A Cutoff Determination of Real-Time Loop-Mediated Isothermal Amplification (LAMP) for End-Point Detection of Campylobacter jejuni in Chicken Meat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Conditions, and DNA Preparation
2.2. Identification of Campylobacter Strains
2.3. LAMP Primers
2.4. LAMP Assay and Condition Optimization
2.5. Receiver Operating Characteristic (ROC) Curve Analysis
2.6. Determination of Specificity, Sensitivity, and Limit of Detection of LAMP Assay
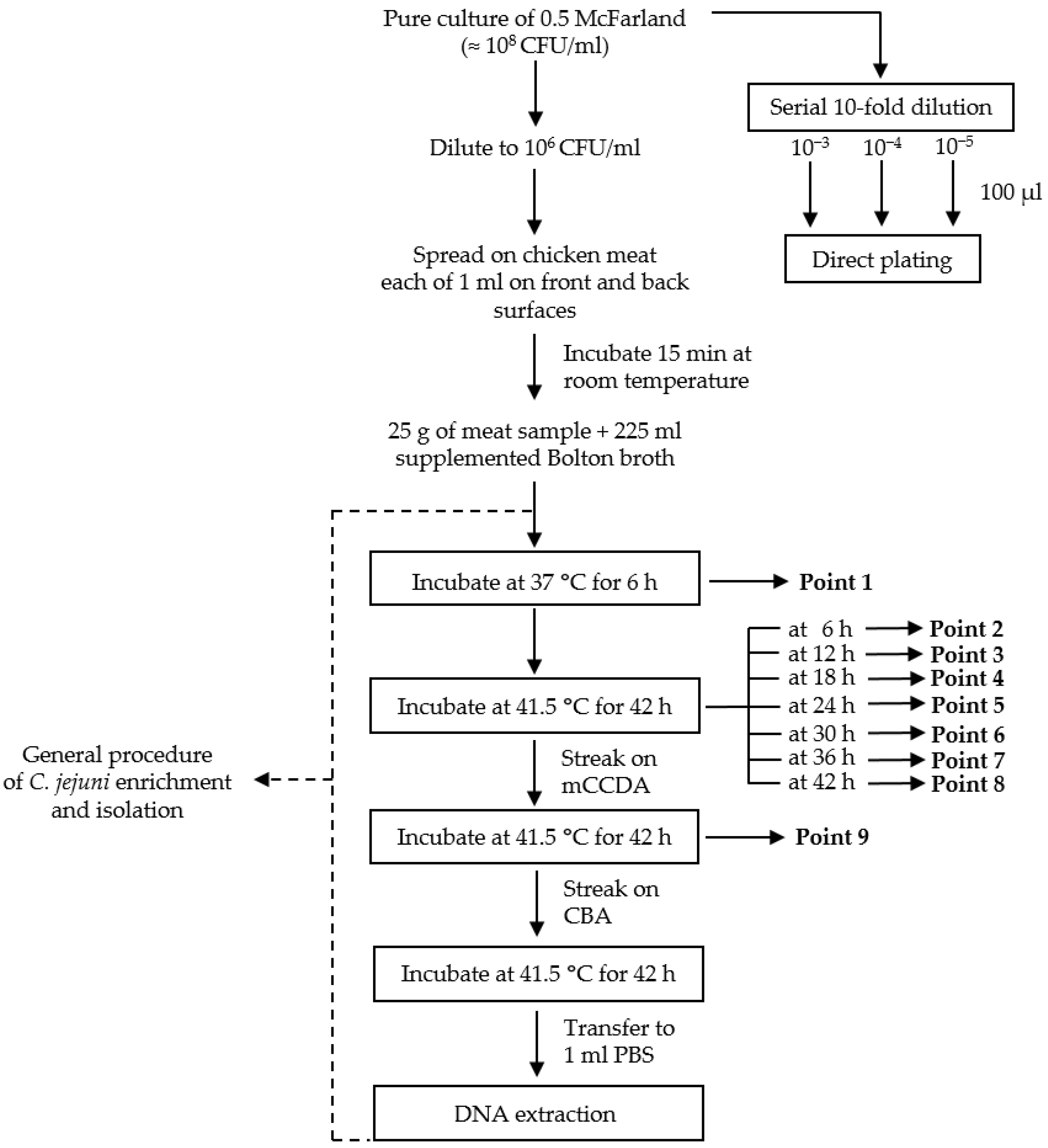
2.7. Detection of C. jejuni in Chicken Meat Samples
2.8. Data Analysis
3. Results
3.1. ROC Curve Analysis
3.2. Specificity, Sensitivity, and Limit of Detection of LAMP Assay
3.3. Detection of C. jejuni in Chicken Meat Samples
4. Discussion
4.1. LAMP Assay Optimization
4.2. ROC Curve Analysis
4.3. Non-Specific Amplification of LAMP Assay
4.4. Campylobacter Species Identification with mPCR Assay
4.5. Specificity and Sensitivity of LAMP Assay
4.6. Limit of Detection of LAMP Assay
4.7. Detection of C. jejuni in Chicken Meat Samples
4.8. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coker, A.O.; Isokpehi, R.D.; Thomas, B.N.; Amisu, K.O.; Obi, C.L. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 2002, 8, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Man, S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahti, E.; Rehn, M.; Ockborn, G.; Hansson, I.; Ågren, J.; Engvall, E.O.; Jernberg, C. Outbreak of campylobacteriosis following a dairy farm visit: Confirmation by genotyping. Foodborne Pathog. Dis. 2017, 14, 326–332. [Google Scholar] [CrossRef]
- Friedman, C.R.; Hoekstra, R.M.; Samuel, M.; Marcus, R.; Bender, J.; Shiferaw, B.; Reddy, S.; Ahuja, S.D.; Helfrick, D.L.; Hardnett, F.; et al. Risk factors for sporadic Campylobacter infection in the United States: A case-control study in FoodNet sites. Clin. Infect. Dis. 2004, 38, S285–S296. [Google Scholar] [CrossRef] [Green Version]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: A review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef] [Green Version]
- Tack, D.M.; Marder, E.P.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Hurd, S.; Scallan, E.; Lathrop, S.; Muse, A.; Ryan, P.; et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2015–2018. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg Goldstein, R.E.; Cruz-Cano, R.; Jiang, C.; Palmer, A.; Blythe, D.; Ryan, P.; Hogan, B.; White, B.; Dunn, J.R.; Libby, T.; et al. Association between community socioeconomic factors, animal feeding operations, and campylobacteriosis incidence rates: Foodborne Diseases Active Surveillance Network (FoodNet), 2004–2010. BMC Infect. Dis. 2016, 16, 354. [Google Scholar] [CrossRef] [Green Version]
- Ravel, A.; Pintar, K.; Nesbitt, A.; Pollari, F. Non food-related risk factors of campylobacteriosis in Canada: A matched case-control study. BMC Public Health 2016, 16, 1016. [Google Scholar] [CrossRef] [Green Version]
- Samosornsuk, W.; Asakura, M.; Yoshida, E.; Taguchi, T.; Eampokalap, B.; Chaicumpa, W.; Yamasaki, S. Isolation and characterization of Campylobacter strains from diarrheal patients in central and suburban Bangkok, Thailand. Jpn. J. Infect. Dis. 2015, 68, 209–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Facciolà, A.; Riso, R.; Avventuroso, E.; Visalli, G.; Delia, S.A.; Laganà, P. Campylobacter: From microbiology to prevention. J. Prev. Med. Hyg. 2017, 58, E79–E92. [Google Scholar] [PubMed]
- Gundogdu, O.; Wren, B.W. Microbe profile: Campylobacter jejuni—Survival instincts. Microbiology 2020, 166, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Golden, C.E.; Mishra, A. Prevalence of Salmonella and Campylobacter spp. in alternative and conventionally produced chicken in the United States: A systematic review and meta-analysis. J. Food Prot. 2020, 83, 1181–1197. [Google Scholar] [CrossRef]
- Osiriphun, S.; Iamtaweejaloen, P.; Kooprasertying, P.; Koetsinchai, W.; Tuitemwong, K.; Erickson, L.E.; Tuitemwong, P. Exposure assessment and process sensitivity analysis of the contamination of Campylobacter in poultry products. Poult. Sci. 2011, 90, 1562–1573. [Google Scholar] [CrossRef]
- Saiyudthong, S.; Phusri, K.; Buates, S. Rapid detection of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari in fresh chicken meat and by-products in Bangkok, Thailand, using modified multiplex PCR. J. Food Prot. 2015, 78, 1363–1369. [Google Scholar] [CrossRef]
- Snelling, W.J.; Matsuda, M.; Moore, J.E.; Dooley, J.S. Campylobacter jejuni. Lett. Appl. Microbiol. 2005, 41, 297–302. [Google Scholar] [CrossRef]
- Denis, M.; Soumet, C.; Rivoal, K.; Ermel, G.; Blivet, D.; Salvat, G.; Colin, P. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 1999, 29, 406–410. [Google Scholar] [CrossRef]
- Yamazaki, W.; Taguchi, M.; Ishibashi, M.; Kitazato, M.; Nukina, M.; Misawa, N.; Inoue, K. Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 2008, 57, 444–451. [Google Scholar] [CrossRef]
- Pavlova, M.R.; Dobreva, E.G.; Ivanova, K.I.; Asseva, G.D.; Ivanov, I.N.; Petrov, P.K.; Velev, V.R.; Tomova, I.I.; Tiholova, M.M.; Kantardjiev, T.V. Multiplex PCR assay for identification and differentiation of Campylobacter jejuni and Campylobacter coli isolates. Folia Med. 2016, 58, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Banowary, B.; Dang, V.T.; Sarker, S.; Connolly, J.H.; Chenu, J.; Groves, P.; Ayton, M.; Raidal, S.; Devi, A.; Vanniasinkam, T.; et al. Differentiation of Campylobacter jejuni and Campylobacter coli using multiplex-PCR and high resolution melt curve analysis. PLoS ONE 2015, 10, e0138808. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Clark, C.G.; Taylor, T.M.; Pucknell, C.; Barton, C.; Price, L.; Woodward, D.L.; Rodgers, F.G. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 2002, 40, 4744–4747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umesha, S.; Manukumar, H.M. Advanced molecular diagnostic techniques for detection of food-borne pathogens: Current applications and future challenges. Crit. Rev. Food Sci. Nutr. 2018, 58, 84–104. [Google Scholar] [CrossRef] [PubMed]
- Sinulingga, T.S.; Aziz, S.A.; Bitrus, A.A.; Zunita, Z.; Abu, J. Occurrence of Campylobacter species from broiler chickens and chicken meat in Malaysia. Trop. Anim. Health Prod. 2020, 52, 151–157. [Google Scholar] [CrossRef]
- Barletta, F.; Mercado, E.H.; Lluque, A.; Ruiz, J.; Cleary, T.G.; Ochoa, T.J. Multiplex real-time PCR for detection of Campylobacter, Salmonella, and Shigella. J. Clin. Microbiol. 2013, 51, 2822–2829. [Google Scholar] [CrossRef] [Green Version]
- de Boer, P.; Rahaoui, H.; Leer, R.J.; Montijn, R.C.; van der Vossen, J.M. Real-time PCR detection of Campylobacter spp.: A comparison to classic culturing and enrichment. Food Microbiol. 2015, 51, 96–100. [Google Scholar] [CrossRef]
- Josefsen, M.H.; Löfström, C.; Hansen, T.B.; Christensen, L.S.; Olsen, J.E.; Hoorfar, J. Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl. Environ. Microbiol. 2010, 76, 5097–5104. [Google Scholar] [CrossRef] [Green Version]
- Regnath, T.; Ignatius, R. Accurate detection of Campylobacter spp. antigens by immunochromatography and enzyme immunoassay in routine microbiological laboratory. Eur. J. Microbiol. Immunol. 2014, 4, 156–158. [Google Scholar] [CrossRef] [Green Version]
- Tissari, P.; Rautelin, H. Evaluation of an enzyme immunoassay-based stool antigen test to detect Campylobacter jejuni and Campylobacter coli. Diagn. Microbiol. Infect. Dis. 2007, 58, 171–175. [Google Scholar] [CrossRef]
- Granato, P.A.; Chen, L.; Holiday, I.; Rawling, R.A.; Novak-Weekley, S.M.; Quinlan, T.; Musser, K.A. Comparison of premier CAMPY enzyme immunoassay (EIA), ProSpecT Campylobacter EIA, and ImmunoCard STAT! CAMPY tests with culture for laboratory diagnosis of Campylobacter enteric infections. J. Clin. Microbiol. 2010, 48, 4022–4027. [Google Scholar] [CrossRef] [Green Version]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.D. Receiver operating characteristic (ROC) curve analysis: A tutorial using body mass index (BMI) as a measure of obesity. J. Phys. Act. Res. 2016, 1, 5–8. [Google Scholar] [CrossRef]
- ISO 10272-2; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for Detection and Enumeration of Campylobacter spp.—Part 2: Colony-Count Technique. International Organization for Standardization: Geneva, Switzerland, 2006.
- Carter, J.V.; Pan, J.; Rai, S.N.; Galandiuk, S. ROC-ing along: Evaluation and interpretation of receiver operating characteristic curves. Surgery 2016, 159, 1638–1645. [Google Scholar] [CrossRef]
- Kreitlow, A.; Becker, A.; Ahmed, M.F.E.; Kittler, S.; Schotte, U.; Plötz, M.; Abdulmawjood, A. Combined loop-mediated isothermal amplification assays for rapid detection and one-step differentiation of Campylobacter jejuni and Campylobacter coli in meat products. Front. Microbiol. 2021, 12, 668824. [Google Scholar] [CrossRef]
- Sabike, I.I.; Uemura, R.; Kirino, Y.; Mekata, H.; Sekiguchi, S.; Okabayashi, T.; Goto, Y.; Yamazaki, W. Use of direct LAMP screening of broiler fecal samples for Campylobacter jejuni and Campylobacter coli in the positive flock identification strategy. Front. Microbiol. 2016, 7, 1582. [Google Scholar] [CrossRef] [Green Version]
- Wang, D. Evaluation and improvement of LAMP assays for detection of Escherichia coli serogroups O26, O45, O103, O111, O121, O145, and O157. Afr. Health Sci. 2017, 17, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.G.; Brewster, J.D.; Paul, M.; Tomasula, P.M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules 2015, 20, 6048–6059. [Google Scholar] [CrossRef] [Green Version]
- Babu, U.S.; Harrison, L.M.; Mammel, M.K.; Bigley, E.C., 3rd; Hiett, K.L.; Balan, K.V. A loop-mediated isothermal amplification (LAMP) assay for the consensus detection of human pathogenic Campylobacter species. J. Microbiol. Methods 2020, 176, 106009. [Google Scholar] [CrossRef]
- Yamazaki, W. Sensitive and rapid detection of Campylobacter jejuni and Campylobacter coli using loop-mediated isothermal amplification. Methods Mol. Biol. 2013, 943, 267–277. [Google Scholar] [CrossRef]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karthik, K.; Rathore, R.; Thomas, P.; Arun, T.R.; Viswas, K.N.; Dhama, K.; Agarwal, R.K. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX 2014, 1, 137–143. [Google Scholar] [CrossRef] [PubMed]
- ISO 10272-1; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for Detection and Enumeration of Campylobacter spp.—Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2006.
- Kabir, S.M.L.; Chowdhury, N.; Asakura, M.; Shiramaru, S.; Kikuchi, K.; Hinenoya, A.; Neogi, S.B.; Yamasaki, S. Comparison of established PCR assays for accurate identification of Campylobacter jejuni and Campylobacter coli. Jpn. J. Infect. Dis. 2019, 72, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asakura, M.; Samosornsuk, W.; Hinenoya, A.; Misawa, N.; Nishimura, K.; Matsuhisa, A.; Yamasaki, S. Development of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the detection and identification of Campylobacter jejuni, Campylobacter coli and Campylobacter fetus. FEMS Immunol. Med. Microbiol. 2008, 52, 260–266. [Google Scholar] [CrossRef] [Green Version]
- Reid, A.N.; Pandey, R.; Palyada, K.; Whitworth, L.; Doukhanine, E.; Stintzi, A. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 2008, 74, 1598–1612. [Google Scholar] [CrossRef] [Green Version]
- On, S.L.; Jordan, P.J. Evaluation of 11 PCR assays for species-level identification of Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 2003, 41, 330–336. [Google Scholar] [CrossRef] [Green Version]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Sridapan, T.; Tangkawsakul, W.; Janvilisri, T.; Luangtongkum, T.; Kiatpathomchai, W.; Chankhamhaengdecha, S. Rapid and simultaneous detection of Campylobacter spp. and Salmonella spp. in chicken samples by duplex loop-mediated isothermal amplification coupled with a lateral flow biosensor assay. PLoS ONE 2021, 16, e0254029. [Google Scholar] [CrossRef]
- Yamazaki, W.; Taguchi, M.; Kawai, T.; Kawatsu, K.; Sakata, J.; Inoue, K.; Misawa, N. Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol. 2009, 75, 1597–1603. [Google Scholar] [CrossRef] [Green Version]
- Quyen, T.L.; Nordentoft, S.; Vinayaka, A.C.; Ngo, T.A.; Engelsmenn, P.; Sun, Y.; Madsen, M.; Bang, D.D.; Wolff, A. A sensitive, specific and simple loop mediated isothermal amplification method for rapid detection of Campylobacter spp. in broiler production. Front. Microbiol. 2019, 10, 2443. [Google Scholar] [CrossRef]
- Lv, R.; Wang, K.; Feng, J.; Heeney, D.D.; Liu, D.; Lu, X. Detection and quantification of viable but non-culturable Campylobacter jejuni. Front. Microbiol. 2020, 10, 2920. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Ma, L.; Lu, X. Rapid determination of viable but non-culturable Campylobacter jejuni in food products by loop-mediated isothermal amplification coupling propidium monoazide treatment. Int. J. Food Microbiol. 2021, 351, 109263. [Google Scholar] [CrossRef] [PubMed]
- Kanki, M.; Sakata, J.; Taguchi, M.; Kumeda, Y.; Ishibashi, M.; Kawai, T.; Kawatsu, K.; Yamasaki, W.; Inoue, K.; Miyahara, M. Effect of sample preparation and bacterial concentration on Salmonella enterica detection in poultry meat using culture methods and PCR assaying of preenrichment broths. Food Microbiol. 2009, 26, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.D.; Simpkin, E.; Lee, R.; Clifton-Hadley, F.A.; Vidal, A.B. Sensitivity of direct culture, enrichment and PCR for detection of Campylobacter jejuni and C. coli in broiler flocks at slaughter. Zoonoses Public Health 2017, 64, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.R.; Cook, N. A rapid lamp-based method for screening poultry samples for Campylobacter without enrichment. Front. Microbiol. 2018, 9, 2401. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, L.; Zhao, M.; Zhu, R.; Deng, J.; Wang, F.; Li, F.; Ding, Y.; Tian, R.; Qian, Y. Four DNA extraction methods used in loop-mediated isothermal amplification for rapid adenovirus detection. J. Virol. Methods 2014, 204, 49–52. [Google Scholar] [CrossRef]

| Target Gene | GenBank Accession No. | Primer | Sequence (5′—3′) * | Gene Location (bp) | Reference |
|---|---|---|---|---|---|
| cj0414 | AL111168 | CJ-FIP | ACAGCACCGCCACCTATAGTAGAAGCTTTTTTAAACTAGGGC (Flc-F2) | 95–76 (Flc), 25–46 (F2) | [19] |
| CJ-BIP | AGGCAGCAGAACTTACGCATTGAGTTTGAAAAAACATTCTACCTCT (B1-B2c) | 101–121 (B1), 181–157 (B2c) | |||
| CJ-F3 | GCAAGACAATATTATTGATCGC (F3) | 3–24 | |||
| CJ-B3 | CTTTCACAGGCTGCACTT (B3c) | 218–201 | |||
| CJ-LF | CTAGCTGCTACTACAGAACCAC (LFc) | 74–53 | |||
| CJ-LB | CATCAAGCTTCACAAGGAAA (LB) | 124–143 |
| Gradient Temperature (°C) | Mean Cq | ||
|---|---|---|---|
| Sample (C. jejuni) | NTC * | Positive Control | |
| 68.0 | 15.64 | 43.99 | |
| 68.4 | 14.49 | 33.74 | |
| 69.2 | 12.95 | 77.28 | |
| 70.4 | 11.38 | 66.96 | |
| 71.8 | 12.89 | 87.60 | |
| 73.0 | 15.43 | 43.73 | |
| 73.7 | 20.62 | 31.69 | |
| 74.0 | 24.31 | 20.85 | 10.43 |
| Analysis of Maximum Likelihood Estimates | |||||
|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error | Wald Chi-Square | Pr > ChiSq |
| Intercept | 1 | 2.3689 | 0.3025 | 61.3305 | <0.0001 |
| Cycle | 1 | −0.0682 | 0.00931 | 53.7099 | <0.0001 |
| ROC Association Statistics | |||||||
|---|---|---|---|---|---|---|---|
| ROC Model | Mann–Whitney | Somers’ D | Gamma | Tau-a | |||
| Area | Standard Error | 95% Wald Confidence Limits | |||||
| Model | 0.9362 | 0.0170 | 0.9030 | 0.9695 | 0.8724 | 0.8724 | 0.4381 |
| ROC1 | 0.5000 | 0 | 0.5000 | 0.5000 | 0 | 0 | |
| ROC Contrast Test Results | |||||||
| Contrast | DF | Chi-Square | Pr > ChiSq | ||||
| Reference = Model | 1 | 661.9499 | <0.0001 | ||||
| Cutoff | Sensitivity | Specificity | YJ |
|---|---|---|---|
| 18.0702 | 0.89655 | 0.91150 | 0.80806 |
| 18.0677 | 0.88793 | 0.91150 | 0.79944 |
| 18.1287 | 0.89655 | 0.90265 | 0.79921 |
| 18.1637 | 0.90517 | 0.89381 | 0.79898 |
| 18.0607 | 0.87931 | 0.91150 | 0.79081 |
| 18.1339 | 0.89655 | 0.89381 | 0.79036 |
| 18.6000 | 0.90517 | 0.88496 | 0.79013 |
| 18.0429 | 0.87069 | 0.91150 | 0.78219 |
| 19.1601 | 0.90517 | 0.87611 | 0.78128 |
| 19.4814 | 0.91379 | 0.86726 | 0.78105 |
| Bacteria | C. jejuni Positives | |
|---|---|---|
| mPCR | LAMP | |
| Campylobacter jejuni | 32/32 | 31/32 |
| Campylobacter coli | 0/8 | 1/8 |
| Salmonella Paratyphi | 8/8 | 0/8 |
| Pseudomonas aeruginosa | 8/8 | 0/8 |
| Staphylococcus aureus | 8/8 | 0/8 |
| Escherichia coli | 8/8 | 1/8 |
| Negative control | 0/8 | 0/8 |
| Assay * | Campylobacter jejuni Concentration (CFU/mL) ** | |||||||
|---|---|---|---|---|---|---|---|---|
| 108 | 107 | 106 | 105 | 104 | 103 | 102 | 101 | |
| LAMP | + | + | + | + | + | + | ± | − |
| mPCR | + | + | + | + | + | + | + | + |
| Media | Temperature (°C) | Incubation Point (h) | C. jejuni Positives (n = 3) * | |||
|---|---|---|---|---|---|---|
| mPCR | LAMP | |||||
| Sample1 | Sample2 | Sample1 | Sample2 | |||
| Bolton broth | 37 | 6 | 3 | 3 | 0 | 0 |
| 41.5 | 12 | 2 | 1 | 1 | 0 | |
| 41.5 | 18 | 2 | 2 | 0 | 0 | |
| 41.5 | 24 | 2 | 1 | 0 | 1 | |
| 41.5 | 30 | 2 | 2 | 0 | 0 | |
| 41.5 | 36 | 1 | 1 | 0 | 1 | |
| 41.5 | 42 | 0 | 0 | 1 | 0 | |
| 41.5 | 48 | 2 | 1 | 0 | 0 | |
| mCCDA | 41.5 | 48 | 3 | 3 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jainonthee, C.; Chaisowwong, W.; Ngamsanga, P.; Wiratsudakul, A.; Meeyam, T.; Pichpol, D. A Cutoff Determination of Real-Time Loop-Mediated Isothermal Amplification (LAMP) for End-Point Detection of Campylobacter jejuni in Chicken Meat. Vet. Sci. 2022, 9, 122. https://doi.org/10.3390/vetsci9030122
Jainonthee C, Chaisowwong W, Ngamsanga P, Wiratsudakul A, Meeyam T, Pichpol D. A Cutoff Determination of Real-Time Loop-Mediated Isothermal Amplification (LAMP) for End-Point Detection of Campylobacter jejuni in Chicken Meat. Veterinary Sciences. 2022; 9(3):122. https://doi.org/10.3390/vetsci9030122
Chicago/Turabian StyleJainonthee, Chalita, Warangkhana Chaisowwong, Phakamas Ngamsanga, Anuwat Wiratsudakul, Tongkorn Meeyam, and Duangporn Pichpol. 2022. "A Cutoff Determination of Real-Time Loop-Mediated Isothermal Amplification (LAMP) for End-Point Detection of Campylobacter jejuni in Chicken Meat" Veterinary Sciences 9, no. 3: 122. https://doi.org/10.3390/vetsci9030122
APA StyleJainonthee, C., Chaisowwong, W., Ngamsanga, P., Wiratsudakul, A., Meeyam, T., & Pichpol, D. (2022). A Cutoff Determination of Real-Time Loop-Mediated Isothermal Amplification (LAMP) for End-Point Detection of Campylobacter jejuni in Chicken Meat. Veterinary Sciences, 9(3), 122. https://doi.org/10.3390/vetsci9030122







