.R.; Mahmoud, M.S.; Hegazi, A.G.; Rahman, E.H.A. Molecular Epidemiological Investigation of Piroplasms and Anaplasmataceae Bacteria in Egyptian Domestic Animals and Associated Ticks. Pathogens 2022, 11, 1194. [Google Scholar] [CrossRef]
- Parvizi, O.; El-Adawy, H.; Melzer, F.; Roesler, U.; Neubauer, H.; Mertens-Scholz, K. Seroprevalence and Molecular Detection of Bovine Anaplasmosis in Egypt. Pathogens 2020, 9, 64. [Google Scholar] [CrossRef]
- Belkahia, H.; Ben, M.; Alberti, A.; Abdi, K.; Issaoui, Z.; Hattab, D.; Gharbi, M.; Messadi, L. First molecular survey and novel genetic variants’ identification of Anaplasma marginale, A. centrale and A. bovis in cattle from Tunisia. Infect. Genet. Evol. 2015, 34, 361–371. [Google Scholar] [CrossRef]
- Seerintra, T.; Saraphol, B.; Thanchomnang, T.; Piratae, S. Molecular prevalence of Anaplasma spp. in cattle and assessment of associated risk factors in Northeast Thailand. Vet. World 2023, 16, 1702–1707. [Google Scholar] [CrossRef]
- Farooqi, S.H.; Ijaz, M.; Rashid, M.I.; Nabi, H.; Islam, S.; Aqib, A.I.; Hussain, K.; Khan, A.; Rizvi, S.N.B.; Mahmood, S.; et al. Molecular epidemiology of bovine anaplasmosis in Khyber Pakhtunkhwa, Pakistan. Trop. Anim. Health Prod. 2018, 50, 1591–1598. [Google Scholar] [CrossRef]
- Stuen, S.; Bergstrom, K.; Petrovec, M.; Van, I.; Schouls, L. Differences in clinical manifestations and hematological and serological responses after experimental infection with genetic variants of Anaplasma phagocytophilum in sheep. Clin. Diagn. Lab. Immunol. 2003, 10, 692–695. [Google Scholar] [CrossRef]
- Stuen, S.; Torsteinbo, W.; Bergstrom, K.; Bardsen, K. Superinfection occurs in Anaplasma phagocytophilum infected sheep irrespective of infection phase and protection status. Acta Vet. Scand. 2009, 51, 41–47. [Google Scholar] [CrossRef][Green Version]
- Rikihisa, Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin. Microbiol. Rev. 2011, 24, 469–489. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Erol, U.; Sahin, O. The first molecular detection of Anaplasma capra in domestic ruminants in the central part of Turkey, with genetic diversity and genotyping of Anaplasma capra. Trop. Anim. Health Prod. 2022, 54, 129. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Erol, U.; Sahin, O.; Aytmirzakizi, A.; Temizel, E.; Aydin, M.; Dumanli, N.; Aktas, M. The detection and phylogenetic analysis of Anaplasma phagocytophilum-like 1, A. ovis and A. capra in sheep: A. capra divides into two genogroups. Vet. Res. Commun. 2022, 46, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Battilani, M.; De Arcangeli, S.; Balboni, A.; Dondi, F. Genetic diversity and molecular epidemiology of Anaplasma. Infect. Genet. Evol. 2017, 49, 195–211. [Google Scholar] [CrossRef]
- Langenwalder, D.; Schmidt, S.; Gilli, U.; Pantchev, N.; Ganter, M.; Silaghi, C.; Aardema, M.; Loewenich, F. Genetic characterization of Anaplasma phagocytophilum strains from goats (Capra aegagrus hircus) and water buffalo (Bubalus bubalis) by 16S rRNA gene, ankA gene and multilocus sequence typing. Ticks Tick Borne Dis. 2019, 10, 101267. [Google Scholar] [CrossRef]
- Jahfari, S.; Coipan, E.; Fonville, M.; Leeuwen, A.; Hengeveld, P.; Heylen, D.; Heyman, P.; Maanen, C.; Butler, C.; Földvári, G.; et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasites Vectors 2014, 7, 365. [Google Scholar] [CrossRef]
- Scharf, W.; Schauer, S.; Freyburger, F.; Petrovec, M.; Schaarschmidt-Kiener, D.; Liebisch, G.; Runge, M.; Ganter, M.; Kehl, A.; Dumler, J.; et al. Distinct Host Species Correlate with Anaplasma phagocytophilum ankA Gene Clusters. J. Clin. Microbiol. 2011, 49, 790–796. [Google Scholar] [CrossRef]
- Silaghi, C.; Hamel, D.; Thiel, C.; Pfister, K.; Passos, L.; Rehbein, S. Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Chastagner, A.; Dugat, T.; Vourc’h, G.; Verheyden, H.; Legrand, L.; Bachy, V.; Chabanne, L.; Joncour, G.; Maillard, R.; Boulouis, H.; et al. Multilocus sequence analysis of Anaplasma phagocytophilum reveals three distinct lineages with different host ranges in clinically ill French cattle. Vet. Res. 2014, 45, 114. [Google Scholar] [CrossRef]
- Huhn, C.; Winter, C.; Wolfsperger, T.; Wüppenhorst, N.; Smrdel, K.; Skuballa, J.; Pfäffle, M.; Petney, T.; Silaghi, C.; Dyachenko, V.; et al. Analysis of the population structure of Anaplasma phagocytophilum using multilocus sequence typing. PLoS ONE 2014, 9, e93725. [Google Scholar] [CrossRef]
- Mukhacheva, T.; Shaikhova, D.; Kovalev, S.; Loewenich, F. Phylogeographical diversity of Anaplasma phagocytophilum in the Asian part of Russia based on multilocus sequence typing and analysis of the ankA gene. Infect. Genet. Evol. 2020, 80, 104234. [Google Scholar] [CrossRef]


| Target Gene | Isolates | Primer Name | Oligonucleotide Sequence (5′–3′) | Amplicon Size (bp) | Annealing | Reference |
|---|---|---|---|---|---|---|
| 16S rRNA | AP and related variants | EE1 | TCCTGGCTCAGAACGAACGCTGGCGGC | 1430 | 55 °C | [35] |
| EE2 | GTCACTGACCCAACCTTAAATGGCTG | |||||
| SSAP2f | GCTGAATGTGGGGATAATTTAT | 641–642 | 55 °C | [23] | ||
| SSAP2r | ATGGCTGCTTCCTTTCGGTTA | |||||
| groEL | AP | EphplgroEL-F | ATGGTATGCAGTTTGATCGC | 55 °C | [36] | |
| EphplgroEL-R | TCTACTCTGTCTTTGCGTTC | 642 | ||||
| EphgroEL-R | TTGAGTACAGCAACACCACCGGAA | 573 | ||||
| AP-like 1 | groEL-1F | TATAGCTAGCATAATTACCCAGAGC | 339 | 53 °C | [37] | |
| groEL-1R | GGTTAGTTCTGCTTTCGATGC | |||||
| groEL-2F | TTATGTCTATGCGCCGTG | 51 °C | ||||
| groEL-2R | CGGACCTTGCCACATTTT | |||||
| AP-like 2 | APHAGOVAR2GROEL_F | TACTCTAGAAGACGCGGTAG | 55 °C | [16] | ||
| APHAGOVAR2GROEL_R1 | ACGAACATTCTTAGCAGTCC | 792 | ||||
| APHAGOVAR2GROEL_R2 | CTTCTATCACCAAATCCTGG |
| Geographic Location | Tested Number | Positive (%) | Co-Infected (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA+ | 95% CI a | AP | 95% CI a | AP-like 1 | 95% CI a | AP-like 2 | 95% CI a | AP/AP-like 1 | 95% CI a | AP-like 1/AP-like 2 | 95% CI a | ||
| Luoyang | 54 | 8 (14.81) | 5.03–24.60 | 2 (3.70) | 0–8.91 | 8 (14.81) | 5.03–24.60 | 2 (3.70) | 0–8.91 | 2 (3.70) | 0–8.91 | 2 (3.70) | 0–8.91 |
| Luohe | 36 | 2 (5.56) | 0–13.42 | 0 | — | 2 (5.56) | 0–13.42 | 0 | — | 0 | — | 0 | — |
| Zhoukou | 38 | 8 (21.05) | 7.47–34.63 | 3 (7.89) | 0–16.88 | 8 (21.05) | 7.47–34.63 | 1 (2.63) | 0–7.96 | 3 (7.89) | 0–16.88 | 1 (2.63) | 0–7.96 |
| Anyang | 54 | 19 (35.19) | 22.03–48.34 | 6 (11.11) | 2.45–19.77 | 19 (35.19) | 22.03–48.34 | 5 (9.26) | 1.27–17.25 | 6 (11.11) | 2.45–19.77 | 5 (9.26) | 1.27–17.25 |
| Puyang | 30 | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — |
| Xinyang | 70 | 17 (24.29) | 14.00–34.58 | 6 (8.57) | 1.85–15.29 | 17 (24.29) | 14.00–34.58 | 8 (11.43) | 3.79–19.07 | 6 (8.57) | 1.85–15.29 | 8 (11.43) | 3.79–19.07 |
| Shangqiu | 100 | 5 (5.00) | 0.65–9.35 | 0 | — | 5 (5.00) | 0.65–9.35 | 0 | — | 0 | — | 0 | — |
| Jiaozuo | 70 | 7 (10.00) | 2.80–17.20 | 2 (2.86) | 0–6.86 | 7 (10.00) | 2.80–17.20 | 5 (7.14) | 0.96–13.33 | 2 (2.86) | 0–6.86 | 5 (7.14) | 0.96–13.33 |
| Zhengzhou | 70 | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — |
| Pingdingshan | 30 | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — |
| Sanmenxia | 70 | 2 (2.86) | 0–6.86 | 0 | — | 2 (2.86) | 0–6.86 | 1 (1.43) | 0–4.28 | — | — | 1 (1.43) | 0–4.28 |
| Nanyang | 40 | 7 (17.50) | 5.19–29.81 | 0 | — | 7 (17.50) | 5.19–29.81 | 0 | — | 0 | — | 0 | — |
| Total | 662 | 75 (11.33) | 8.91–13.75 | 19 (2.87) | 1.59–4.15 | 75 (11.33) | 8.91–13.75 | 22 (3.32) | 1.95–4.69 | 19 (2.87) | 1.59–4.15 | 22 (3.32) | 1.95–4.69 |
| Group | Tested | Positive (%) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA+ | 95% CI a | p-Value b | OR | AP | 95% CI a | p-Value b | OR | AP-like 1 | 95% CI a | p-value b | OR | AP-like 2 | 95% CI a | p-Value b | OR | |||
| Sex | Female | 607 | 60 (9.88) | 0.15–0.56 | p < 0.01 | 0.29 | 15 (2.47) | 0.10–1.01 | 0.01 < p < 0.05 | 0.32 | 60 (9.88) | 0.15–0.56 | p < 0.01 | 0.29 | 20 (3.29) | 0.21–4.00 | p > 0.05 | 0.90 |
| Male | 55 | 15 (27.27) | — | — | 1 | 4 (7.27) | — | 1 | 15 (27.27) | — | — | 1 | 2 (3.64) | — | — | 1 | ||
| Age | <2 | 181 | 31 (17.13) | 1.14–3.07 | 0.01 < p < 0.05 | 1.87 | 10 (5.52) | 1.12–7.04 | 0.01 < p < 0.05 | 2.81 | 31 (17.13) | 1.14–3.07 | 0.01 < p < 0.05 | 1.87 | 10 (5.52) | 0.89–4.94 | p > 0.05 | 2.10 |
| 2–5 | 442 | 44 (9.95) | — | — | 1 | 9 (2.04) | — | — | 1 | 44 (9.95) | — | — | 1 | 12 (2.71) | — | — | 1 | |
| >5 | 39 | 0 | — | 0.01 < p < 0.05 | — | 0 | — | p > 0.05 | — | 0 | — | p > 0.05 | 0.9 | 0 | — | p > 0.05 | — | |
| Feeding habits | Grazing | 82 | 45 (54.88) | 12.62–39.40 | p < 0.01 | 22.30 | 11 (13.41) | 4.3–28.46 | p < 0.01 | 11.08 | 45 (54.88) | 12.62–39.40 | p < 0.01 | 22.30 | 16 (19.51) | 8.77–61.32 | p < 0.01 | 23.19 |
| Household | 580 | 30 (5.17) | — | — | 1 | 8 (1.38) | — | — | 1 | 30 (5.17) | — | — | 1 | 6 (1.03) | 0–2.0 | — | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Wang, Y.; Cui, Y.; Wang, J.; Fan, S.; Ning, C. Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum and Related Strains in Cattle from Henan, China. Vet. Sci. 2025, 12, 252. https://doi.org/10.3390/vetsci12030252
Yan Y, Wang Y, Cui Y, Wang J, Fan S, Ning C. Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum and Related Strains in Cattle from Henan, China. Veterinary Sciences. 2025; 12(3):252. https://doi.org/10.3390/vetsci12030252
Chicago/Turabian StyleYan, Yaqun, Yongli Wang, Yanyan Cui, Jin Wang, Shuhua Fan, and Changshen Ning. 2025. "Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum and Related Strains in Cattle from Henan, China" Veterinary Sciences 12, no. 3: 252. https://doi.org/10.3390/vetsci12030252
APA StyleYan, Y., Wang, Y., Cui, Y., Wang, J., Fan, S., & Ning, C. (2025). Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum and Related Strains in Cattle from Henan, China. Veterinary Sciences, 12(3), 252. https://doi.org/10.3390/vetsci12030252





