Molecular Detection and Differentiation of Arthropod, Fungal, Protozoan, Bacterial and Viral Pathogens of Honeybees
Abstract
:1. Introduction
2. Description of Main Bee Pathogens and Molecular Methods for Their Detection
2.1. Arthropoda
2.1.1. Overview
2.1.2. Varroa destructor and V. jacobsoni, Causative Agents of Varoosis
2.1.3. Acarapsis woodi Causing Acarapiosis
2.1.4. Tropilaelaps mercedesae and T. clareae
2.1.5. Aethina tumida or Small Hive Beetle (SHB)
| Type of Reaction | Species or Genotypes | Target | Size of Amplicon or RFLP Pattern (nt) or Number of Markers | Accession Number | Ref. |
|---|---|---|---|---|---|
| Varroa | |||||
| RAPD | V. destructor a/V. jacobsoni | fingerprints | variable | n.a. | [125] |
| PCR-amplicon sequencing | V. destructor V. jacobsoni V. rindereri V. underwoodi | cox1 | 458 | AJ493124 | [12] |
| V. destructor haplogroups: K1, V1, C1, C2, C3, J1 | cox1 | 929 | [56] | ||
| haplotypes: K1-1/2,3,4; V1-1,2,3,4; C1-1,2; C2-1; C3-1; J1-1,2,3,4,5,6 | atp6-cox3 | 818 | |||
| cytb | 985 | ||||
| V. destructor haplogroups: K1, K2, J2, C1, C4 | cox1 | 821 | [64] | ||
| haplotypes: K1-1/2,3,5,6,7,8,9,10,11,12,13,14,15; K2-1; J2-1; C1-2,C4-1 | cytb | 985 | |||
| V. destructor haplogroups: K1, J1, C1-1 | cox1 cytb cox3 | various | AJ493124 AY163547 | [62] | |
| V. destructor haplotypes: KArg-N1, KArg-N2 heteroplasmy KArg-N1/N2 | ND4-ND4L | 839 | [68] | ||
| ARMS-PCR | V. destructor haplotype: K, S1; heteroplasmy: KS1 | cox1 | variable | AF106899 | [66] |
| PCR-RFLP | V. destructor haplotype J and K | cox1 | 68,129 and 273 for haplotype J; 129,341 for haplotype K (EcoNI) 230 and 340 for J haplotype; no cut in K haplotype (SacI) | AJ493124 | [71] |
| V. destructor haplotype J and K | cytb | 128/124 and 252/256 for J haplotype, 376 for K haplotype (SacI) | AJ493124 AJ784872 | [60] | |
| V. destructor haplotype K and P1 heteroplasmy: KP1 | 226 for haplotype K; 58,168 for haplotype P1 (VspI) | JX970945 | [66] | ||
| V. destructor tau-fluvalinate resistant vs. non-resistant variant | NaVCh | 603 for Tau-fluvalinate resistant; 270,333 for Tau-fluvalinate susceptible homozygotes (SacI) | KC152656 | [76] | |
| Microsatellites | V. destructor multilocus genotypes | tandem repeats | 20 markers | AF229974-77 AF229979-85 | [73] |
| 16 and 13 markers | AJ558164-79 | [60,126] | |||
| 4 markers | several | [118] | |||
| several potential markers | several | [63] | |||
| Acarapis | |||||
| PCR | A. externus A. dorsalis A. woodi | cox1 | 377 | GQ916565 | [84] |
| 247 | AB638409 AB638410 | [85] | |||
| 162 | EU190886 FJ603294 FJ603296 GQ916565 | [80] | |||
| A. woodi, not A. exernus; A. dorsalis not confirmed | 180 | AB634837 | [86] | ||
| qPCR | A. woodi | cox1 | 113 | EU190886 | [87] |
| ITS2 | 94 | HQ25966-7 HQ259670 FJ603297 FJ603298 | [87] | ||
| Tropilaelaps | |||||
| RAPD | T. clareae, T. koenigerum | fingerprints | variable | n.a. | [99] |
| PCR-RFLP | T. clareae T. mercedesae T. thaii T. koenigerum | ITS | 63,260,280 or 140,150,310 for T. clareae; 270,330 or 285,310 for T. koemigerum (MseI and Sau3AI) | n.a. (universal primers [98]) | [99] |
| ITS1-5.8S-ITS2 | distinguishes the 4 species (Bme1580I, PsiI, and RsaI) | n.a. (universal primers [98]) | [93] | ||
| cox1 | distinguishes the 4 species (FauI, BsrI, BstYI and SwaI) | n.a. (universal primers [100]) | |||
| qPCR | T. clareae | cox1 | various | msa b | [101] |
| Aethina | |||||
| PCR | A. tumida | cox1 | 1080 | KT380626 | [118] |
| qPCR | 109 | AF227645-54 AF522354–58 | [116] | ||
| 396 | KT380625-6 AF227647 | [117] | |||
| LAMP | variable | msa b | [120] | ||
| Microsatellites | tandem repeats | 15 markers | several | [121] | |
2.2. Fungi
2.2.1. Overview
2.2.2. Nosema apis and N. ceranae, Causative Agents of Nosemosis
2.2.3. Ascosphaera apis, Causative Agent of Chalkbrood
2.2.4. Aspergillus sp.
| Type of Reaction | Species | Target | Size of Amplicon (nt) | Accession Number | Ref. |
|---|---|---|---|---|---|
| Nosema | |||||
| PCR | 16S rRNA | 222 | AY741110 U97150 U26533 | [131] | |
| 208,212 | U97150 DQ486027 | [142] | |||
| 488 | U97150 | [211] | |||
| Nosema apis | |||||
| PCR | 16S rRNA | 209 | 857487 | [212] | |
| 240 | U26534 | [130] | |||
| 325 | U97150 | [213] | |||
| 401 | [142] | ||||
| PCR-RFLP | 16S rRNA | 91,136,175 (MspI, NdeI) | U97150 | [131] | |
| 18S rRNA | 433 | [214] | |||
| qPCR | 16 rRNA, ITS, 18S rRNA | 269 | U97150 | [177] | |
| 16S rRNA | 104 | [167] | |||
| 278 | [178] | ||||
| 103 | [180] | ||||
| 312 | DQ235446 | [215] (improvement on [170]) | |||
| rpb1 | 71 | DQ996230 | [146] | ||
| LAMP | 16S rRNA | variable | JQ639306 | [185] | |
| Nosema ceranae | |||||
| PCR | 16S rRNA | 252 | U26533 | [130] | |
| 250 | DQ486027 | [142] | |||
| PCR-RFLP | 16S rRNA | 104,116,177 (MspI, PacI) | DQ078785 | [131] | |
| 18S rRNA | 175 and 262 | [214] | |||
| qPCR | 16S rRNA, ITS, 18SrRNA | 250 | DQ486027 | [177] | |
| 16S rRNA | 142 | [167] | |||
| 18 rRNA | 316 | [178] | |||
| 16S rRNA | 92 | [180] | |||
| hsp70 | 65 | XM_002995382 | [181] | ||
| 16S rRNA | 221 | DQ329034 | [215] (improvement on [170]) | ||
| ptp3 | 90 | XM_002996713 | [146] | ||
| 5S rRNA, 16S rRNA, ITS | 216 | JX205151 | [182] | ||
| 5S rRNA | 76 | EF091879 | [183] | ||
| LAMP | 16S rRNA | variable | DQ078785 | [185] | |
| variable | DQ486027 | [186] | |||
| ptp3 | variable | XM_024473556 | [148] | ||
| Nosemaapis and N. ceranae | |||||
| PCR | ITS region | 118–122 | AY741110 | [213] | |
| multiplex PCR | 16S rRNA | 218–219 | DQ486027 | [170] | |
| 321 | DQ329034 U26533 DQ078785 DQ286728 | [216] | |||
| N. apis | 16S rRNA | 321 | DQ329034 | [171] | |
| N. ceranae | 218 | DQ486027 | |||
| A. mellifera | RPS5 gene | 115 | XM_006570237 | ||
| N. apis | 16S rRNA | 224 | U97150 | [15] | |
| N. ceranae | 143 | DQ486027 | |||
| N. apis | rpb1 | 297 | DQ996230 | [14] | |
| N. ceranae | 662 | M_002995356 | |||
| Ascosphaera apis | |||||
| PCR | 5.8S rRNA | 136 | U68313 U18362 | [195] | |
| ITS1-5.8S rRNA-ITS2 | 525,439 | GQ867766 | [194] | ||
| 486 | [193] | ||||
| rep-PCR | ERIC, BOX, or REP elements | variable | n.a. | [196] | |
| Aspergillus | |||||
| LAMP | A. flavus | 18S rRNA | variable | D63696 | [210] |
| PCR | Aspergillus spp. | ITS region | variable | n.k. | [98] |
| β-tubulin | variable | n.k. | [209] | ||
2.3. Protozoa
2.3.1. Overview
2.3.2. Crithidia mellificae and Lotmaria passim
2.3.3. Apicystis bombi
2.3.4. Malpighamoeba mellificae
| Type of Reaction | Species | Target | Size of Amplicon (nt) | Accession Number | Ref. |
|---|---|---|---|---|---|
| Lotmaria passim | |||||
| PCR | cytb | 247 | KJ684960 | [225] | |
| 18S rRNA | 459 | KM066228 KJ713376 KJ71337 | [224] | ||
| GAPDH | 402 | M066224 KJ713349 KJ71335 | |||
| 18S rRNA | 163 | KM066244 | [227] | ||
| qPCR | cytb | 146 | KJ684960 | [164] | |
| Crithidia mellificae | |||||
| PCR | cytb | 140 | KJ684951 | [225] | |
| GAPDH | 140 | KJ713345 | [227] | ||
| Lotmaria and Crithidia | |||||
| qPCR a | C. mellificae (and L. passim) | 18S rRNA | 123/125 | KX953204/MN879795 | [162] a |
| multiplex PCR | L. passim | rpb1 | 254 | LT976800-2 | [226] |
| C. mellificae | GAPDH | 177 | |||
| C. bombi | TOPII | 133 | |||
| multiplex qPCR | L. passim | cytb | 184 | MG494247 KJ684969 | [228] |
| C. mellificae | 146 | ||||
| Apicystis bombi | |||||
| PCR | 18S rRNA | 850 | FN546182 | [236] | |
| Malpighamoeba mellificae | |||||
| qPCR | 18S rRNA | 137 | OL757386 | [243] | |
2.4. Bacteria
2.4.1. Overview
2.4.2. Paenibacillus larvae, the Causative Agent of American Foulbrood
2.4.3. Melissococcus plutonius, Causing European Foulbrood
2.4.4. Spiroplasma apis and S. melliferum Causatives Agents of May Disease
2.4.5. Serratia marcescens Causing Honeybee Sepsis
| Type of Reaction | Species, Strains or Genotype | Target | Amplicon Size (nt), Sequence Types and Profiles | Accession Number and Amplification Primers | Ref. |
|---|---|---|---|---|---|
| Paenibacillus larvae | |||||
| PCR | 16S rRNA | 973 | X60619 | [259] | |
| 1106 | AY030079 | [260] | |||
| 700 | [261] | ||||
| 237,374,451,695,739 | [256] | ||||
| 665,965 | [262] | ||||
| metalloproteinase | 155,242,342 | AF111421 | [256] | ||
| 273 | [262] | ||||
| ERIC I + II | ERIC amplicon of 970bp | 550 | n.a. | [318] | |
| nPCR | 16S rRNA | 572 (final amplicon) | AY030079 | [263] | |
| qPCR | 16S rRNA | 233 | U85263 | [264] | |
| 74 | X60619 | [265] | |||
| 380 | AY030079 | [266] | |||
| 130 | CP019687, locus tag BXP28_01730 | [258] | |||
| 167 | [267] | ||||
| multiplex qPCR | ERIC I + II | 16S rRNA | 249 | AY530294 | [271,319] |
| ERIC I + II | ItuC | 209 | CP019717 | ||
| ERIC I | plx1 | 176 | KC456421 | ||
| rep-PCR | ERIC I-V genotype | ERIC sequences | ERIC profiles I-V | ERIC1R-ERIC2 primer pair | [320] |
| MLST | ST (sequence type) 1-21 + 24,25 | clpC, ftsA, glpF, glpT, Natrans, rpoB, sigF | ST1–21 ST1-7, 13-21: ERIC I; ST10-12,24: ERIC II; ST8-9: ERIC III/IV; ST25:ERIC V | HG530076-109 of target genes and alleles | [20] |
| ST 1-15 | ilvD, tri, purH, recF, pyre, SucC, glpF | ST1–15 ST1-2,7-15: ERIC I; ST4: ERIC II; ST6: ERIC III, ST3,5: ERIC IV | KY673263-528 of target genes and alleles | [273] | |
| MLVA | MLVA type 1-23 | VNTRs | MLVA1-23; MLVA1-17: ERIC I MLVA18-21: ERIC II MLVA22: ERICIII; MLVA23: ERIV IV | VNTR primer pairs A-E | [274] |
| solid wgMLST | wgMLST types | reference whole genome including 5745 loci derived from comparison of 179 genomes | much improved discrimination compared to traditional MLST | n.a. | [21] |
| Melissococcus plutonius | |||||
| PCR | 16S rRNA | 812 | X75752 | [321] | |
| melissotoxin A | 1360 | KMT29105 | [284] | ||
| multiplex PCR | typical strains (CC3, CC13) | napA | 187 | AB778538 | [299] |
| atypical strains (C12) | Fur | 424 | BAL62104 | ||
| seminested PCR | 16S rRNA | 486,276 | X757551 | [292,322] | |
| qPCR | sodA | 79 | EF666055 | [291] | |
| 16S rRNA | 69 | AJ301842 | [323] | ||
| LAMP | gyrB | variable | AP012200, locus_tag MPTP_0005 | [294] | |
| MLST | STs cluster into clonal complexes CC3, 13 and CC12 | argE, galK, gbpB, purR | ST1-27; ST1, 4,8,9,13,14, 15,17,18,20,26: CC13; ST2,3,4,5,6,7,11,12,22,23,24: CC3 ST10,12,16,19,21,25, 27: CC12 | HF569117-42 | [19,295] [296] [297] |
| Nanoparticle-based detection | cell wall-associated protease gene | n.a. | NC_015516 | [293] | |
| P. larvae and M. plutonius | |||||
| multiplex qPCR | P. larvae | tnp60 | 87 | CP003355 | [300] |
| M. plutonius | napA | 92 | AB778538 | ||
| a A. mellifera | actin | 87 | AB023025 | ||
| multiplex PCR | P. larvae | 16S rRNA | 973 | [195,259] | |
| M. plutonius | 281 | M. plutonius: b msa based on AY862507, AJ301842, X75751-2 | |||
| c Ascosphaera apis | 5.8S rRNA | 136 | A. apis: b msa. based on U68313, U18362 | ||
| Spiroplasma apis and S. melliferum | |||||
| multiplex PCR | S. apis and S. melliferum | 16S rRNA | 976 | JN628939 | [308] |
| S. apis | rpoB | 636 | DQ313816 | ||
| S. melliferum | spiralin | 160 | M59366 | ||
| multiplex qPCR | S. apis | 16S rRNA-ITS1 | 190 | AY736030 | [302] |
| S. melliferum | spiralin | 160 | M59366 | ||
| A. mellifera | RPS5 gene | 115 | GB11132 | ||
2.5. Virus
2.5.1. Overview
2.5.2. Molecular Detection of Common Bee Viruses and Their Variants
2.5.3. Application of Molecular Diagnostics for the Detection of Multiple Viruses
2.5.4. Recently Identified Bee Viruses by Metagenomics
| Type of Reaction | Species or Genotypes | Target | Size of Amplicon (nt) | Accession Number | Ref. |
|---|---|---|---|---|---|
| Acute Bee Paralysis Virus (ABPV) | |||||
| RT-PCR | VP1 | 900 | AF150629 | [390] | |
| intergenic region, VP2, VP4, VP3, VP3, VP1, VP1 | 722,788,686,619, 398,858,687 | [373] | |||
| RdRP | 452 | [326] | |||
| VP3 | 618 | [388] | |||
| RT-qPCR (SYBR Green) | RdRP | 66 | AF150629 | [418] | |
| 178 | NC_002548 | [378] | |||
| 177 | AF150629 | [324] | |||
| VP1 | 197 | AF150629 | [389] | ||
| RT-qPCR (TaqMan) | VP3 | 67 | AF263733 | [328] | |
| ORF2 | nk | AF126050 | [419] | ||
| Aphid lethal paralysis virus strain Brookings (ALP-Br) | |||||
| RT-PCR | helicase | 464 | Q871932 | [162] | |
| RT-qPCR (SYBR Green) | helicase | 141 | Q871932 | [162] | |
| Apis iridiscent virus | |||||
| RT-qPCR (TaqMan) | major capsid protein | 95 | AF042340 | [328] | |
| Big Sioux River virus (BSRV) | |||||
| RT-PCR | protease | 519 | GF423195 | [162] | |
| RT-qPCR (SYBR Green) | 5′UTR | 281 | n.k. | [162] | |
| Black Queen Cell Virus (BQCV) | |||||
| RT-PCR | ORF2 | 700 | NC_003784 | [390] | |
| RdRP | 424 | AF183905 | [326] | ||
| 5′UTR | 472 | AF125252 | [388] | ||
| capsid/3′UTR | 700 | NC003784 | [420] | ||
| RT-qPCR (SYBR Green) | helicase | 107 | AF125252 | [418] | |
| ORF2 | 141 | NC_003784 | [162] | ||
| 294 | [389] | ||||
| RT-qPCR (TaqMan) | capsid polyprotein | 71 | NC003784 | [328] | |
| Chronic Bee Paralysis Virus (CBPV) | |||||
| RT-PCR | RdRP | 445 | AF375659 | [398] | |
| 5′UTR | 315 | AF375659 | [388] | ||
| RT-qPCR (SYBR Green) | RdRP | 97 | AF375659 | [418] | |
| RdRP | 148 | EU122229 | [394] | ||
| RT-qPCR (TaqMan) | RdRP | 101 | EU122229 | [371] | |
| 57 | FJ345309 | [421] | |||
| Deformed-Wing Virus (DWV) | |||||
| RT-PCR | structural polyprotein | 194 | AY292384 | [401] | |
| polyprotein | 434 | AJ489744 | [388] | ||
| helicase | 174 | [334] | |||
| non-structural proteins | 205 | [422] | |||
| DWV A | structural and non-structural proteins | variable | AJ489744 | [423] | |
| DWV B | variable | NC_006494 | |||
| DWV A | capsid | 424 | NC004830 | [420] | |
| DWV B | 528 | ||||
| DWV C | 446 | ||||
| RT-qPCR (SYBR Green) | DWV A | 3Cpro | 136 | n.k. | [389] |
| DWV B | L | 413 | n.k. | ||
| DWV A and DWV B | helicase | 179 | AY292384 | [341] | |
| DWV A | VP2 | 211 | AY292384 | ||
| DWV B | IRES (internal ribosome entry site) | 116 | AY251269 | ||
| 69 | [418] | ||||
| DWV A | RdRP | 155 | NC_004830 | [331] | |
| DWV B | 155 | AY_251269 | |||
| DWV C | 152 | CEND01000001 | |||
| DWV A | IRES | 118 | AJ489744 | [423] | |
| DWV B | 117 | NC_006494 | |||
| DWV A | structural proteins | 97 | AJ489744 | ||
| DWV B | 97 | NC_006494 | |||
| DWV A | non-structural proteins | 101 | AJ489744 | ||
| DWV B | 101 | NC_006494 | |||
| DWV A | helicase | 186 | AY292384 | [422] | |
| DWV B | 189 | AY292384 | |||
| RT-qPCR (TaqMan) | helicase | 702 | NC_004830 | [396] | |
| RdRP | 114 | [328] | |||
| polyprotein | 67 | HM067437 | [421] | ||
| DWV A | VP3 | 72 | AY292384 | [406] | |
| DWV B | 73 | AY251269 | |||
| Israeli Acute Paralysis Virus (IAPV) | |||||
| RT-PCR | 3′UTR | 475 | NC_009025 | [381] | |
| capsid | 840 | NC009025 | [420] | ||
| RT-PCR | intergenic region, poliprotein | 185 | EU218534 | [383] | |
| RT-qPCR (SYBR Green) | VP3 | 226 | EF219380 | [374] | |
| RdRP | 137 | ||||
| ORF2 | 114 | ||||
| 203 | n.k. | [389] | |||
| 114 | NC_009025 | [368] | |||
| RT-qPCR (SYBR Green) | ORF2 RdRP | 226 137 | n.k. | [374] | |
| Multi-point PCR (SYBR Green) | VP3 VP1 RdRP | 298 225 219 | KC690270 | [387] | |
| RT-qPCR (TaqMan) | RNApol | 63 | EU436450 | [421] | |
| Kashmir Bee Virus (KBV) | |||||
| RT-PCR | RdRP | 417 | NC_004807 | [397] | |
| 683 | AY275710 | [424] | |||
| 3Cpro | 290 | [361] | |||
| ORF2 | 395 | [388] | |||
| capsid | 625 | NC004807 | [420] | ||
| RT-qPCR (SYBR Green) | 3Cpro | 69 | AY275710 | [418] | |
| 122 | [376] | ||||
| ORF2 | 200 | [389] | |||
| RT-qPCR (TaqMan) | RdRP | 63 | AY275710 | [421] | |
| VP3 | 69 | AF263725 | [328] | ||
| Lake Sinai Virus (LSV) | |||||
| RT-PCR | capsid polyprotein | 365 | NC_032433 | [369] | |
| 205 | NC_032433 | [162] | |||
| LSV1 | RdRP | 672 | HQ871931 | ||
| LSV2 | capsid protein | 558 | HQ888865 | ||
| LSV3 | RdRP | 243 | JQ480620 | [368] | |
| LSV4 | 379 | JX878492 | |||
| LSV5 | 190 | KC880124 | |||
| RT-qPCR (SYBR Green) | LSV universal | RdRP | 188 | NC_032433 | [162] |
| LSV1 | 153 | HQ871931 | |||
| LSV2 | 225 | HQ888865 | |||
| RT-qPCR (TaqMan) | LSV1, 2, 3, 4 universal | RdRP | 152 | n.k. | [368] |
| LSV3 | 123 | KY465717 | [405] | ||
| Moku virus (MV) | |||||
| RT-qPCR (SYBR Green) | RdRP | 93 | KU645789 | [365] | |
| Slow Bee Paralysis Virus (SBPV) | |||||
| RT-qPCR (SYBR Green) | n.k. | 226 | NC_014137 KY243931 | [389] | |
| Sacbrood Virus (SBV) | |||||
| RT-PCR | helicase | 823 | AF092924 | [351] | |
| 123 | AF092924 | [368] | |||
| 824 | AF092924 | [334] | |||
| SBV genome | variable | [425] | |||
| 5′UTR | 487 | [388] | |||
| structural proteins | 816 | [426] | |||
| 211 | [361] | ||||
| RdRP | 426 | [326] | |||
| capsid | 693 | [420] | |||
| RT-qPCR (SYBR Green) | RdRP | 70 | NC002066 | [418] | |
| VP3 | 335 | AF092924 | [389] | ||
| RT-qPCR (TaqMan) | polyprotein | 70 | AF092924 | [363] | |
| 106 | MG545287 | [405] | |||
| RdRP | 70 | NC002066 | [328] | ||
| VP3 | 103 | AF092924 | [376] | ||
| ABPV, BQCV, CBPV, DWV, IAPV, SBV | |||||
| RT-PCR multiplex | ABPV | forward primer: intergenic region of Aparavirus (generic primer of both ABPV and IAPV); reverse primer: polyprotein | 460 | AF486073 | [23] |
| BQCV | polyprotein | 536 | EF517520/7762 | ||
| CBPV | RdRP | 774 | EU122229 | ||
| DWV | structural polyprotein | 269 | GU109335 | ||
| IAPV | intergenic region/polyprotein | 158 | HQ897161 | ||
| SBV | polyprotein | 342 | AF092924 | ||
| ABPV, BQCV, SBV | |||||
| RT-PCR multiplex | ABPV | ORF2 | 202 | NC_002548 | [22] |
| BQCV | ORF1 | 322 | AF183905: 379-700 | ||
| SBV | ORF | 487 | NC_002066: 221-708 | ||
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lautenbach, S.; Seppelt, R.; Liebscher, J.; Dormann, C.F. Spatial and Temporal Trends of Global Pollination Benefit. PLoS ONE 2012, 7, e35954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding Pollinators and Their Values to Human Well-Being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Pasho, D.J.M.; Applegate, J.R.; Hopkins, D.I. Diseases and Pests of Honey Bees (Apis mellifera). Vet. Clin. N. Am. Food Anim. 2021, 37, 401–412. [Google Scholar] [CrossRef] [PubMed]
- MacAfee, A. The Surprisingly Sophisticated Ways That Honey Bees Fight Disease. Available online: https://americanbeejournal.com/the-surprisingly-sophisticated-ways-that-honey-bees-fight-disease/ (accessed on 19 January 2022).
- Tehel, A.; Brown, M.J.; Paxton, R.J. Impact of Managed Honey Bee Viruses on Wild Bees. Curr. Opin. Virol. 2016, 19, 16–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanetti, A.; Bortolotti, L.; Cilia, G. Pathogens Spillover from Honey Bees to Other Arthropods. Pathogens 2021, 10, 1044. [Google Scholar] [CrossRef]
- Forfert, N.; Natsopoulou, M.E.; Frey, E.; Rosenkranz, P.; Paxton, R.J.; Moritz, R.F.A. Parasites and Pathogens of the Honeybee (Apis mellifera) and Their Influence on Inter-Colonial Transmission. PLoS ONE 2015, 10, e0140337. [Google Scholar] [CrossRef] [PubMed]
- Shimanuki, H.; Knox, D.A. Bee Health and International Trade. Rev. Sci. Tech. OIE 1997, 16, 172–176. [Google Scholar] [CrossRef] [Green Version]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 Years Postdetection Perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef] [Green Version]
- Traynor, K.; Mondet, F.; de Miranda, J.; Techer, M.; Kowallik, V.; Oddie, M.; Chantawannakul, P.; McAfee, A. Varroa destructor: A Complex Parasite, Crippling Honeybees Worldwide. Preprints 2020, 36, 592–606. [Google Scholar] [CrossRef]
- Morfin, N.; Anguiano-Baez, R.; Guzman-Novoa, E. Honey Bee (Apis mellifera) Immunity. Vet. Clin. N. Am. Food Anim. 2021, 37, 521–533. [Google Scholar] [CrossRef]
- Anderson, D.L.; Trueman, J.W.H. Varroa jacobsoni (Acari: Varroidae) Is More than One Species. Exp. Appl. Acarol. 2000, 24, 165–189. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Bauchan, G.R.; Murphy, C.A.; Ravoet, J.; de Graaf, D.C.; Evans, J.D. Characterization of Two Species of Trypanosomatidae from the Honey Bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. Gen., n. sp. J. Eukaryot. Microbiol. 2015, 62, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Gisder, S.; Genersch, E. Molecular Differentiation of Nosema apis and Nosema ceranae Based on Species–Specific Sequence Differences in a Protein Coding Gene. J. Invert. Pathol. 2013, 113, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fries, I.; Chauzat, M.-P.; Chen, Y.-P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard Methods for Nosema Research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Tokarev, Y.S.; Huang, W.-F.; Solter, L.F.; Malysh, J.M.; Becnel, J.J.; Vossbrinck, C.R. A Formal Redefinition of the Genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and Reassignment of Species Based on Molecular Phylogenetics. J. Invert. Pathol. 2020, 169, 107279. [Google Scholar] [CrossRef]
- Cameron, T.C.; Wiles, D.; Beddoe, T. Current Status of Loop-Mediated Isothermal Amplification Technologies for the Detection of Honey Bee Pathogens. Front. Vet. Sci. 2021, 8, 345. [Google Scholar] [CrossRef]
- Roudel, M.; Tottey, J.; Corbara, B.; Delbac, F.; Blot, N. New Insights on the Genetic Diversity of the Honeybee Parasite Nosema ceranae Based on Multilocus Sequence Analysis. Parasitology 2013, 140, 1346–1356. [Google Scholar] [CrossRef]
- Haynes, E.; Helgason, T.; Young, J.P.W.; Thwaites, R.; Budge, G.E. A Typing Scheme for the Honeybee Pathogen Melissococcus plutonius Allows Detection of Disease Transmission Events and a Study of the Distribution of Variants. Environ. Microbiol. Rep. 2013, 5, 525–529. [Google Scholar] [CrossRef]
- Morrissey, B.J.; Helgason, T.; Poppinga, L.; Fünfhaus, A.; Genersch, E.; Budge, G.E. Biogeography of Paenibacillus larvae, the Causative Agent of American Foulbrood, Using a New Multilocus Sequence Typing Scheme. Environ. Microbiol. 2015, 17, 1414–1424. [Google Scholar] [CrossRef] [Green Version]
- Papić, B.; Diricks, M.; Kušar, D. Analysis of the Global Population Structure of Paenibacillus larvae and Outbreak Investigation of American Foulbrood Using a Stable WgMLST Scheme. Front. Vet. Sci. 2021, 8, 582677. [Google Scholar] [CrossRef]
- Grabensteiner, E.; Bakonyi, T.; Ritter, W.; Pechhacker, H.; Nowotny, N. Development of a Multiplex RT-PCR for the Simultaneous Detection of Three Viruses of the Honeybee (Apis mellifera L.): Acute Bee Paralysis Virus, Black Queen Cell Virus and Sacbrood Virus. J. Invert. Pathol. 2007, 94, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Sguazza, G.H.; Reynaldi, F.J.; Galosi, C.M.; Pecoraro, M.R. Simultaneous Detection of Bee Viruses by Multiplex PCR. J. Virol. Methods 2013, 194, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E.; Aubert, M. Emerging and Re-Emerging Viruses of the Honey Bee (Apis mellifera L.). Vet. Res. 2010, 41, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantawannakul, P.; Ramsey, S.; vanEngelsdorp, D.; Khongphinitbunjong, K.; Phokasem, P. Tropilaelaps Mite: An Emerging Threat to European Honey Bee. Curr. Opin. Insect. Sci. 2018, 26, 69–75. [Google Scholar] [CrossRef]
- Sammataro, D.; Gerson, U.; Needham, G. Parasitic Mites of Honey Bees: Life History, Implications, and Impact. Annu. Rev. Entomol. 2000, 45, 519–548. [Google Scholar] [CrossRef] [PubMed]
- vanEngelsdorp, D.; Hayes, J.; Underwood, R.M.; Pettis, J. A Survey of Honey Bee Colony Losses in the U.S., Fall 2007 to Spring 2008. PLoS ONE 2008, 3, e4071. [Google Scholar] [CrossRef] [PubMed]
- Currie, R.W.; Pernal, S.F.; Guzmán-Novoa, E. Honey Bee Colony Losses in Canada. J. Apic. Res. 2010, 49, 104–106. [Google Scholar] [CrossRef]
- Dahle, B. The Role of Varroa destructor for Honey Bee Colony Losses in Norway. J. Apic. Res. 2010, 49, 124–125. [Google Scholar] [CrossRef]
- Genersch, E.; von der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; et al. The German Bee Monitoring Project: A Long Term Study to Understand Periodically High Winter Losses of Honey Bee Colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef] [Green Version]
- Le Conte, Y.; Ellis, M.; Ritter, W. Varroa Mites and Honey Bee Health: Can Varroa Explain Part of the Colony Losses? Apidologie 2010, 41, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and Control of Varroa destructor. J. Invert. Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Novoa, E.; Eccles, L.; Calvete, Y.; Mcgowan, J.; Kelly, P.G.; Correa-Benítez, A. Varroa destructor Is the Main Culprit for the Death and Reduced Populations of Overwintered Honey Bee (Apis mellifera) Colonies in Ontario, Canada. Apidologie 2010, 41, 443–450. [Google Scholar] [CrossRef] [Green Version]
- Oudemans, A.C. On a New Genus and Species of Parasitic Acari. Notes Leyden Mus. 1904, 24, 216–222. [Google Scholar]
- Chantawannakul, P.; de Guzman, L.I.; Li, J.; Williams, G.R. Parasites, Pathogens, and Pests of Honeybees in Asia. Apidologie 2016, 47, 301–324. [Google Scholar] [CrossRef]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D.L.; Locke, B.; Delaplane, K.S.; Wauquiez, Q.; Tannahill, C.; Frey, E.; Ziegelmann, B.; et al. Standard Methods for Varroa Research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef] [Green Version]
- Delaplane, K.S.; Hood, W.M. Economic Threshold for Varroa jacobsoni Oud. in the Southeastern USA. Apidologie 1999, 30, 383–395. [Google Scholar] [CrossRef] [Green Version]
- Fries, I.; Hansen, H.; Imdorf, A.; Rosenkranz, P. Swarming in Honey Bees (Apis mellifera) and Varroa destructor Population Development in Sweden. Apidologie 2003, 34, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Rosenkranz, P.; Kirsch, R.; Renz, R. Population Dynamics of Honey Bee Colonies and Varroa Tolerance: A Comparison between Uruguay and Germany. In Proceedings of the 7th Encontro Sobre Abelhas, Ribeirão Preto, Brazil, 12–15 July 2006. [Google Scholar]
- Griffiths, D.A.; Needham, G.R.; Page Jr, R.E.; Delfinado-Baker, M.; Bowman, C.E. Functional Morphology of the Mouthparts of Varroa jacobsoni and Tropilaelaps clareae as a Basis for the Interpretation of Their Life-Styles; Ellis Horwood: Chichester, UK, 1988; pp. 479–489. [Google Scholar]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor Feeds Primarily on Honey Bee Fat Body Tissue and Not Hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef] [Green Version]
- Boecking, O.; Genersch, E. Varroosis—The Ongoing Crisis in Bee Keeping. J. Verbr. Lebensm. 2008, 3, 221–228. [Google Scholar] [CrossRef]
- Santillán-Galicia, M.T.; Ball, B.V.; Clark, S.J.; Alderson, P.G. Transmission of Deformed Wing Virus and Slow Paralysis Virus to Adult Bees (Apis mellifera L.) by Varroa destructor. J. Apic. Res. 2010, 49, 141–148. [Google Scholar] [CrossRef]
- Martin, S.J.; Brettell, L.E. Deformed Wing Virus in Honeybees and Other Insects. Annu. Rev. Virol. 2019, 6, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Miranda, J.R.; Cordoni, G.; Budge, G. The Acute Bee Paralysis Virus–Kashmir Bee Virus–Israeli Acute Paralysis Virus Complex. J. Invert. Pathol. 2010, 103, S30–S47. [Google Scholar] [CrossRef] [PubMed]
- Giacobino, A.; Molineri, A.I.; Pacini, A.; Fondevila, N.; Pietronave, H.; Rodríguez, G.; Palacio, A.; Bulacio Cagnolo, N.; Orellano, E.; Salto, C.E.; et al. Varroa destructor and Viruses Association in Honey Bee Colonies under Different Climatic Conditions. Environ. Microbiol. Rep. 2016, 8, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Siede, R. Honey Bee Viruses. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2007; Volume 70, pp. 33–80. ISBN 978-0-12-373728-1. [Google Scholar]
- Thaduri, S.; Locke, B.; Granberg, F.; de Miranda, J.R. Temporal Changes in the Viromes of Swedish Varroa-Resistant and Varroa-Susceptible Honeybee Populations. PLoS ONE 2018, 13, e0206938. [Google Scholar] [CrossRef]
- Tantillo, G.; Bottaro, M.; Di Pinto, A.; Martella, V.; Di Pinto, P.; Terio, V. Virus Infections of Honeybees Apis mellifera. Ital. J. Food Safety 2015, 4, 5364. [Google Scholar] [CrossRef] [Green Version]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.-P.; vanEngelsdorp, D. Drivers of Colony Losses. Curr. Opin. Insect. Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef]
- OIE—World Organisation for Animal Health. Varroosis of Honey Bees (Infestarion of Honey Bees with Varroa spp.). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Online Version); OIE: Paris, France, 2021; pp. 1–13. [Google Scholar]
- Macedo, P.; Wu, J.; Ellis, M. Using Inert Dusts to Detect and Assess Varroa Infestation in Honey Bee Colonies. J. Apic. Res. 2002, 41, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.V.; Moon, R.D.; Burkness, E.C.; Hutchison, W.D.; Spivak, M. Practical Sampling Plans for Varroa destructor (Acari: Varroidae) in Apis mellifera (Hymenoptera: Apidae) Colonies and Apiaries. J. Econ. Entomol. 2010, 103, 1039–1050. [Google Scholar] [CrossRef]
- Anderson, D.L.; Fuchs, S. Two Genetically Distinct Populations of Varroa jacobsoni with Contrasting Reproductive Abilities on Apis mellifera. J. Apic. Res. 1998, 37, 69–78. [Google Scholar] [CrossRef]
- Techer, M.A.; Rane, R.V.; Grau, M.L.; Roberts, J.M.K.; Sullivan, S.T.; Liachko, I.; Childers, A.K.; Evans, J.D.; Mikheyev, A.S. Divergent Evolutionary Trajectories Following Speciation in Two Ectoparasitic Honey Bee Mites. Commun. Biol. 2019, 2, 357. [Google Scholar] [CrossRef] [Green Version]
- Navajas, M.; Anderson, D.L.; de Guzman, L.I.; Huang, Z.Y.; Clement, J.; Zhou, T.; Le Conte, Y. New Asian Types of Varroa destructor: A Potential New Threat for World Apiculture. Apidologie 2010, 41, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Navajas, M.; Le Conte, Y.; Solignac, M.; Cros-Arteil, S.; Cornuet, J.-M. The Complete Sequence of the Mitochondrial Genome of the Honeybee Ectoparasite Mite Varroa destructor (Acari: Mesostigmata). Mol. Biol. Evol. 2002, 19, 2313–2317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, J.D.; Lopez, D.L. Complete Mitochondrial DNA Sequence of the Important Honey Bee Pest, Varroa destructor (Acari: Varroidae). Exp. Appl. Acarol. 2002, 27, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Anderson, D.L.; Huang, Z.Y.; Huang, S.; Yao, J.; Ken, T.; Zhang, Q. Identification of Varroa Mites (Acari: Varroidae) Infesting Apis Cerana and Apis mellifera in China. Apidologie 2004, 35, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Solignac, M.; Cornuet, J.; Vautrin, D.; Le Conte, Y.; Anderson, D.; Evans, J.; Cros-Arteil, S.; Navajas, M. The Invasive Korea and Japan Types of Varroa destructor, Ectoparasitic Mites of the Western Honeybee (Apis mellifera), Are Two Partly Isolated Clones. Proc. R. Soc. B 2005, 272, 411–419. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, I.; Garrido-Bailón, E.; Martín-Hernández, R.; Meana, A.; Higes, M.; De la Rúa, P. Genetic Profile of Varroa destructor Infesting Apis mellifera Iberiensis Colonies. J. Apic. Res. 2008, 47, 310–313. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Schiavo, G.; Ribani, A.; Bertolini, F.; Bovo, S.; Fontanesi, L. A next Generation Sequencing Approach for Targeted Varroa destructor (Acari: Varroidae) Mitochondrial DNA Analysis Based on Honey Derived Environmental DNA. J. Invert. Pathol. 2019, 161, 47–53. [Google Scholar] [CrossRef]
- Cornman, R.S.; Schatz, M.C.; Johnston, J.S.; Chen, Y.-P.; Pettis, J.; Hunt, G.; Bourgeois, L.; Elsik, C.; Anderson, D.; Grozinger, C.M.; et al. Genomic Survey of the Ectoparasitic Mite Varroa destructor, a Major Pest of the Honey Bee Apis mellifera. BMC Genom. 2010, 11, 602. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Wang, S.; Neumann, P.; Chen, G.; Page, P.; Li, L.; Hu, F.; Zheng, H.; Dietemann, V. Population Genetics and Host Specificity of Varroa destructor Mites Infesting Eastern and Western Honeybees. J. Pest. Sci. 2021, 94, 1487–1504. [Google Scholar] [CrossRef]
- Roberts, J.M.K.; Anderson, D.L.; Tay, W.T. Multiple Host Shifts by the Emerging Honeybee Parasite, Varroa jacobsoni. Mol. Ecol. 2015, 24, 2379–2391. [Google Scholar] [CrossRef]
- Gajić, B.; Stevanović, J.; Radulović, Ž.; Kulišić, Z.; Vejnović, B.; Glavinić, U.; Stanimirović, Z. Haplotype Identification and Detection of Mitochondrial DNA Heteroplasmy in Varroa destructor Mites Using ARMS and PCR–RFLP Methods. Exp. Appl. Acarol. 2016, 70, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Gajić, B.; Muñoz, I.; De la Rúa, P.; Stevanović, J.; Lakić, N.; Kulišić, Z.; Stanimirović, Z. Coexistence of Genetically Different Varroa destructor in Apis mellifera Colonies. Exp. Appl. Acarol. 2019, 78, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Muntaabski, I.; Russo, R.M.; Liendo, M.C.; Palacio, M.A.; Cladera, J.L.; Lanzavecchia, S.B.; Scannapieco, A.C. Genetic Variation and Heteroplasmy of Varroa destructor Inferred from ND4 MtDNA Sequences. Parasitol. Res. 2020, 119, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Strapazzon, R.; Carneiro, F.E.; Guerra, J.C.V., Jr.; Moretto, G. Genetic Characterization of the Mite Varroa destructor (Acari: Varroidae) Collected from Honey Bees Apis mellifera (Hymenoptera, Apidae) in the State of Santa Catarina, Brazil. Genet. Mol. Res. 2009, 8, 990–997. [Google Scholar] [CrossRef]
- de Guzman, L.I.; Rinderer, T.E.; Stelzer, J.A. Occurrence of Two Genotypes of Varroa jacobsoni Oud. in North America. Apidologie 1999, 30, 31–36. [Google Scholar] [CrossRef]
- Rodríguez, A.; Yadró, C.A.; Pérez, A.; Invernizzi, C.; Tomasco, I. Characterization of Varroa destructor Mites in Cuba Using Mitochondrial and Nuclear Markers. J. Apic. Sci. 2020, 64, 335–343. [Google Scholar] [CrossRef]
- Little, S. Amplification-refractory Mutation System (ARMS) Analysis of Point Mutations. Curr. Protoc. Hum. Genet. 1995, 7, 9.8.1–9.8.12. [Google Scholar] [CrossRef]
- Evans, J.D. Microsatellite Loci in the Honey Bee Parasitic Mite Varroa jacobsoni. Mol. Ecol. 2000, 9, 1436–1438. [Google Scholar] [CrossRef]
- Kelomey, A.E.; Paraiso, A.; Sina, H.; Legout, H.; Garnery, L.; Baba-Moussa, L. Genetic Characterization of the Honeybee Ectoparasitic Mite Varroa destructor from Benin (West Africa) Using Mitochondrial and Microsatellite Markers. Exp. Appl. Acarol. 2017, 72, 61–67. [Google Scholar] [CrossRef]
- Strachecka, A.; Borsuk, G.; Olszewski, K.; Paleolog, J. A New Detection Method for a Newly Revealed Mechanism of Pyrethroid Resistance Development in Varroa destructor. Parasitol. Res. 2015, 114, 3999–4004. [Google Scholar] [CrossRef] [Green Version]
- Stara, J.; Pekar, S.; Nesvorna, M.; Erban, T.; Vinsova, H.; Kopecky, J.; Doskocil, I.; Kamler, M.; Hubert, J. Detection of Tau-Fluvalinate Resistance in the Mite Varroa destructor Based on the Comparison of Vial Test and PCR–RFLP of Kdr Mutation in Sodium Channel Gene. Exp. Appl. Acarol. 2019, 77, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Delfinado-Baker, M.; Baker, E.W. Notes on Honey Bee Mites of the Genus Acarapis Hirst (Acari: Tarsonemidae). Int. J. Acarol. 1982, 8, 211–226. [Google Scholar] [CrossRef]
- Ahn, A.-J.; Ahn, K.-S.; Noh, J.-H.; Kim, Y.-H.; Yoo, M.-S.; Kang, S.-W.; Yu, D.-H.; Shin, S.S. Molecular Prevalence of Acarapis Mite Infestations in Honey Bees in Korea. Korean J. Parasitol. 2015, 53, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Eischen, F.A. Overwintering Performance of Honey Bee Colonies Heavily Infested with Acarapis woodi (Rennie). Apidologie 1987, 18, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Bailón, E.; Bartolomé, C.; Prieto, L.; Botías, C.; Martínez-Salvador, A.; Meana, A.; Martín-Hernández, R.; Higes, M. The Prevalence of Acarapis woodi in Spanish Honey Bee (Apis mellifera) Colonies. Exp. Parasitol. 2012, 132, 530–536. [Google Scholar] [CrossRef]
- Quintana, S.; Szawarski, N.; Sarlo, G.; Medici, S.; Rivero, M.; Eguaras, M.; Maggi, M. Comparison of QPCR and Morphological Methods for Detection of Acarapis woodi in Honey Bee Samples. J. Apic. Sci. 2019, 63, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Grant, G.A.; Nelson, D.L.; Olsen, P.E.; Rice, W.A. The “Elisa” Detection of Tracheal Mites in Whole Honey Bee Samples. Am. Bee J. 1993, 133, 652–655. [Google Scholar]
- Shimanuki, H.; Knox, D.A. Diagnosis of Honey Bee Diseases; No. AH–690; U.S. Department of Agriculture, Agriculture Handbook: Washington, DC, USA, 2000.
- Evans, J.D.; Pettis, J.S.; Smith, I.B. A Diagnostic Genetic Test for the Honey Bee Tracheal Mite, Acarapis woodi. J. Apic. Res. 2007, 46, 195–197. [Google Scholar] [CrossRef]
- Kojima, Y.; Yoshiyama, M.; Kimura, K.; Kadowaki, T. PCR-Based Detection of a Tracheal Mite of the Honey Bee Acarapis woodi. J. Invert. Pathol. 2011, 108, 135–137. [Google Scholar] [CrossRef]
- Cepero, A.; Martín-Hernández, R.; Prieto, L.; Gómez-Moracho, T.; Martínez-Salvador, A.; Bartolomé, C.; Maside, X.; Meana, A.; Higes, M. Is Acarapis woodi a Single Species? A New PCR Protocol to Evaluate Its Prevalence. Parasitol. Res. 2015, 114, 651–658. [Google Scholar] [CrossRef]
- Delmiglio, C.; Fan, Q.H.; George, S.; Ward, L.; Budge, G.; Flynn, A.; Kumarasinghe, L. Development and Evaluation of a Real-Time PCR Assay for the Detection of Acarapis woodi (Tracheal Mites) in Apis mellifera. Apidologie 2016, 47, 691–702. [Google Scholar] [CrossRef]
- Takashima, S.; Ohari, Y.; Itagaki, T. The Prevalence and Molecular Characterization of Acarapis woodi and Varroa destructor Mites in Honeybees in the Tohoku Region of Japan. Parasitol. Int. 2020, 75, 102052. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Fontanesi, L. Honey as a Source of Environmental DNA for the Detection and Monitoring of Honey Bee Pathogens and Parasites. Vet. Sci. 2020, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- OIE—World Organisation for Animal Health. Infestation of Honey Bees with Tropilaelaps spp. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2018; pp. 765–776. ISBN 978-92-95108-18-9. [Google Scholar]
- Woyke, J. Survivas and Prophylactic Control of Tropilaelaps Clarear Infesting Apis mellifera Colonies in Afghanistan. Apidologie 1984, 15, 421–434. [Google Scholar] [CrossRef] [Green Version]
- Thapa, R.; Wongsiri, S.; Manandhar, D. Current Status of Predators and Disease of Honey Bees in Nepal. In Proceedings of the Seventh International Conference on Tropical Bees: Management and Diversity, Chiang Mai, Thailand, 19–25 March 2000; International Bee Research Association: Cardiff, UK, 2000; pp. 221–226. [Google Scholar]
- Anderson, D.L.; Morgan, M.J. Genetic and Morphological Variation of Bee-Parasitic Tropilaelaps Mites (Acari: Laelapidae): New and Re-Defined Species. Exp. Appl. Acarol. 2007, 43, 1–24. [Google Scholar] [CrossRef] [PubMed]
- de Guzman, L.I.; Williams, G.R.; Khongphinitbunjong, K.; Chantawannakul, P. Ecology, Life History, and Management of Tropilaelaps Mites. J. Econ. Entomol. 2017, 110, 319–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khongphinitbunjong, K.; Neumann, P.; Chantawannakul, P.; Williams, G.R. The Ectoparasitic Mite Tropilaelaps Mercedesae Reduces Western Honey Bee, Apis mellifera, Longevity and Emergence Weight, and Promotes Deformed Wing Virus Infections. J. Invert. Pathol. 2016, 137, 38–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dainat, B.; Ken, T.; Berthoud, H.; Neumann, P. The Ectoparasitic Mite Tropilaelaps Mercedesae (Acari, Laelapidae) as a Vector of Honeybee Viruses. Insects. Soc. 2009, 56, 40–43. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.L.; Roberts, J.M.K. Standard Methods for Tropilaelaps Mites Research. J. Apic. Res. 2013, 52, 1–16. [Google Scholar] [CrossRef] [Green Version]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In Pcr Protocols: A Guide to Methods and Applications; Academic Press, Elsevier: Cambridge, MA, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Tangjingjai, W.; Verakalasa, P.; Sittipraneed, S.; Klinbunga, S.; Lekprayoon, C. Genetic Differences between Tropilaelaps clareae and Tropilaelaps Koenigerum in Thailand Based on ITS and RAPD Analyses. Apidologie 2003, 34, 513–524. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.L.; Gibbs, A.J.; Gibson, N.L. Identification and Phylogeny of Spore-Cyst Fungi (Ascosphaera spp.) Using Ribosomal DNA Sequences. Mycol. Res. 1998, 102, 541–547. [Google Scholar] [CrossRef]
- Del Cont, A.; De Georges, B.; Huleux, A.; Duquesne, V. Rapid Identification of Tropilaelaps Mite (Mesostigmata: Laelapidae) Species Using a COI Barcode-HRM. J. Econ. Entomol. 2021, 114, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Murray, A. List of Coleoptera Received from Old Calabar, on the West Coast of Africa. Ann. Magaz. Nat. Hist. 1867, 19, 334–340. [Google Scholar] [CrossRef]
- Lundie, A.E. The Small Hive Beetle, Aethina tumida; Entomological Series 3; Department of Agriculture and Forestry: Pretoria, South Africa, 1940; Volume 220, 30p. [Google Scholar]
- Neumann, P.; Evans, J.D.; Pettis, J.S.; Pirk, C.W.W.; Schäfer, M.O.; Tanner, G.; Ellis, J.D. Standard Methods for Small Hive Beetle Research. J. Apic. Res. 2013, 52, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Neumann, P.; Elzen, P.J. The Biology of the Small Hive Beetle (Aethina tumida, Coleoptera: Nitidulidae): Gaps in Our Knowledge of an Invasive Species. Apidologie 2004, 35, 229–247. [Google Scholar] [CrossRef] [Green Version]
- Hepburn, H.R.; Radloff, S.E. Honeybees of Africa; Springer: Berlin/Heidelberg, Germany, 1998; ISBN 978-3-642-08389-1. [Google Scholar]
- Hood, W.M. The Small Hive Beetle, Aethina tumida: A Review. Bee World 2004, 85, 51–59. [Google Scholar] [CrossRef]
- Ellis, J.; Munn, P. The Worldwide Health Status of Honey Bees. Bee World 2015, 86, 88–101. [Google Scholar] [CrossRef]
- Neumann, P.; Ellis, J.D. The Small Hive Beetle (Aethina tumida Murray, Coleoptera: Nitidulidae): Distribution, Biology and Control of an Invasive Species. J. Apic. Res. 2008, 47, 181–183. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, S.; Liu, J.; Huang, W.; Ji, C.; Ren, Q.; Xia, X.; Hou, C. First Detection of Small Hive Beetle Aethina tumida Murray (Coleoptera: Nitidulidae) Infesting Eastern Honeybee, Apis Cerana Fabricius (Hymenoptera: Apidae), in China. Sociobiology 2020, 67, 126–128. [Google Scholar] [CrossRef]
- Cervancia, C.R.; de Guzman, L.I.; Polintan, E.A.; Dupo, A.L.B.; Locsin, A.A. Current Status of Small Hive Beetle Infestation in the Philippines. J. Apic. Res. 2016, 55, 74–77. [Google Scholar] [CrossRef]
- Elzen, P.J.; Baxter, J.R.; Westervelt, D.; Randall, C.; Delaplane, K.S.; Cutts, L.; Wilson, W.T. Field Control and Biology Studies of a New Pest Species, Aethina tumida Murray (Coleoptera, Nitidulidae), Attacking European Honey Bees in the Western Hemisphere. Apidologie 1999, 30, 361–366. [Google Scholar] [CrossRef] [Green Version]
- de Landa, G.F.; Porrini, M.P.; Revainera, P.; Porrini, D.P.; Farina, J.; Correa-Benítez, A.; Maggi, M.D.; Eguaras, M.J.; Quintana, S. Pathogens Detection in the Small Hive Beetle (Aethina tumida (Coleoptera: Nitidulidae)). Neotrop. Entomol. 2021, 50, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Nanetti, A.; Ellis, J.D.; Cardaio, I.; Cilia, G. Detection of Lotmaria passim, Crithidia mellificae and Replicative Forms of Deformed Wing Virus and Kashmir Bee Virus in the Small Hive Beetle (Aethina tumida). Pathogens 2021, 10, 372. [Google Scholar] [CrossRef]
- Cilia, G.; Cardaio, I.; dos Santos, P.E.J.; Ellis, J.D.; NANETTI, A. The First Detection of Nosema ceranae (Microsporidia) in the Small Hive Beetle, Aethina tumida Murray (Coleoptera: Nitidulidae). Apidologie 2018, 49, 619–624. [Google Scholar] [CrossRef] [Green Version]
- Ward, L.; Brown, M.; Neumann, P.; Wilkins, S.; Pettis, J.; Boonham, N. A DNA Method for Screening Hive Debris for the Presence of Small Hive Beetle (Aethina tumida). Apidologie 2007, 38, 272–280. [Google Scholar] [CrossRef] [Green Version]
- Silacci, P.; Biolley, C.; Jud, C.; Charrière, J.-D.; Dainat, B. An Improved DNA Method to Unambiguously Detect Small Hive Beetle Aethina tumida, an Invasive Pest of Honeybee Colonies: DNA Method to Detect Small Hive Beetle Aethina tumida. Pest. Manag. Sci. 2018, 74, 2667–2670. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Pettis, J.S.; Shimanuki, H. Mitochondrial DNA Relationships in an Emergent Pest of Honey Bees: Aethina tumida (Coleoptera: Nitidulidae) from the United States and Africa. Ann. Entomol. Soc. Am. 2000, 93, 415–420. [Google Scholar] [CrossRef]
- Cepero, A.; Higes, M.; Martínez-Salvador, A.; Meana, A.; Martín-Hernández, R. A Two Year National Surveillance for Aethina tumida Reflects Its Absence in Spain. BMC Res. Notes 2014, 7, 878. [Google Scholar] [CrossRef] [Green Version]
- Ponting, S.; Tomkies, V.; Stainton, K. Rapid Identification of the Invasive Small Hive Beetle (Aethina tumida) Using LAMP. Pest. Manag. Sci. 2021, 77, 1476–1481. [Google Scholar] [CrossRef]
- Evans, J.D.; Spiewok, S.; Teixeira, E.W.; Neumann, P. Microsatellite Loci for the Small Hive Beetle, Aethina tumida, a Nest Parasite of Honey Bees. Mol. Ecol. Res. 2008, 8, 698–700. [Google Scholar] [CrossRef]
- Lounsberry, Z.; Spiewok, S.; Pernal, S.F.; Sonstegard, T.S.; Hood, W.M.; Pettis, J.; Neumann, P.; Evans, J.D. Worldwide Diaspora of Aethina tumida (Coleoptera: Nitidulidae), a Nest Parasite of Honey Bees. Ann. Entomol. Soc. Am. 2010, 103, 671–677. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; McKenna, D.; Scully, E.; Cook, S.C.; Dainat, B.; Egekwu, N.; Grubbs, N.; Lopez, D.; Lorenzen, M.D.; Reyna, S.M.; et al. Genome of the Small Hive Beetle (Aethina tumida, Coleoptera: Nitidulidae), a Worldwide Parasite of Social Bee Colonies, Provides Insights into Detoxification and Herbivory. GigaScience 2018, 7, giy138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, J.; Okuyama, H.; Martin, S.J. Complete Mitochondrial DNA Sequence of the Small Hive Beetle Aethina tumida (Insecta: Coleoptera) from Hawaii. Mitochondrial DNA B Resour. 2019, 4, 1522–1523. [Google Scholar] [CrossRef] [Green Version]
- Kraus, B.; Hunt, G. Differentiation of Varroa jacobsoni Oud Populations by Random Amplification of Polymorphic DNA (RAPD). Apidologie 1995, 26, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Solignac, M.; Vautrin, D.; Pizzo, A.; Navajas, M.; Le Conte, Y.; Cornuet, J.-M. Characterization of Microsatellite Markers for the Apicultural Pest Varroa destructor (Acari: Varroidae) and Its Relatives. Mol. Ecol. Notes 2003, 3, 556–559. [Google Scholar] [CrossRef]
- Nicoletti, R.; Becchimanzi, A. Ecological and Molecular Interactions between Insects and Fungi. Microorganisms 2022, 10, 96. [Google Scholar] [CrossRef]
- Islam, W.; Adnan, M.; Shabbir, A.; Naveed, H.; Abubakar, Y.S.; Qasim, M.; Tayyab, M.; Noman, A.; Nisar, M.S.; Khan, K.A.; et al. Insect-Fungal-Interactions: A Detailed Review on Entomopathogenic Fungi Pathogenicity to Combat Insect Pests. Microb. Pathog. 2021, 159, 105122. [Google Scholar] [CrossRef]
- Jensen, A.B.; Aronstein, K.; Flores, J.M.; Vojvodic, S.; Palacio, M.A.; Spivak, M. Standard Methods for Fungal Brood Disease Research. J. Apic. Res. 2013, 52, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a New Microsporidian Parasite in Honeybees in Europe. J. Invert. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef]
- Klee, J.; Besana, A.M.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.Q.; Chinh, T.X.; Puerta, F.; Ruz, J.M.; Kryger, P.; et al. Widespread Dispersal of the Microsporidian Nosema ceranae, an Emergent Pathogen of the Western Honey Bee, Apis mellifera. J. Invert. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef]
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae Has Infected Apis mellifera in Europe since at Least 1998 and May Be More Virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- Huang, W.-F.; Jiang, J.-H.; Chen, Y.-W.; Wang, C.-H. A Nosema ceranae Isolate from the Honeybee Apis mellifera. Apidologie 2007, 38, 30–37. [Google Scholar] [CrossRef]
- Han, B.; Weiss, L.M. Microsporidia: Obligate Intracellular Pathogens within the Fungal Kingdom. Microbiol. Spectr. 2017, 5, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A Higher-Level Phylogenetic Classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed]
- Corradi, N.; Selman, M. Latest Progress in Microsporidian Genome Research. J. Eukaryot. Microbiol. 2013, 60, 309–312. [Google Scholar] [CrossRef]
- Fries, I.; Feng, F.; da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), Morphological and Molecular Characterization of a Microsporidian Parasite of the Asian Honey Bee Apis Cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Botías, C.; Anderson, D.L.; Meana, A.; Garrido-Bailón, E.; Martín-Hernández, R.; Higes, M. Further Evidence of an Oriental Origin for Nosema ceranae (Microsporidia: Nosematidae). J. Invert. Pathol. 2012, 110, 108–113. [Google Scholar] [CrossRef]
- Guerrero-Molina, C.; Correa-Benítez, A.; Md. Hamiduzzaman, M.; Guzman-Novoa, E. Nosema ceranae Is an Old Resident of Honey Bee (Apis mellifera) Colonies in Mexico, Causing Infection Levels of One Million Spores per Bee or Higher during Summer and Fall. J. Invert. Pathol. 2016, 141, 38–40. [Google Scholar] [CrossRef]
- Teixeira, E.W.; dos Santos, L.G.; Sattler, A.; Message, D.; Alves, M.L.T.M.F.; Martins, M.F.; Grassi-Sella, M.L.; Francoy, T.M. Nosema ceranae Has Been Present in Brazil for More than Three Decades Infecting Africanized Honey Bees. J. Invert. Pathol. 2013, 114, 250–254. [Google Scholar] [CrossRef]
- Ferroglio, E.; Zanet, S.; Peraldo, N.; Tachis, E.; Trisciuoglio, A.; Laurino, D.; Porporato, M. Nosema ceranae Has Been Infecting Honey Bees Apis mellifera in Italy since at Least 1993. J. Apic. Res. 2013, 52, 60–61. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae Is a Long-Present and Wide-Spread Microsporidian Infection of the European Honey Bee (Apis mellifera) in the United States. J. Invert. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.; Leal, G.; Conget, P. Nosema ceranae an Emergent Pathogen of Apis mellifera in Chile. Parasitol. Res. 2012, 111, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Ivgin Tunca, R.; Oskay, D.; Gosterit, A.; Tekin, O.K. Does Nosema ceranae Wipe out Nosema apis in Turkey? Iran J. Parasitol. 2016, 11, 259–264. [Google Scholar]
- Papini, R.; Mancianti, F.; Canovai, R.; Cosci, F.; Rocchigiani, G.; Benelli, G.; Canale, A. Prevalence of the Microsporidian Nosema ceranae in Honeybee (Apis mellifera) Apiaries in Central Italy. Saudi J. Biol. Sci. 2017, 24, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Urbieta-Magro, A.; Higes, M.; Meana, A.; Gómez-Moracho, T.; Rodríguez-García, C.; Barrios, L.; Martín-Hernández, R. The Levels of Natural Nosema spp. Infection in Apis mellifera Iberiensis Brood Stages. Int. J. Parasitol. 2019, 49, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Matović, K.; Vidanović, D.; Manić, M.; Stojiljković, M.; Radojičić, S.; Debeljak, Z.; Šekler, M.; Ćirić, J. Twenty-Five-Year Study of Nosema spp. in Honey Bees (Apis mellifera) in Serbia. Saudi J. Biol. Sci. 2020, 27, 518–523. [Google Scholar] [CrossRef]
- Lannutti, L.; Mira, A.; Basualdo, M.; Rodriguez, G.; Erler, S.; Silva, V.; Gisder, S.; Genersch, E.; Florin-Christensen, M.; Schnittger, L. Development of a Loop-Mediated Isothermal Amplification (LAMP) and a Direct LAMP for the Specific Detection of Nosema ceranae, a Parasite of Honey Bees. Parasitol. Res. 2020, 119, 3947–3956. [Google Scholar] [CrossRef]
- Wang, Q.; Dai, P.; Guzman-Novoa, E.; Wu, Y.; Hou, C.; Diao, Q. Nosema ceranae, the Most Common Microsporidium Infecting Apis mellifera in the Main Beekeeping Regions of China since at Least 2005. J. Apic. Res. 2019, 58, 562–566. [Google Scholar] [CrossRef]
- Rubanov, A.; Russell, K.A.; Rothman, J.A.; Nieh, J.C.; McFrederick, Q.S. Intensity of Nosema ceranae Infection Is Associated with Specific Honey Bee Gut Bacteria and Weakly Associated with Gut Microbiome Structure. Sci. Rep. 2019, 9, 3820. [Google Scholar] [CrossRef]
- Milbrath, M.O.; van Tran, T.; Huang, W.-F.; Solter, L.F.; Tarpy, D.R.; Lawrence, F.; Huang, Z.Y. Comparative Virulence and Competition between Nosema apis and Nosema ceranae in Honey Bees (Apis mellifera). J. Invert. Pathol. 2015, 125, 9–15. [Google Scholar] [CrossRef]
- Gisder, S.; Schüler, V.; Horchler, L.L.; Groth, D.; Genersch, E. Long-Term Temporal Trends of Nosema spp. Infection Prevalence in Northeast Germany: Continuous Spread of Nosema ceranae, an Emerging Pathogen of Honey Bees (Apis mellifera), but No General Replacement of Nosema apis. Front. Cell. Infect. Microbiol. 2017, 7, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsen, B.; De la Mora, A.; Lacey, B.; Eccles, L.; Kelly, P.G.; Medina-Flores, C.A.; Petukhova, T.; Morfin, N.; Guzman-Novoa, E. Seasonality of Nosema ceranae Infections and Their Relationship with Honey Bee Populations, Food Stores, and Survivorship in a North American Region. Vet. Sci. 2020, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Ostroverkhova, N.V.; Konusova, O.L.; Kucher, A.N.; Kireeva, T.N.; Rosseykina, S.A. Prevalence of the Microsporidian Nosema spp. in Honey Bee Populations (Apis mellifera) in Some Ecological Regions of North Asia. Vet. Sci. 2020, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Branchiccela, B.; Arredondo, D.; Higes, M.; Invernizzi, C.; Martín-Hernández, R.; Tomasco, I.; Zunino, P.; Antúnez, K. Characterization of Nosema ceranae Genetic Variants from Different Geographic Origins. Microb. Ecol. 2017, 73, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Chemurot, M.; De Smet, L.; Brunain, M.; De Rycke, R.; de Graaf, D.C. Nosema Neumanni n. sp. (Microsporidia, Nosematidae), a New Microsporidian Parasite of Honeybees, Apis mellifera in Uganda. Eur. J. Protistol. 2017, 61, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Ohari, Y.; Itagaki, T. Prevalence of Nosema Species Infections in Apis Cerana Japonica and Apis mellifera Honeybees in the Tohoku Region of Japan. Parasitol. Int. 2021, 83, 102361. [Google Scholar] [CrossRef]
- Gisder, S.; Möckel, N.; Linde, A.; Genersch, E. A Cell Culture Model for Nosema ceranae and Nosema apis Allows New Insights into the Life Cycle of These Important Honey Bee-Pathogenic Microsporidia: Nosema spp. Infection in Cell Culture. Environ. Microbiol. 2011, 13, 404–413. [Google Scholar] [CrossRef]
- Roberts, K.E.; Evison, S.E.F.; Baer, B.; Hughes, W.O.H. The Cost of Promiscuity: Sexual Transmission of Nosema Microsporidian Parasites in Polyandrous Honey Bees. Sci. Rep. 2015, 5, 10982. [Google Scholar] [CrossRef]
- Castelli, L.; Branchiccela, B.; Garrido, M.; Invernizzi, C.; Porrini, M.; Romero, H.; Santos, E.; Zunino, P.; Antúnez, K. Impact of Nutritional Stress on Honeybee Gut Microbiota, Immunity, and Nosema ceranae Infection. Microb. Ecol. 2020, 80, 908–919. [Google Scholar] [CrossRef]
- Costa, C.; Tanner, G.; Lodesani, M.; Maistrello, L.; Neumann, P. Negative Correlation between Nosema ceranae Spore Loads and Deformed Wing Virus Infection Levels in Adult Honey Bee Workers. J. Invert. Pathol. 2011, 108, 224–225. [Google Scholar] [CrossRef]
- Runckel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; DeRisi, J.L. Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia. PLoS ONE 2011, 6, e20656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goblirsch, M. Nosema ceranae Disease of the Honey Bee (Apis mellifera). Apidologie 2018, 49, 131–150. [Google Scholar] [CrossRef] [Green Version]
- Vejnovic, B.; Stevanovic, J.; Schwarz, R.S.; Aleksic, N.; Mirilovic, M.; Jovanovic, N.M.; Stanimirovic, Z. Quantitative PCR Assessment of Lotmaria passim in Apis mellifera Colonies Co-Infected Naturally with Nosema ceranae. J. Invert. Pathol. 2018, 151, 76–81. [Google Scholar] [CrossRef] [PubMed]
- OIE—World Organisation for Animal Health. Nosemosis of Honey Bees. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2018; pp. 744–749. ISBN 978-92-95108-18-9. [Google Scholar]
- Snow, J.W.; Ceylan Koydemir, H.; Karinca, D.K.; Liang, K.; Tseng, D.; Ozcan, A. Rapid Imaging, Detection, and Quantification of Nosema ceranae Spores in Honey Bees Using Mobile Phone-Based Fluorescence Microscopy. Lab Chip 2019, 19, 789–797. [Google Scholar] [CrossRef]
- Bourgeois, A.L.; Rinderer, T.E.; Beaman, L.D.; Danka, R.G. Genetic Detection and Quantification of Nosema apis and Nosema ceranae in the Honey Bee. J. Invert. Pathol. 2010, 103, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Erler, S.; Lommatzsch, S.; Lattorff, H.M.G. Comparative Analysis of Detection Limits and Specificity of Molecular Diagnostic Markers for Three Pathogens (Microsporidia, Nosema spp.) in the Key Pollinators Apis mellifera and Bombus Terrestris. Parasitol. Res. 2012, 110, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Galajda, R.; Valenčáková, A.; Sučik, M.; Kandráčová, P. Nosema Disease of European Honey Bees. J. Fungi 2021, 7, 714. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of Colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Md. Hamiduzzaman, M.; Guzman-Novoa, E.; Goodwin, P.H. A Multiplex PCR Assay to Diagnose and Quantify Nosema Infections in Honey Bees (Apis mellifera). J. Invert. Pathol. 2010, 105, 151–155. [Google Scholar] [CrossRef]
- Sagastume, S.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Ribosomal Gene Polymorphism in Small Genomes: Analysis of Different 16S RRNA Sequences Expressed in the Honeybee Parasite Nosema ceranae (Microsporidia). J. Eukaryot. Microbiol. 2014, 61, 42–50. [Google Scholar] [CrossRef]
- Cilia, G.; Luchetti, G.; Nanetti, A. Polymorphism of 16s RRNA Gene: Any Effect on the Biomolecular Quantitation of the Honey Bee (Apis mellifera L., 1758) Pathogen Nosema ceranae? Appl. Sci. 2022, 12, 422. [Google Scholar] [CrossRef]
- Cornman, R.S.; Chen, Y.P.; Schatz, M.C.; Street, C.; Zhao, Y.; Desany, B.; Egholm, M.; Hutchison, S.; Pettis, J.S.; Lipkin, W.I.; et al. Genomic Analyses of the Microsporidian Nosema ceranae, an Emergent Pathogen of Honey Bees. PLoS Pathog. 2009, 5, e1000466. [Google Scholar] [CrossRef] [PubMed]
- Pelin, A.; Selman, M.; Aris-Brosou, S.; Farinelli, L.; Corradi, N. Genome Analyses Suggest the Presence of Polyploidy and Recent Human-Driven Expansions in Eight Global Populations of the Honeybee Pathogen Nosema ceranae: Genome Diversity in the Honeybee Pathogen Nosema ceranae. Environ. Microbiol. 2015, 17, 4443–4458. [Google Scholar] [CrossRef]
- Huang, Q.; Wu, Z.H.; Li, W.F.; Guo, R.; Xu, J.S.; Dang, X.Q.; Ma, Z.G.; Chen, Y.P.; Evans, J.D. Genome and Evolutionary Analysis of Nosema ceranae: A Microsporidian Parasite of Honey Bees. Front. Microbiol. 2021, 12, 645353. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical Coexistence of Nosema ceranae and Nosema apis in Honey Bees. J. Invert. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef]
- Forsgren, E.; Fries, I. Comparative Virulence of Nosema ceranae and Nosema apis in Individual European Honey Bees. Vet. Pathol. 2010, 170, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Burgher-MacLellan, K.L.; Williams, G.R.; Shutler, D.; MacKenzie, K.; Rogers, R.E.L. Optimization of Duplex Real-Time PCR with Melting-Curve Analysis for Detecting the Microsporidian Parasites Nosema apis and Nosema ceranae in Apis mellifera. Can. Entomol. 2010, 142, 271–283. [Google Scholar] [CrossRef]
- Traver, B.E.; Fell, R.D. Prevalence and Infection Intensity of Nosema in Honey Bee (Apis mellifera L.) Colonies in Virginia. J. Invert. Pathol. 2011, 107, 43–49. [Google Scholar] [CrossRef]
- Cilia, G.; Cabbri, R.; Maiorana, G.; Cardaio, I.; Dall’Olio, R.; Nanetti, A. A Novel TaqMan ® Assay for Nosema ceranae Quantification in Honey Bee, Based on the Protein Coding Gene Hsp70. Eur. J. Protistol. 2018, 63, 44–50. [Google Scholar] [CrossRef]
- Truong, A.-T.; Sevin, S.; Kim, S.; Yoo, M.-S.; Cho, Y.S.; Yoon, B. Rapidly Quantitative Detection of Nosema ceranae in Honeybees Using Ultra-Rapid Real-Time Quantitative PCR. J. Vet. Sci. 2021, 22, e40. [Google Scholar] [CrossRef]
- Xing, W.; Zhou, D.; Long, Q.; Sun, M.; Guo, R.; Wang, L. Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation. Insects 2021, 12, 728. [Google Scholar] [CrossRef]
- Cilia, G.; Sagona, S.; Giusti, M.; Jarmela dos Santos, P.E.; Nanetti, A.; Felicioli, A. Nosema ceranae Infection in Honeybee Samples from Tuscanian Archipelago (Central Italy) Investigated by Two QPCR Methods. Saudi J. Biol. Sci. 2019, 26, 1553–1556. [Google Scholar] [CrossRef] [PubMed]
- Ptaszyńska, A.A.; Borsuk, G.; Woźniakowski, G.; Gnat, S.; Małek, W. Loop-Mediated Isothermal Amplification (LAMP) Assays for Rapid Detection and Differentiation of Nosema apis and N. Ceranae in Honeybees. FEMS Microbiol. Lett. 2014, 357, 40–48. [Google Scholar] [CrossRef]
- Chupia, V.; Patchanee, P.; Krutmuang, P.; Pikulkaew, S. Development and Evaluation of Loop-Mediated Isothermal Amplification for Rapid Detection of Nosema ceranae in Honeybee. Asian Pac. J. Trop. Dis. 2016, 6, 952–956. [Google Scholar] [CrossRef]
- Maside, X.; Gómez-Moracho, T.; Jara, L.; Martín-Hernández, R.; De la Rúa, P.; Higes, M.; Bartolomé, C. Population Genetics of Nosema apis and Nosema ceranae: One Host (Apis mellifera) and Two Different Histories. PLoS ONE 2015, 10, e0145609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Moracho, T.; Bartolome, C.; Bello, X.; Martín-Hernández, R.; Higes, M.; Maside, X. Recent Worldwide Expansion of Nosema ceranae (Microsporidia) in Apis mellifera Populations Inferred from Multilocus Patterns of Genetic Variation. Infect. Genet. Evol. 2015, 31, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Aronstein, K.A.; Murray, K.D. Chalkbrood Disease in Honey Bees. J. Invert. Pathol. 2010, 103, S20–S29. [Google Scholar] [CrossRef]
- Evison, S.E. Chalkbrood: Epidemiological Perspectives from the Host–Parasite Relationship. Curr. Opin. Insect. Sci. 2015, 10, 65–70. [Google Scholar] [CrossRef]
- Gilliam, M.; Vandenberg, J.D. Fungi. In Honey Bee Pests, Predators, and Diseases; Morse, R., Flottum, K., Eds.; A I Root Co.: Medina, WA, USA, 1997; pp. 81–110. [Google Scholar]
- Hornitzky, M.A.Z. Literature Review of Chalkbrood, a Fungal Disease of Honeybees: A Report for the Rural Industries Research and Development Corporation; Rural Industries Research and Development Corporation: Barton, Australia, 2001; ISBN 978-0-642-58370-3. [Google Scholar]
- Murray, K.D.; Aronstein, K.A.; Jones, W.A. A Molecular Diagnostic Method for Selected Ascosphaera Species Using PCR Amplification of Internal Transcribed Spacer Regions of RDNA. J. Apic. Res. 2005, 44, 61–64. [Google Scholar] [CrossRef]
- James, R.R.; Skinner, J.S. PCR Diagnostic Methods for Ascosphaera Infections in Bees. J. Invert. Pathol. 2005, 90, 98–103. [Google Scholar] [CrossRef]
- Garrido-Bailón, E.; Higes, M.; Martínez-Salvador, A.; Antúnez, K.; Botías, C.; Meana, A.; Prieto, L.; Martín-Hernández, R. The Prevalence of the Honeybee Brood Pathogens Ascosphaera Apis, Paenibacillus larvae and Melissococcus plutonius in Spanish Apiaries Determined with a New Multiplex PCR Assay. Microb. Biotechnol. 2013, 6, 731–739. [Google Scholar] [CrossRef]
- Reynaldi, F.J.; López, A.C.; Albo, G.N.; Alippi, A.M. Differentiation of Ascosphaera Apis Isolates by Rep-PCR Fingerprinting and Determination of Chalkbrood Incidence in Argentinean Honey Samples. J. Apic. Res. 2003, 42, 68–76. [Google Scholar] [CrossRef]
- Qin, X.; Evans, J.D.; Aronstein, K.A.; Murray, K.D.; Weinstock, G.M. Genome Sequences of the Honey Bee Pathogens Paenibacillus larvae and Ascosphaera Apis. Insect Mol. Biol. 2006, 15, 715–718. [Google Scholar] [CrossRef] [Green Version]
- Rehner, S.A.; Evans, J.D. Microsatellite Loci for the Fungus Ascosphaera Apis: Cause of Honey Bee Chalkbrood Disease. Mol. Ecol. Res. 2009, 9, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.M.; McGee, P.A.; Oldroyd, B.P. Variable Virulence among Isolates of Ascosphaera Apis: Testing the Parasite-Pathogen Hypothesis for the Evolution of Polyandry in Social Insects. Naturwissenschaften 2013, 100, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Somerville, D.; Frese, M.; Nayudu, M. Environmental Gut Bacteria in European Honey Bees (Apis mellifera) from Australia and Their Relationship to the Chalkbrood Disease. PLoS ONE 2020, 15, e0238252. [Google Scholar] [CrossRef] [PubMed]
- Dharampal, P.S.; Diaz-Garcia, L.; Haase, M.A.B.; Zalapa, J.; Currie, C.R.; Hittinger, C.T.; Steffan, S.A. Microbial Diversity Associated with the Pollen Stores of Captive-Bred Bumble Bee Colonies. Insects 2020, 11, 250. [Google Scholar] [CrossRef]
- Getachew, A.; Abejew, T.A.; Wu, J.; Xu, J.; Yu, H.; Tan, J.; Wu, P.; Tu, Y.; Kang, W.; Wang, Z.; et al. Transcriptome Profiling Reveals Insertional Mutagenesis Suppressed the Expression of Candidate Pathogenicity Genes in Honeybee Fungal Pathogen, Ascosphaera Apis. Sci. Rep. 2020, 10, 7532. [Google Scholar] [CrossRef]
- Schubert, M.; Spiegel, H.; Schillberg, S.; Nölke, G. Aspergillus-Specific Antibodies—Targets and Applications. Biotechnol. Adv. 2018, 36, 1167–1184. [Google Scholar] [CrossRef]
- Goldman, G.H.; Osmani, S.A. (Eds.) The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods, 1st ed.; CRC Press: Boca Raton, FL, USA, 2007; Volume 26, ISBN 978-0-429-12916-2. [Google Scholar]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of Aspergillosis: Insights into the Pathogenic Potency of Aspergillus Fumigatus and Some Other Aspergillus Species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef] [Green Version]
- Foley, K.; Fazio, G.; Jensen, A.B.; Hughes, W.O.H. The Distribution of Aspergillus spp. Opportunistic Parasites in Hives and Their Pathogenicity to Honey Bees. Vet. Microbiol. 2014, 169, 203–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Mazcorro, J.F.; Kawas, J.R.; Marroquin-Cardona, A.G. Descriptive Bacterial and Fungal Characterization of Propolis Using Ultra-High-Throughput Marker Gene Sequencing. Insects 2019, 10, 402. [Google Scholar] [CrossRef] [Green Version]
- Vojvodic, S.; Jensen, A.B.; James, R.R.; Boomsma, J.J.; Eilenberg, J. Temperature Dependent Virulence of Obligate and Facultative Fungal Pathogens of Honeybee Brood. Vet. Microbiol. 2011, 149, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-S.; Luong, G.; Lee, B.-S. Development of In-Field-Diagnosis of Aspergillus Flavus by Loop-Mediated Isothermal Amplification in Honeybee. J. Apic. 2016, 31, 25. [Google Scholar] [CrossRef]
- Stevanovic, J.; Stanimirovic, Z.; Genersch, E.; Kovacevic, S.R.; Ljubenkovic, J.; Radakovic, M.; Aleksic, N. Dominance of Nosema ceranae in Honey Bees in the Balkan Countries in the Absence of Symptoms of Colony Collapse Disorder. Apidologie 2011, 42, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Webster, T.C.; Pomper, K.W.; Hunt, G.; Thacker, E.M.; Jones, S.C. Nosema apis Infection in Worker and Queen Apis mellifera. Apidologie 2004, 35, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Klee, J.; Tek Tay, W.; Paxton, R.J. Specific and Sensitive Detection of Nosema Bombi (Microsporidia: Nosematidae) in Bumble Bees (Bombus spp.; Hymenoptera: Apidae) by PCR of Partial RRNA Gene Sequences. J. Invert. Pathol. 2006, 91, 98–104. [Google Scholar] [CrossRef]
- Tapaszti, Z.; Forgách, P.; Kővágó, C.; Békési, L.; Bakonyi, T.; Rusvai, M. First Detection and Dominance of Nosema ceranae in Hungarian Honeybee Colonies. Acta Vet. Hung. 2009, 57, 383–388. [Google Scholar] [CrossRef] [Green Version]
- González, S.A.C.; Valencia, G.L.; Cabrera, C.O.; Gómez Gómez, S.D.; Torres, K.M.; Blandón, K.O.E.; Guerrero Velázquez, J.G.; Paz, L.E.S.; Trasviña Muñoz, E.; Monge Navarro, F.J. Prevalence and Geographical Distribution of Nosema apis and Nosema ceranae in Apiaries of Northwest Mexico Using a Duplex Real-Time PCR with Melting-Curve Analysis. J. Apic. Res. 2019, 59, 195–203. [Google Scholar] [CrossRef]
- Carletto, J.; Blanchard, P.; Aurélie, G.; Schurr, F.; Chauzat, M.-P.; Ribière, M. Improving Molecular Discrimination of Nosema apis and Nosema ceranae. J. Invert. Pathol. 2013, 113, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, A.; Ellis, J.; Stark, D.; Barratt, J. The Evolution of Trypanosomatid Taxonomy. Parasit Vectors 2017, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Bartolomé, C.; Buendía-Abad, M.; Benito, M.; Sobrino, B.; Amigo, J.; Carracedo, A.; Martín-Hernández, R.; Higes, M.; Maside, X. Longitudinal Analysis on Parasite Diversity in Honeybee Colonies: New Taxa, High Frequency of Mixed Infections and Seasonal Patterns of Variation. Sci. Rep. 2020, 10, 10454. [Google Scholar] [CrossRef]
- Langridge, D.F. Flagellated Protozoa (Fam. Trypanosomidae) in the Honeybee, Apis mellifera, in Australia. J. Invert. Pathol 1966, 8, 124–126. [Google Scholar] [CrossRef]
- Langridge, D.F.; McGhee, R.B. Crithidia mellificae n. sp. an Acidophilic Trypanosomatid of the Honey Bee Apis mellifera. J. Protoz. 1967, 14, 485–487. [Google Scholar] [CrossRef]
- Ravoet, J.; Schwarz, R.S.; Descamps, T.; Yañez, O.; Tozkar, C.O.; Martin-Hernandez, R.; Bartolomé, C.; De Smet, L.; Higes, M.; Wenseleers, T.; et al. Differential Diagnosis of the Honey Bee Trypanosomatids Crithidia mellificae and Lotmaria passim. J. Invert. Pathol. 2015, 130, 21–27. [Google Scholar] [CrossRef]
- Castelli, L.; Branchiccela, B.; Invernizzi, C.; Tomasco, I.; Basualdo, M.; Rodriguez, M.; Zunino, P.; Antúnez, K. Detection of Lotmaria passim in Africanized and European Honey Bees from Uruguay, Argentina and Chile. J. Invert. Pathol. 2019, 160, 95–97. [Google Scholar] [CrossRef]
- Gómez-Moracho, T.; Buendía-Abad, M.; Benito, M.; García-Palencia, P.; Barrios, L.; Bartolomé, C.; Maside, X.; Meana, A.; Jiménez-Antón, M.D.; Olías-Molero, A.I.; et al. Experimental Evidence of Harmful Effects of Crithidia mellificae and Lotmaria passim on Honey Bees. Int. J. Parasitol. 2020, 50, 1117–1124. [Google Scholar] [CrossRef]
- Arismendi, N.; Bruna, A.; Zapata, N.; Vargas, M. PCR-Specific Detection of Recently Described Lotmaria passim (Trypanosomatidae) in Chilean Apiaries. J. Invert. Pathol. 2016, 134, 1–5. [Google Scholar] [CrossRef]
- Stevanovic, J.; Schwarz, R.S.; Vejnovic, B.; Evans, J.D.; Irwin, R.E.; Glavinic, U.; Stanimirovic, Z. Species-Specific Diagnostics of Apis mellifera Trypanosomatids: A Nine-Year Survey (2007–2015) for Trypanosomatids and Microsporidians in Serbian Honey Bees. J. Invert. Pathol. 2016, 139, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Bartolomé, C.; Buendía, M.; Benito, M.; De la Rúa, P.; Ornosa, C.; Martín-Hernández, R.; Higes, M.; Maside, X. A New Multiplex PCR Protocol to Detect Mixed Trypanosomatid Infections in Species of Apis and Bombus. J. Invert. Pathol. 2018, 154, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Galuppi, R.; Fontanesi, L. Analysis of Honey Environmental DNA Indicates That the Honey Bee (Apis mellifera L.) Trypanosome Parasite Lotmaria passim Is Widespread in the Apiaries of the North of Italy. J. Invert. Pathol. 2021, 184, 107628. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Palmer-Young, E.; Skyrm, K.; Daly, T.; Sylvia, M.; Averill, A.; Rich, S. Triplex Real-Time PCR for Detection of Crithidia mellificae and Lotmaria passim in Honey Bees. Parasitol. Res. 2018, 117, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Runckel, C.; DeRisi, J.; Flenniken, M.L. A Draft Genome of the Honey Bee Trypanosomatid Parasite Crithidia mellificae. PLoS ONE 2014, 9, e95057. [Google Scholar] [CrossRef] [Green Version]
- Schmid-Hempel, P. On the Evolutionary Ecology of Host-Parasite Interactions: Addressing the Question with Regard to Bumblebees and Their Parasites. Naturwissenschaften 2001, 88, 147–158. [Google Scholar] [CrossRef]
- Schoonvaere, K.; Brunain, M.; Baeke, F.; De Bruyne, M.; De Rycke, R.; de Graaf, D.C. Comparison between Apicystis Cryptica sp. n. and Apicystis Bombi (Arthrogregarida, Apicomplexa): Gregarine Parasites That Cause Fat Body Hypertrophism in Bees. Eur. J. Protistol. 2020, 73, 125688. [Google Scholar] [CrossRef]
- Plischuk, S.; Meeus, I.; Smagghe, G.; Lange, C.E. Apicystis Bombi (Apicomplexa: Neogregarinorida) Parasitizing Apis mellifera and Bombus Terrestris (Hymenoptera: Apidae) in Argentina. Environ. Microbiol. Rep. 2011, 3, 565–568. [Google Scholar] [CrossRef]
- Morimoto, T.; Kojima, Y.; Yoshiyama, M.; Kimura, K.; Yang, B.; Peng, G.; Kadowaki, T. Molecular Detection of Protozoan Parasites Infecting Apis mellifera Colonies in Japan. Environ. Microbiol. Rep. 2013, 5, 74–77. [Google Scholar] [CrossRef]
- Gamboa, V.; Ravoet, J.; Brunain, M.; Smagghe, G.; Meeus, I.; Figueroa, J.; Riaño, D.; de Graaf, D.C. Bee Pathogens Found in Bombus Atratus from Colombia: A Case Study. J. Invert. Pathol. 2015, 129, 36–39. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Peng, G.; Li, T.; Kadowaki, T. Molecular and Phylogenetic Characterization of Honey Bee Viruses, Nosema Microsporidia, Protozoan Parasites, and Parasitic Mites in China. Ecol. Evol. 2013, 3, 298–311. [Google Scholar] [CrossRef]
- Meeus, I.; De Graaf, D.C.; Jans, K.; Smagghe, G. Multiplex PCR Detection of Slowly-Evolving Trypanosomatids and Neogregarines in Bumblebees Using Broad-Range Primers. J. Appl. Microbiol. 2010, 109, 107–115. [Google Scholar] [CrossRef] [Green Version]
- King, R.L.; Taylor, A.B. Malpighamoeba Locustae, n. sp. (Amoebidae), a Protozoan Parasitic in the Malpighian Tubes of Grasshoppers. Trans. Am. Microsc. Soc. 1936, 55, 6–10. [Google Scholar] [CrossRef]
- Rossi, M.; Ott, S.R.; Niven, J.E. Malpighamoeba Infection Compromises Fluid Secretion and P-Glycoprotein Detoxification in Malpighian Tubules. Sci. Rep. 2020, 10, 15953. [Google Scholar] [CrossRef] [PubMed]
- Mehlhorn, H. Malphighamoeba mellificae, Chapter 1839. In Encyclopedia of Parasitology; Mehlhorn, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; p. 1577. ISBN 978-3-662-43977-7. [Google Scholar]
- Dittes, J.; Aupperle-Lellbach, H.; Schäfer, M.O.; Mülling, C.K.W.; Emmerich, I.U. Veterinary Diagnostic Approach of Common Virus Diseases in Adult Honeybees. Vet. Sci. 2020, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- Aydin, L.; Gulegen, E.; Cakmak, I.; Girisgin, A.; Wells, H. Relation between Nosema and Chalkbrood Diseases, and Its Implication for an Apiary Management Model. Bull. Vet. Inst. Pulawy 2006, 50, 471. [Google Scholar]
- Solomon, S.; Degu, T.; Fesseha, H.; Mathewos, M. Study on Major Parasitic Diseases of Adult Honeybees in Three Districts of Kaffa Zone, Southern Ethiopia. Vet. Med. Int. 2021, 2021, e6346703. [Google Scholar] [CrossRef]
- Schäfer, M.O.; Horenk, J.; Wylezich, C. Molecular Detection of Malpighamoeba Mellificae in Honey Bees. Vet. Sci. 2022, 9, 148. [Google Scholar] [CrossRef]
- Brødsgaard, C.J.; Ritter, W.; Hansen, H. Response of in Vitro Reared Honey Bee Larvae to Various Doses of Paenibacillus larvae larvae Spores. Apidologie 1998, 29, 569–578. [Google Scholar] [CrossRef] [Green Version]
- Ebeling, J.; Knispel, H.; Hertlein, G.; Fünfhaus, A.; Genersch, E. Biology of Paenibacillus larvae, a Deadly Pathogen of Honey Bee Larvae. Appl. Microbiol. Biotechnol. 2016, 100, 7387–7395. [Google Scholar] [CrossRef]
- Woodrow, A.; Gochnauer, T. Susceptibility of Honeybee Larvae to American Foulbrood. Bee Cult. 1941, 69, 148–151. [Google Scholar]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial Pathogens of Bees. Curr. Opin. Insect. Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef]
- Hansen, H.; Brødsgaard, C.J. American Foulbrood: A Review of Its Biology, Diagnosis and Control. Bee World 1999, 80, 5–23. [Google Scholar] [CrossRef]
- Genersch, E. American Foulbrood in Honeybees and Its Causative Agent, Paenibacillus larvae. J. Invert. Pathol. 2010, 103, S10–S19. [Google Scholar] [CrossRef]
- OIE—World Organisation for Animal Health. American Foulbrood of Honey Bees (Infection of Honey Bees with Paenibacillus larvae). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2018; pp. 719–735. ISBN 978-92-95108-18-9. [Google Scholar]
- Gende, L.; Satta, A.; Ligios, V.; Ruiu, L.; Buffa, F.; Fernández, N.; Churio, S.; Eguaras, M.; Fiori, M.; Floris, I. Searching for an American Foulbrood Early Detection Threshold by the Determination of Paenibacillus larvae Spore Load in Worker Honey Bees. Bull. Insectol. 2011, 64, 229–233. [Google Scholar]
- de Graaf, D.C.; Alippi, A.M.; Antúnez, K.; Aronstein, K.A.; Budge, G.; De Koker, D.; De Smet, L.; Dingman, D.W.; Evans, J.D.; Foster, L.J.; et al. Standard Methods for American Foulbrood Research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, O.S.; Fisher, N.A. Growth and Laboratory Maintenance of Paenibacillus larvae. Curr. Microbiol. 2018, 48, 9E.1.1–9E.1.6. [Google Scholar] [CrossRef]
- Rusenova, N.V.; Parvanov, P.; Stanilova, S. Development of Multiplex PCR for Fast Detection of Paenibacillus larvae in Putrid Masses and in Isolated Bacterial Colonies. Appl. Biochem. Microbiol. 2013, 49, 79–84. [Google Scholar] [CrossRef]
- Andrade, V.D.M.; Flores, J.L.H.; López, M.A.R.; Hernández, A.C.; Gómez, S.R.; Calvillo, R.P.M.; Martínez, A.G.E.; Pérez, J.C.; Hernández, I.A.; Hidalgo, E.Á.; et al. Evaluation of the Presence of Paenibacillus larvae in Commercial Bee Pollen Using PCR Amplification of the Gene for TRNACys. Braz. J. Microbiol. 2019, 50, 471–480. [Google Scholar] [CrossRef]
- Bakonyi, T.; Derakhshifar, I.; Grabensteiner, E.; Nowotny, N. Development and Evaluation of PCR Assays for the Detection of Paenibacillus larvae in Honey Samples: Comparison with Isolation and Biochemical Characterization. Appl. Environ. Microbiol. 2003, 69, 1504–1510. [Google Scholar] [CrossRef] [Green Version]
- Ryba, S.; Titera, D.; Haklova, M.; Stopka, P. A PCR Method of Detecting American Foulbrood (Paenibacillus larvae) in Winter Beehive Wax Debris. Vet. Microbiol. 2009, 139, 193–196. [Google Scholar] [CrossRef]
- Rossi, F.; Amadoro, C.; Ruberto, A.; Ricchiuti, L. Evaluation of Quantitative PCR (QPCR) Paenibacillus larvae Targeted Assays and Definition of Optimal Conditions for Its Detection/Quantification in Honey and Hive Debris. Insects 2018, 9, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govan, V.A.; Allsopp, M.H.; Davison, S. A PCR Detection Method for Rapid Identification of Paenibacillus larvae. Appl. Environ. Microbiol. 1999, 65, 2243–2245. [Google Scholar] [CrossRef] [Green Version]
- Dobbelaere, W.; de Graaf, D.C.; Peeters, J.E. Development of a Fast and Reliable Diagnostic Method for American Foulbrood Disease (Paenibacillus larvae subsp. larvae) Using a 16S RRNA Gene Based PCR. Apidologie 2001, 32, 363–370. [Google Scholar] [CrossRef] [Green Version]
- Piccini, C.; D’Alessandro, B.; Antúnez, K.; Zunino, P. Detection of Paenibacillus larvae Subspecies larvae Spores in Naturally Infected Bee Larvae and Artificially Contaminated Honey by PCR. World J. Microbiol. Biotechnol. 2002, 18, 761–765. [Google Scholar] [CrossRef]
- Neuendorf, S.; Hedtke, K.; Tangen, G.; Genersch, E. Biochemical Characterization of Different Genotypes of Paenibacillus larvae subsp. larvae, a Honey Bee Bacterial Pathogen. Microbiology 2004, 150, 2381–2390. [Google Scholar] [CrossRef] [Green Version]
- Lauro, F.M.; Favaretto, M.; Covolo, L.; Rassu, M.; Bertoloni, G. Rapid Detection of Paenibacillus larvae from Honey and Hive Samples with a Novel Nested PCR Protocol. Int. J. Food. Microbiol. 2003, 81, 195–201. [Google Scholar] [CrossRef]
- Han, S.-H.; Lee, D.-B.; Lee, D.-W.; Kim, E.-H.; Yoon, B.-S. Ultra-Rapid Real-Time PCR for the Detection of Paenibacillus larvae, the Causative Agent of American Foulbrood (AFB). J. Invert. Pathol. 2008, 99, 8–13. [Google Scholar] [CrossRef]
- Chagas, S.; Vaucher, R.; Brandelli, A. Detection of Paenibacillus larvae by Real-Time PCR. Acta Sci. Vet. 2010, 38, 251–256. [Google Scholar] [CrossRef]
- Martínez, J.; Simon, V.; Gonzalez, B.; Conget, P. A Real-Time PCR-Based Strategy for the Detection of Paenibacillus larvae Vegetative Cells and Spores to Improve the Diagnosis and the Screening of American Foulbrood. Lett. Appl. Microbiol. 2010, 50, 603–610. [Google Scholar] [CrossRef]
- Quintana, S.; Fernández, N.J.; Pagnuco, I.A.; Medici, S.K.; Eguaras, M.J.; Gende, L.B. Report of a Real-Time Pcr Assay for Paenibacillus larvae Dna Detection from Spores of Scale Samples. Rev. Arg. Prod. Anim. 2017, 37, 83–88. [Google Scholar]
- Kušar, D.; Papić, B.; Zajc, U.; Zdovc, I.; Golob, M.; Žvokelj, L.; Knific, T.; Avberšek, J.; Ocepek, M.; Pislak Ocepek, M. Novel TaqMan PCR Assay for the Quantification of Paenibacillus larvae Spores in Bee-Related Samples. Insects 2021, 12, 1034. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E.; Forsgren, E.; Pentikäinen, J.; Ashiralieva, A.; Rauch, S.; Kilwinski, J.; Fries, I. Reclassification of Paenibacillus larvae subsp. Pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without Subspecies Differentiation. Int. J. Syst. Evol. 2006, 56, 501–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beims, H.; Bunk, B.; Erler, S.; Mohr, K.I.; Spröer, C.; Pradella, S.; Günther, G.; Rohde, M.; von der Ohe, W.; Steinert, M. Discovery of Paenibacillus larvae ERIC V: Phenotypic and Genomic Comparison to Genotypes ERIC I-IV Reveal Different Inventories of Virulence Factors Which Correlate with Epidemiological Prevalences of American Foulbrood. Int. J. Med. Microbiol. 2020, 310, 151394. [Google Scholar] [CrossRef]
- Beims, H.; Janke, M.; von der Ohe, W.; Steinert, M. Rapid Identification and Genotyping of the Honeybee Pathogen Paenibacillus larvae by Combining Culturing and Multiplex Quantitative PCR. Open Vet. J. 2020, 10, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Ashiralieva, A.; Hedtke, K.; Genersch, E. Negative Correlation between Individual-Insect-Level Virulence and Colony-Level Virulence of Paenibacillus larvae, the Etiological Agent of American Foulbrood of Honeybees. Appl. Environ. Microbiol. 2009, 75, 3344–3347. [Google Scholar] [CrossRef] [Green Version]
- Krongdang, S.; Evans, J.D.; Pettis, J.S.; Chantawannakul, P. Multilocus Sequence Typing, Biochemical and Antibiotic Resistance Characterizations Reveal Diversity of North American Strains of the Honey Bee Pathogen Paenibacillus larvae. PLoS ONE 2017, 12, e0176831. [Google Scholar] [CrossRef]
- Descamps, T.; De Smet, L.; Stragier, P.; De Vos, P.; de Graaf, D.C. Multiple Locus Variable Number of Tandem Repeat Analysis: A Molecular Genotyping Tool for Paenibacillus larvae. Microb. Biotechnol. 2016, 9, 772–781. [Google Scholar] [CrossRef] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole Genome Sequencing Options for Bacterial Strain Typing and Epidemiologic Analysis Based on Single Nucleotide Polymorphism versus Gene-by-Gene-Based Approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Ågren, J.; Schäfer, M.O.; Forsgren, E. Using Whole Genome Sequencing to Study American Foulbrood Epidemiology in Honeybees. PLoS ONE 2017, 12, e0187924. [Google Scholar] [CrossRef]
- Žugelj, A.; Papić, B.; Zdovc, I.; Zajc, U.; Golob, M.; Avberšek, J.; Kušar, D. ERIC and WGS Typing of Paenibacillus larvae in Slovenia: Investigation of ERIC I Outbreaks. Insects 2021, 12, 362. [Google Scholar] [CrossRef]
- White, G.F. The Cause of European Foulbrood; U.S. Government Printing Office: Washington, DC, USA, 1912.
- Bailey, L.; Collins, M.D. Reclassification of ‘Streptococcus Pluton’ (White) in a New Genus Melissococcus, as Melissococcus Pluton Nom. Rev.; Comb. Nov. J. Appl. Bacteriol. 1982, 53, 215–217. [Google Scholar] [CrossRef]
- Trüper, H.G.; de’ Clari, L. 1998 Taxonomic Note: Erratum and Correction of Further Specific Epithets Formed as Substantives (Nouns) ‘in Apposition’. Int. J. Syst. Evol 1998, 48, 615. [Google Scholar] [CrossRef] [Green Version]
- Forsgren, E.; Budge, G.E.; Charrière, J.-D.; Hornitzky, M.A.Z. Standard Methods for European Foulbrood Research. J. Apic. Res. 2013, 52, 1–14. [Google Scholar] [CrossRef]
- Djukic, M.; Erler, S.; Leimbach, A.; Grossar, D.; Charrière, J.-D.; Gauthier, L.; Hartken, D.; Dietrich, S.; Nacke, H.; Daniel, R.; et al. Comparative Genomics and Description of Putative Virulence Factors of Melissococcus plutonius, the Causative Agent of European Foulbrood Disease in Honey Bees. Genes 2018, 9, 419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossar, D.; Kilchenmann, V.; Forsgren, E.; Charrière, J.-D.; Gauthier, L.; Chapuisat, M.; Dietemann, V. Putative Determinants of Virulence in Melissococcus plutonius, the Bacterial Agent Causing European Foulbrood in Honey Bees. Virulence 2020, 11, 554–567. [Google Scholar] [CrossRef]
- McKee, B.A.; David Goodman, R.; Alan Hornitzky, M. The Transmission of European Foulbrood (Melissococcus plutonius) to Artificially Reared Honey Bee Larvae (Apis mellifera). J. Apic. Res. 2004, 43, 93–100. [Google Scholar] [CrossRef]
- Giersch, T.; Barchia, I.; Hornitzky, M. Can Fatty Acids and Oxytetracycline Protect Artificially Raised Larvae from Developing European Foulbrood? Apidologie 2010, 41, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Okumura, K.; Arai, R.; Okura, M.; Kirikae, T.; Takamatsu, D.; Osaki, M.; Miyoshi-Akiyama, T. Complete Genome Sequence of Melissococcus plutonius DAT561, a Strain That Shows an Unusual Growth Profile and Is Representative of an Endemic Cluster in Japan. J. Bacteriol. 2012, 194, 3014. [Google Scholar] [CrossRef] [Green Version]
- Takamatsu, D.; Arai, R.; Miyoshi-Akiyama, T.; Okumura, K.; Okura, M.; Kirikae, T.; Kojima, A.; Osaki, M. Identification of Mutations Involved in the Requirement of Potassium for Growth of Typical Melissococcus plutonius Strains. Appl. Environ. Microbiol. 2013, 79, 3882–3886. [Google Scholar] [CrossRef] [Green Version]
- Sopko, B.; Zitek, J.; Nesvorna, M.; Markovic, M.; Kamler, M.; Titera, D.; Erban, T.; Hubert, J. Detection and Quantification of Melissococcus plutonius in Honey Bee Workers Exposed to European Foulbrood in Czechia through Conventional PCR, QPCR, and Barcode Sequencing. J. Apic. Res. 2020, 59, 503–514. [Google Scholar] [CrossRef]
- Biová, J.; Charrière, J.-D.; Dostálková, S.; Škrabišová, M.; Petřivalský, M.; Bzdil, J.; Danihlík, J. Melissococcus plutonius Can Be Effectively and Economically Detected Using Hive Debris and Conventional PCR. Insects 2021, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Roetschi, A.; Berthoud, H.; Kuhn, R.; Imdorf, A. Infection Rate Based on Quantitative Real-Time PCR of Melissococcus plutonius, the Causal Agent of European Foulbrood, in Honeybee Colonies before and after Apiary Sanitation. Apidologie 2008, 39, 362–371. [Google Scholar] [CrossRef]
- McKee, B.A.; Djordjevic, S.P.; Goodman, R.D.; Hornitzky, M.A. The Detection of Melissococcus Pluton in Honey Bees (Apis mellifera) and Their Products Using a Hemi-Nested PCR. Apidologie 2003, 34, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Saleh, M.; Soliman, H.; Sørum, H.; Fauske, A.K.; El-Matbouli, M. A Novel Gold Nanoparticles-Based Assay for Rapid Detection of Melissococcus plutonius, the Causative Agent of European Foulbrood. Vet. Rec. 2012, 171, 400. [Google Scholar] [CrossRef]
- Nguyen, P.V.; Lee, B.; Yoo, M.-S.; Yoon, B.-S. Development and clinical validation of a DNA gyrase subunit B gene based loop-mediated isothermal amplification method for detection of Melissococcus plutonius. J. Apic. 2012, 27, 51–58. [Google Scholar]
- Budge, G.E.; Shirley, M.D.F.; Jones, B.; Quill, E.; Tomkies, V.; Feil, E.J.; Brown, M.A.; Haynes, E.G. Molecular Epidemiology and Population Structure of the Honey Bee Brood Pathogen Melissococcus plutonius. ISME J. 2014, 8, 1588–1597. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, D.; Morinishi, K.; Arai, R.; Sakamoto, A.; Okura, M.; Osaki, M. Typing of Melissococcus plutonius Isolated from European and Japanese Honeybees Suggests Spread of Sequence Types across Borders and between Different Apis Species. Vet. Microbiol. 2014, 171, 221–226. [Google Scholar] [CrossRef]
- Nakamura, K.; Yamazaki, Y.; Shiraishi, A.; Kobayashi, S.; Harada, M.; Yoshiyama, M.; Osaki, M.; Okura, M.; Takamatsu, D. Virulence Differences among Melissococcus plutonius Strains with Different Genetic Backgrounds in Apis mellifera Larvae under an Improved Experimental Condition. Sci. Rep. 2016, 6, 33329. [Google Scholar] [CrossRef] [Green Version]
- Arai, R.; Tominaga, K.; Wu, M.; Okura, M.; Ito, K.; Okamura, N.; Onishi, H.; Osaki, M.; Sugimura, Y.; Yoshiyama, M.; et al. Diversity of Melissococcus plutonius from Honeybee Larvae in Japan and Experimental Reproduction of European Foulbrood with Cultured Atypical Isolates. PLoS ONE 2012, 7, e33708. [Google Scholar] [CrossRef] [Green Version]
- Arai, R.; Miyoshi-Akiyama, T.; Okumura, K.; Morinaga, Y.; Wu, M.; Sugimura, Y.; Yoshiyama, M.; Okura, M.; Kirikae, T.; Takamatsu, D. Development of Duplex PCR Assay for Detection and Differentiation of Typical and Atypical Melissococcus plutonius Strains. J. Vet. Sci. 2014, 76, 491–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dainat, B.; Grossar, D.; Ecoffey, B.; Haldemann, C. Triplex Real-Time PCR Method for the Qualitative Detection of European and American Foulbrood in Honeybee. J. Microbiol. Mets. 2018, 146, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Mouches, C.; Bové, J.M.; Albisetti, J.; Clark, T.B.; Tully, J.G. A Spiroplasma of Serogroup IV Causes a May-Disease-like Disorder of Honeybees in Southwestern France. Microb. Ecol. 1982, 8, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Teixeira, É.W.; Tauber, J.P.; Birke, J.M.; Martins, M.F.; Fonseca, I.; Evans, J.D. Honey Bee Colonies Act as Reservoirs for Two Spiroplasma Facultative Symbionts and Incur Complex, Multiyear Infection Dynamics. MicrobiologyOpen 2014, 3, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Mouches, C.; Bové, J.M.; Tully, J.G.; Rose, D.L.; McCoy, R.E.; Carle-Junca, P.; Garnier, M.; Saillard, C. Spiroplasma Apis, a New Species from the Honey-Bee Apis mellifera. Ann. Inst. Pasteur. Microbiol. 1983, 134, 383–397. [Google Scholar] [CrossRef]
- Clark, T.B. Honeybee Spiroplasmosis, a New Problem for Beekeepers. Am. Bee J. 1978, 118, 18–23. [Google Scholar]
- Zheng, H.-Q.; Chen, Y.P. Detection of Spiroplasma Melliferum in Honey Bee Colonies in the US. J. Invert. Pathol. 2014, 119, 47–49. [Google Scholar] [CrossRef]
- Ku, C.; Lo, W.-S.; Chen, L.-L.; Kuo, C.-H. Complete Genomes of Two Dipteran-Associated Spiroplasmas Provided Insights into the Origin, Dynamics, and Impacts of Viral Invasion in Spiroplasma. Genome Biol. Evol. 2013, 5, 1151–1164. [Google Scholar] [CrossRef] [Green Version]
- Lo, W.-S.; Gasparich, G.E.; Kuo, C.-H. Found and Lost: The Fates of Horizontally Acquired Genes in Arthropod-Symbiotic Spiroplasma. Genome Biol. Evol. 2015, 7, 2458–2472. [Google Scholar] [CrossRef] [Green Version]
- Meeus, I.; Vercruysse, V.; Smagghe, G. Molecular Detection of Spiroplasma Apis and Spiroplasma Melliferum in Bees. J. Invert. Pathol. 2012, 109, 172–174. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Grimont, F. The Genus Serratia. Annu. Rev. Microbiol. 1978, 32, 221–248. [Google Scholar] [CrossRef] [PubMed]
- da Mota, F.F.; Castro, D.P.; Vieira, C.S.; Gumiel, M.; de Albuquerque, J.P.; Carels, N.; Azambuja, P. In Vitro Trypanocidal Activity, Genomic Analysis of Isolates, and in vivo Transcription of Type VI Secretion System of Serratia marcescens Belonging to the Microbiota of Rhodnius prolixus Digestive Tract. Front. Microbiol. 2019, 9, 3205. [Google Scholar] [CrossRef] [PubMed]
- El Sanousi, S.M.; El Sarag, M.S.A.; Mohamed, S.E.Y. 1987 Properties of Serratia marcescens Isolated from Diseased Honeybee (Apis mellifera) Larvae. Microbiol. 1986, 133, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Burritt, N.L.; Foss, N.J.; Neeno-Eckwall, E.C.; Church, J.O.; Hilger, A.M.; Hildebrand, J.A.; Warshauer, D.M.; Perna, N.T.; Burritt, J.B. Sepsis and Hemocyte Loss in Honey Bees (Apis mellifera) Infected with Serratia marcescens Strain Sicaria. PLoS ONE 2016, 11, e0167752. [Google Scholar] [CrossRef] [Green Version]
- Burnside, C.E. A Septicemic Condition of Adult Bees. J. Econ. Entomol. 1928, 21, 379–386. [Google Scholar] [CrossRef]
- Raymann, K.; Coon, K.L.; Shaffer, Z.; Salisbury, S.; Moran, N.A. Pathogenicity of Serratia marcescens Strains in Honey Bees. mBio 2018, 9, e01649-18. [Google Scholar] [CrossRef] [Green Version]
- Masry, S.H.D.; Taha, T.H.; Botros, W.A.; Mahfouz, H.; Al-Kahtani, S.N.; Ansari, M.J.; Hafez, E.E. Antimicrobial Activity of Camphor Tree Silver Nano-Particles against Foulbrood Diseases and Finding out New Strain of Serratia marcescens via DGGE-PCR, as a Secondary Infection on Honeybee Larvae. Saudi J. Biol. Sci. 2021, 28, 2067–2075. [Google Scholar] [CrossRef]
- Hubert, J.; Erban, T.; Kamler, M.; Kopecky, J.; Nesvorna, M.; Hejdankova, S.; Titera, D.; Tyl, J.; Zurek, L. Bacteria Detected in the Honeybee Parasitic Mite Varroa destructor Collected from Beehive Winter Debris. J. Appl. Microbiol. 2015, 119, 640–654. [Google Scholar] [CrossRef]
- Erban, T.; Ledvinka, O.; Kamler, M.; Nesvorna, M.; Hortova, B.; Tyl, J.; Titera, D.; Markovic, M.; Hubert, J. Honeybee (Apis mellifera)-Associated Bacterial Community Affected by American Foulbrood: Detection of Paenibacillus larvae via Microbiome Analysis. Sci. Rep. 2017, 7, 5084. [Google Scholar] [CrossRef]
- Alippi, A.M.; Lopez, A.C.; Aguilar, O.M. A PCR-Based Method That Permits Specific Detection of Paenibacillus larvae subsp. larvae, the Cause of American Foulbrood of Honey Bees, at the Subspecies Level. Lett. Appl. Microbiol. 2004, 39, 25–33. [Google Scholar] [CrossRef]
- Djukic, M.; Brzuszkiewicz, E.; Fünfhaus, A.; Voss, J.; Gollnow, K.; Poppinga, L.; Liesegang, H.; Garcia-Gonzalez, E.; Genersch, E.; Daniel, R. How to Kill the Honey Bee Larva: Genomic Potential and Virulence Mechanisms of Paenibacillus larvae. PLoS ONE 2014, 9, e90914. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E.; Otten, C. The Use of Repetitive Element PCR Fingerprinting (Rep-PCR) for Genetic Subtyping of German Field Isolates of Paenibacillus larvae subsp. larvae. Apidologie 2003, 34, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Govan, V.A.; Brözel, V.; Allsopp, M.H.; Davison, S. A PCR Detection Method for Rapid Identification of Melissococcus Pluton in Honeybee Larvae. Appl. Environ. Microbiol. 1998, 64, 1983–1985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djordjevic, S.P.; Noone, K.; Smith, L.; Hornitzky, M.A.Z. Development of a Hemi-Nested PCR Assay for the Specific Detection of Melissococcus Pluton. J. Apic. Res. 1998, 37, 165–174. [Google Scholar] [CrossRef]
- Budge, G.E.; Barrett, B.; Jones, B.; Pietravalle, S.; Marris, G.; Chantawannakul, P.; Thwaites, R.; Hall, J.; Cuthbertson, A.G.S.; Brown, M.A. The Occurrence of Melissococcus plutonius in Healthy Colonies of Apis mellifera and the Efficacy of European Foulbrood Control Measures. J. Invert. Pathol. 2010, 105, 164–170. [Google Scholar] [CrossRef]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noël, L.M.-L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tentcheva, D.; Gauthier, L.; Jouve, S.; Canabady-Rochelle, L.; Dainat, B.; Cousserans, F.; Colin, M.E.; Ball, B.V.; Bergoin, M. Polymerase Chain Reaction Detection of Deformed Wing Virus (DWV) in Apis mellifera and Varroa destructor. Apidologie 2004, 35, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and Seasonal Variations of Six Bee Viruses in Apis mellifera L. and Varroa destructor Mite Populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.J. The Role of Varroa and Viral Pathogens in the Collapse of Honeybee Colonies: A Modelling Approach. J. Appl. Ecol. 2001, 38, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Chantawannakul, P.; Ward, L.; Boonham, N.; Brown, M. A Scientific Note on the Detection of Honeybee Viruses Using Real-Time PCR (TaqMan) in Varroa Mites Collected from a Thai Honeybee (Apis mellifera) Apiary. J. Invert. Pathol. 2006, 91, 69–73. [Google Scholar] [CrossRef]
- Hartmann, U.; Forsgren, E.; Charrière, J.-D.; Neumann, P.; Gauthier, L. Dynamics of Apis mellifera Filamentous Virus (AmFV) Infections in Honey Bees and Relationships with Other Parasites. Viruses 2015, 7, 2654–2667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, C.; Li, B.; Deng, S.; Chu, Y.; Diao, Q. Diagnosis and Distribution of the Apis mellifera Filamentous Virus (AmFV) in Honey Bees (Apis mellifera) in China. Insects. Soc. 2017, 64, 597–603. [Google Scholar] [CrossRef]
- Kevill, J.L.; Highfield, A.; Mordecai, G.J.; Martin, S.J.; Schroeder, D.C. ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees. Viruses 2017, 9, 314. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, D.A.; Fuller, Z.L.; Ray, A.M.; Brockmann, A.; Frazier, M.; Gikungu, M.W.; Martinez, J.F.I.; Kapheim, K.M.; Kerby, J.T.; Kocher, S.D.; et al. Investigating the Viral Ecology of Global Bee Communities with High-Throughput Metagenomics. Sci. Rep. 2018, 8, 8879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Miranda, J. Diagnostic Techniques for Virus Detection in Honey Bees. In Virology and the Honey Bee; European Research Commission: Brussels, Belgium, 2008; pp. 121–232. ISBN 92-79-00586-3. [Google Scholar]
- Huang, S.; Li, J.; Zhang, Y.; Li, Z.; Evans, J.D.; Rose, R.; Gilligan, T.M.; LeBrun, A.; He, N.; Zheng, T.; et al. A Novel Method for the Detection and Diagnosis of Virus Infections in Honey Bees. J. Virol. Methods. 2021, 293, 114163. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.-P.; Gauthier, L.; Genersch, E.; de Graaf, D.C.; et al. Standard Methods for Virus Research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.P.; Budge, G.; Cornman, R.S.; De la Rua, P.; de Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard Methods for Molecular Research in Apis mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef] [Green Version]
- Kevill, J.L.; de Souza, F.S.; Sharples, C.; Oliver, R.; Schroeder, D.C.; Martin, S.J. DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US. Viruses 2019, 11, 426. [Google Scholar] [CrossRef] [Green Version]
- Posada-Florez, F.; Childers, A.K.; Heerman, M.C.; Egekwu, N.I.; Cook, S.C.; Chen, Y.; Evans, J.D.; Ryabov, E.V. Deformed Wing Virus Type A, a Major Honey Bee Pathogen, Is Vectored by the Mite Varroa destructor in a Non-Propagative Manner. Sci. Rep. 2019, 9, 12445. [Google Scholar] [CrossRef] [Green Version]
- de Miranda, J.R.; Genersch, E. Deformed Wing Virus. J. Invert. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Chen, Y.; Madella, S.; Nessa, A.; vanEngelsdorp, D.; Evans, J.D. Recent Spread of Varroa destructor Virus-1, a Honey Bee Pathogen, in the United States. Sci. Rep. 2017, 7, 17447. [Google Scholar] [CrossRef] [Green Version]
- Bradford, E.L.; Christie, C.R.; Campbell, E.M.; Bowman, A.S. A Real-Time PCR Method for Quantification of the Total and Major Variant Strains of the Deformed Wing Virus. PLoS ONE 2017, 12, e0190017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tehel, A.; Vu, Q.; Bigot, D.; Gogol-Döring, A.; Koch, P.; Jenkins, C.; Doublet, V.; Theodorou, P.; Paxton, R. The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees. Viruses 2019, 11, 114. [Google Scholar] [CrossRef] [Green Version]
- Anguiano-Baez, R.; Guzman-Novoa, E.; Md. Hamiduzzaman, M.; Espinosa-Montaño, L.G.; Correa-Benítez, A. Varroa destructor (Mesostigmata: Varroidae) Parasitism and Climate Differentially Influence the Prevalence, Levels, and Overt Infections of Deformed Wing Virus in Honey Bees (Hymenoptera: Apidae). J. Insect Sci. 2016, 16, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global Honey Bee Viral Landscape Altered by a Parasitic Mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Ding, G.; Fondevila, N.; Palacio, M.A.; Merke, J.; Martinez, A.; Camacho, B.; Aignasse, A.; Figini, E.; Rodriguez, G.; Lv, L.; et al. Prevalence of Honeybee Viruses in Different Regions of China and Argentina. Rev. Sci. Tech. 2016, 35, 825–833. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a Honey Bee Pathogen: First Report of a Third Master Variant of the Deformed Wing Virus Quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasesco, C.; Quintana, S.; Gerónimo, V.D.; García, M.L.G.; Sguazza, G.; Bravi, M.E.; Fargnoli, L.; Reynaldi, F.J.; Eguaras, M.; Maggi, M. Deformed Wing Virus Type a and b in Managed Honeybee Colonies of Argentina. Bull. Entomol. Res. 2021, 111, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Tibatá, V.M.; Sanchez, A.; Palmer-Young, E.; Junca, H.; Solarte, V.M.; Madella, S.; Ariza, F.; Figueroa, J.; Corona, M. Africanized Honey Bees in Colombia Exhibit High Prevalence but Low Level of Infestation of Varroa Mites and Low Prevalence of Pathogenic Viruses. PLoS ONE 2021, 16, e0244906. [Google Scholar] [CrossRef]
- Dalmon, A.; Desbiez, C.; Coulon, M.; Thomasson, M.; Le Conte, Y.; Alaux, C.; Vallon, J.; Moury, B. Evidence for Positive Selection and Recombination Hotspots in Deformed Wing Virus (DWV). Sci. Rep. 2017, 7, 41045. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Lim, S.-J.; Kim, S.; Kim, M.; Kim, B.; Tai, T.A.; Kim, S.; Yoon, B. Rapid Detection of Deformed Wing Virus in Honeybee Using Ultra-Rapid QPCR and a DNA-Chip. J. Vet. Sci. 2020, 21, e4. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Hammond, J.; Hsu, H.; Evans, J.; Feldlaufer, M. Multiple Virus Infections in the Honey Bee and Genome Divergence of Honey Bee Viruses. J. Invert. Pathol. 2004, 87, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Meeus, I.; Smagghe, G.; Siede, R.; Jans, K.; de Graaf, D.C. Multiplex RT-PCR with Broad-Range Primers and an Exogenous Internal Amplification Control for the Detection of Honeybee Viruses in Bumblebees. J. Invert. Pathol. 2010, 105, 200–203. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Lopez, D.; Grubbs, K.; Posada-Florez, F.; Weaver, D.; Girten, W.; vanEngelsdorp, D.; Chen, Y.; Evans, J.D. Dynamic Evolution in the Key Honey Bee Pathogen Deformed Wing Virus: Novel Insights into Virulence and Competition Using Reverse Genetics. PLoS Biol. 2019, 17, e3000502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, J.M.K.; Simbiken, N.; Dale, C.; Armstrong, J.; Anderson, D.L. Tolerance of Honey Bees to Varroa Mite in the Absence of Deformed Wing Virus. Viruses 2020, 12, 575. [Google Scholar] [CrossRef]
- Brettell, L.E.; Mordecai, G.J.; Schroeder, D.C.; Jones, I.M.; da Silva, J.R.; Vicente-Rubiano, M.; Martin, S.J. A Comparison of Deformed Wing Virus in Deformed and Asymptomatic Honey Bees. Insects 2017, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or in vitro, Transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terio, V.; Martella, V.; Camero, M.; Decaro, N.; Testini, G.; Bonerba, E.; Tantillo, G.; Buonavoglia, C. Detection of a Honeybee Iflavirus with Intermediate Characteristics between Kakugo Virus and Deformed Wing Virus. New Microbiol. 2008, 31, 439–444. [Google Scholar] [PubMed]
- Fujiyuki, T.; Matsuzaka, E.; Nakaoka, T.; Takeuchi, H.; Wakamoto, A.; Ohka, S.; Sekimizu, K.; Nomoto, A.; Kubo, T. Distribution of Kakugo Virus and Its Effects on the Gene Expression Profile in the Brain of the Worker Honeybee Apis mellifera L. J. Virol. 2009, 83, 11560–11568. [Google Scholar] [CrossRef] [Green Version]
- Fujiyuki, T.; Ohka, S.; Takeuchi, H.; Ono, M.; Nomoto, A.; Kubo, T. Prevalence and Phylogeny of Kakugo Virus, a Novel Insect Picorna-like Virus That Infects the Honeybee (Apis mellifera L.), under Various Colony Conditions. J. Virol. 2006, 80, 11528–11538. [Google Scholar] [CrossRef] [Green Version]
- Molineri, A.I.; Pacini, A.; Giacobino, A.; Bulacio-Cagnolo, N.; Aignasse, A.; Zago, L.; Fondevila, N.; Ferrufino, C.; Merke, J.; Orellano, E.; et al. Prevalence of honey bee (Apis mellifera) viruses in temperate and subtropical regions from Argentina. Rev. Arg. Microbiol. 2017, 49, 166–173. [Google Scholar] [CrossRef]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate Transmission Routes and Interactions between Picorna-like Viruses (Kashmir Bee Virus and Sacbrood Virus) with the Honeybee Host and the Parasitic Varroa Mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Freiberg, M.; De Jong, D.; Message, D.; Cox-Foster, D. First Report of Sacbrood Virus in Honey Bee (Apis mellifera) Colonies in Brazil. Genet. Mol. Res. 2012, 11, 3310–3314. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.-P.; Thiéry, R.; Ribière, M. Development and Validation of a Real-Time Two-Step RT-QPCR TaqMan(®) Assay for Quantitation of Sacbrood Virus (SBV) and Its Application to a Field Survey of Symptomatic Honey Bee Colonies. J. Virol. Methods. 2014, 197, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Antúnez, K.; Anido, M.; Branchiccela, B.; Harriet, J.; Campa, J.; Invernizzi, C.; Santos, E.; Higes, M.; Martín-Hernández, R.; Zunino, P. Seasonal Variation of Honeybee Pathogens and Its Association with Pollen Diversity in Uruguay. Microb. Ecol. 2015, 70, 522–533. [Google Scholar] [CrossRef]
- Highfield, A.; Kevill, J.; Mordecai, G.; Hunt, J.; Henderson, S.; Sauvard, D.; Feltwell, J.; Martin, S.J.; Sumner, S.; Schroeder, D.C. Detection and Replication of Moku Virus in Honey Bees and Social Wasps. Viruses 2020, 12, 607. [Google Scholar] [CrossRef]
- Glenny, W.; Cavigli, I.; Daughenbaugh, K.F.; Radford, R.; Kegley, S.E.; Flenniken, M.L. Honey Bee (Apis mellifera) Colony Health and Pathogen Composition in Migratory Beekeeping Operations Involved in California Almond Pollination. PLoS ONE 2017, 12, e0182814. [Google Scholar] [CrossRef] [PubMed]
- Bigot, D.; Dalmon, A.; Roy, B.; Hou, C.; Germain, M.; Romary, M.; Deng, S.; Diao, Q.; Weinert, L.A.; Cook, J.M.; et al. The Discovery of Halictivirus Resolves the Sinaivirus Phylogeny. J. Gen. Virol. 2017, 98, 2864–2875. [Google Scholar] [CrossRef] [PubMed]
- Daughenbaugh, K.F.; Martin, M.; Brutscher, L.M.; Cavigli, I.; Garcia, E.; Lavin, M.; Flenniken, M.L. Honey Bee Infecting Lake Sinai Viruses. Viruses 2015, 7, 3285–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwanowicz, D.D.; Wu-Smart, J.Y.; Olgun, T.; Smart, A.H.; Otto, C.R.V.; Lopez, D.; Evans, J.D.; Cornman, R. An Updated Genetic Marker for Detection of Lake Sinai Virus and Metagenetic Applications. PeerJ 2020, 8, e9424. [Google Scholar] [CrossRef]
- Blanchard, P.; Ribière, M.; Celle, O.; Lallemand, P.; Schurr, F.; Olivier, V.; Iscache, A.L.; Faucon, J.P. Evaluation of a Real-Time Two-Step RT-PCR Assay for Quantitation of Chronic Bee Paralysis Virus (CBPV) Genome in Experimentally-Infected Bee Tissues and in Life Stages of a Symptomatic Colony. J. Virol. Methods. 2007, 141, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, P.; Regnault, J.; Schurr, F.; Dubois, E.; Ribière, M. Intra-Laboratory Validation of Chronic Bee Paralysis Virus Quantitation Using an Accredited Standardised Real-Time Quantitative RT-PCR Method. J. Virol. Methods. 2012, 180, 26–31. [Google Scholar] [CrossRef]
- Celle, O.; Blanchard, P.; Olivier, V.; Schurr, F.; Cougoule, N.; Faucon, J.-P.; Ribière, M. Detection of Chronic Bee Paralysis Virus (CBPV) Genome and Its Replicative RNA Form in Various Hosts and Possible Ways of Spread. Virus Res. 2008, 133, 280–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakonyi, T.; Grabensteiner, E.; Kolodziejek, J.; Rusvai, M.; Topolska, G.; Ritter, W.; Nowotny, N. Phylogenetic Analysis of Acute Bee Paralysis Virus Strains. Appl. Environ. Microbiol. 2002, 68, 6446–6450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios, G.; Hui, J.; Quan, P.L.; Kalkstein, A.; Honkavuori, K.S.; Bussetti, A.V.; Conlan, S.; Evans, J.; Chen, Y.P.; vanEngelsdorp, D.; et al. Genetic Analysis of Israel Acute Paralysis Virus: Distinct Clusters Are Circulating in the United States. J. Virol. 2008, 82, 6209–6217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Miranda, J.R.; Drebot, M.; Tyler, S.; Shen, M.; Cameron, C.E.; Stoltz, D.B.; Camazine, S.M. Complete Nucleotide Sequence of Kashmir Bee Virus and Comparison with Acute Bee Paralysis Virus. J. Gen. Virol. 2004, 85, 2263–2270. [Google Scholar] [CrossRef] [PubMed]
- Cox-Foster, D.; Conlan, S.; Holmes, E.; Palacios, G.; Evans, J.; Moran, N.; Quan, P.-L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Kukielka, D.; Perez, A.M.; Higes, M.; del Carmen Bulboa, M.; Sánchez-Vizcaíno, J.M. Analytical Sensitivity and Specificity of a RT-PCR for the Diagnosis and Characterization of the Spatial Distribution of Three Apis mellifera Viral Diseases in Spain. Apidologie 2008, 39, 607–617. [Google Scholar] [CrossRef] [Green Version]
- Siede, R.; König, M.; Büchler, R.; Failing, K.; Thiel, H.-J. A Real-Time PCR Based Survey on Acute Bee Paralysis Virus in German Bee Colonies. Apidologie 2008, 39, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D. Beepath: An Ordered Quantitative-PCR Array for Exploring Honey Bee Immunity and Disease. J. Invert. Pathol. 2006, 93, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Rivkin, H.; Slabezki, Y.; Chejanovsky, N. Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder. Viruses 2014, 6, 2012–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maori, E.; Lavi, S.; Mozes-Koch, R.; Gantman, Y.; Peretz, Y.; Edelbaum, O.; Tanne, E.; Sela, I. Isolation and Characterization of Israeli Acute Paralysis Virus, a Dicistrovirus Affecting Honeybees in Israel: Evidence for Diversity Due to Intra- and Inter-Species Recombination. J. Gen. Virol. 2007, 88, 3428–3438. [Google Scholar] [CrossRef]
- Soroker, V.; Hetzroni, A.; Yakobson, B.; David, D.; David, A.; Voet, H.; Slabezki, Y.; Efrat, H.; Levski, S.; Kamer, Y.; et al. Evaluation of Colony Losses in Israel in Relation to the Incidence of Pathogens and Pests. Apidologie 2011, 42, 192–199. [Google Scholar] [CrossRef] [Green Version]
- Reynaldi, F.J.; Sguazza, G.H.; Tizzano, M.A.; Fuentealba, N.; Galosi, C.M.; Pecoraro, M.R. First Report of Israeli Acute Paralysis Virus in Asymptomatic Hives of Argentina. Rev. Arg. Microbiol. 2011, 43, 84–86. [Google Scholar]
- Chen, Y.P.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-Virus Interaction in Collapsing Honey Bee Colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Yang, S.; Zhao, H.; Diao, Q.; Hou, C. IAPV-Induced Paralytic Symptoms Associated with Tachypnea via Impaired Tracheal System Function. Int. J. Molec. Sci. 2021, 22, 10078. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.-T.; Kim, B.; Kim, S.; Kim, M.; Kim, J.; Kim, S.; Yoon, B. Rapid Detection of Israeli Acute Paralysis Virus Using Multi-Point Ultra-Rapid Real-Time PCR (UR-QPCR). J. Apic. Res. 2019, 58, 746–753. [Google Scholar] [CrossRef]
- Berényi, O.; Bakonyi, T.; Derakhshifar, I.; Köglberger, H.; Nowotny, N. Occurrence of Six Honeybee Viruses in Diseased Austrian Apiaries. Appl. Environ. Microbiol. 2006, 72, 2414–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, B.; Forsgren, E.; Fries, I.; de Miranda, J.R. Acaricide Treatment Affects Viral Dynamics in Varroa destructor-Infested Honey Bee Colonies via Both Host Physiology and Mite Control. Appl. Environ. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Benjeddou, M.; Leat, N.; Allsopp, M.; Davison, S. Detection of Acute Bee Paralysis Virus and Black Queen Cell Virus from Honeybees by Reverse Transcriptase PCR. Appl. Environ. Microbiol. 2001, 67, 2384–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moharrami, M.; Modirrousta, H. Molecular Identification of Six Honeybee Viruses in Iranian Apiaries. Arch. Razi Inst. 2018, 73, 311–318. [Google Scholar] [CrossRef] [PubMed]
- vanEngelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony Collapse Disorder: A Descriptive Study. PLoS ONE 2009, 4, e6481. [Google Scholar] [CrossRef] [PubMed]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D.; vanEngelsdorp, D. Multiyear Survey Targeting Disease Incidence in US Honey Bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef] [Green Version]
- Faurot-Daniels, C.; Glenny, W.; Daughenbaugh, K.F.; McMenamin, A.J.; Burkle, L.A.; Flenniken, M.L. Longitudinal Monitoring of Honey Bee Colonies Reveals Dynamic Nature of Virus Abundance and Indicates a Negative Impact of Lake Sinai Virus 2 on Colony Health. PLoS ONE 2020, 15, e0237544. [Google Scholar] [CrossRef]
- Cavigli, I.; Daughenbaugh, K.F.; Martin, M.; Lerch, M.; Banner, K.; Garcia, E.; Brutscher, L.M.; Flenniken, M.L. Pathogen Prevalence and Abundance in Honey Bee Colonies Involved in Almond Pollination. Apidologie 2016, 47, 251–266. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Pettis, J.S.; Feldlaufer, M.F. Detection of Multiple Viruses in Queens of the Honey Bee Apis mellifera L. J. Invert. Pathol. 2005, 90, 118–121. [Google Scholar] [CrossRef]
- Stoltz, D.; Shen, X.-R.; Boggis, C.; Sisson, G. Molecular Diagnosis of Kashmir Bee Virus Infection. J. Apic. Res. 1995, 34, 153–160. [Google Scholar] [CrossRef]
- Ribière, M.; Triboulot, C.; Mathieu, L.; Aurières, C.; Faucon, J.-P.; Pépin, M. Molecular Diagnosis of Chronic Bee Paralysis Virus Infection. Apidologie 2002, 33, 339–351. [Google Scholar] [CrossRef]
- D’Alvise, P.; Seeburger, V.; Gihring, K.; Kieboom, M.; Hasselmann, M. Seasonal Dynamics and Co-Occurrence Patterns of Honey Bee Pathogens Revealed by High-Throughput RT-QPCR Analysis. Ecol. Evol. 2019, 9, 10241–10252. [Google Scholar] [CrossRef] [Green Version]
- Papp, T.; Spann, D.; Marschang, R. Development and Use of a Real-Time Polymerase Chain Reaction for the Detection of Group II Invertebrate Iridoviruses in Pet Lizards and Prey Insects. J. Zoo. Wildl. Med. 2014, 45, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Smith, I.B.; Collins, A.M.; Pettis, J.S.; Feldlaufer, M.F. Detection of Deformed Wing Virus Infection in Honey Bees, Apis mellifera L., in the United States. Am. Bee J. 2004, 144, 557–559. Available online: https://www.ars.usda.gov/research/publications/publication/?seqNo115=156038 (accessed on 15 March 2022).
- Topley, E.; Davison, S.; Leat, N.; Benjeddou, M. Detection of Three Honeybee Viruses Simultaneously by a Single Multiplex Reverse Transcriptase PCR. African J. Biotechnol. 2005, 4, 763–767. [Google Scholar]
- Teixeira, E.W.; Chen, Y.; Message, D.; Pettis, J.; Evans, J.D. Virus Infections in Brazilian Honey Bees. J. Invert. Pathol. 2008, 99, 117–119. [Google Scholar] [CrossRef]
- Glover, R.H.; Adams, I.P.; Budge, G.; Wilkins, S.; Boonham, N. Detection of Honey Bee (Apis mellifera) Viruses with an Oligonucleotide Microarray. J. Invert. Pathol. 2011, 107, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Šimenc, L.; Knific, T.; Toplak, I. The Comparison of Honeybee Viral Loads for Six Honeybee Viruses (ABPV, BQCV, CBPV, DWV, LSV3 and SBV) in Healthy and Clinically Affected Honeybees with TaqMan Quantitative Real-Time RT-PCR Assays. Viruses 2021, 13, 1340. [Google Scholar] [CrossRef]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.-P.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of Quantitative Real-Time RT-PCR Assays for the Detection of Six Honeybee Viruses. J. Virol. Methods 2019, 270, 70–78. [Google Scholar] [CrossRef]
- Castelli, L.; Genchi García, M.L.; Dalmon, A.; Arredondo, D.; Antúnez, K.; Invernizzi, C.; Reynaldi, F.J.; Le Conte, Y.; Beaurepaire, A. Intra-Colonial Viral Infections in Western Honey Vees (Apis mellifera). Microorganisms 2021, 9, 1087. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; vanEngelsdorp, D.; Evans, J.D. Pathogen Webs in Collapsing Honey Bee Colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef] [Green Version]
- McMahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A Sting in the Spit: Widespread Cross-Infection of Multiple RNA Viruses across Wild and Managed Bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the Invertebrate RNA Virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Brutscher, L.M.; McMenamin, A.J.; Flenniken, M.L. The Buzz about Honey Bee Viruses. PLoS Pathog. 2016, 12, e1005757. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee Viruses: Ecology, Pathogenicity, and Impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef]
- Deboutte, W.; Beller, L.; Yinda, C.K.; Shi, C.; Smets, L.; Vanmechelen, B.; Conceição-Neto, N.; Dallmeier, K.; Maes, P.; de Graaf, D.C.; et al. Hymenoptera Associated Eukaryotic Virome Lacks Host Specificity. bioRxiv, 2020. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Cornman, R.S.; Evans, J.D.; Semberg, E.; Haddad, N.; Neumann, P.; Gauthier, L. Genome Characterization, Prevalence and Distribution of a Macula-like Virus from Apis mellifera and Varroa destructor. Viruses 2015, 7, 3586–3602. [Google Scholar] [CrossRef] [Green Version]
- Remnant, E.J.; Shi, M.; Buchmann, G.; Blacquière, T.; Holmes, E.C.; Beekman, M.; Ashe, A. A Diverse Range of Novel RNA Viruses in Geographically Distinct Honey Bee Populations. J. Virol. 2017, 91, e00158-17. [Google Scholar] [CrossRef] [Green Version]
- McMenamin, A.J.; Flenniken, M.L. Recently Identified Bee Viruses and Their Impact on Bee Pollinators. Curr. Opin. Insect. Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.; Sela, N.; Erez, T.; Nestel, D.; Pettis, J.; Neumann, P.; Chejanovsky, N. New Viruses from the Ectoparasite Mite Varroa destructor Infesting Apis mellifera and Apis Cerana. Viruses 2019, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, L.; Tentcheva, D.; Tournaire, M.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Viral Load Estimation in Asymptomatic Honey Bee Colonies Using the Quantitative RT-PCR Technique. Apidologie 2007, 38, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Jamnikar Ciglenečki, U.; Toplak, I. Development of a Real-Time RT-PCR Assay with TaqMan Probe for Specific Detection of Acute Bee Paralysis Virus. J. Virol. Methods 2012, 184, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; vanEngelsdorp, D.; Lipkin, W.I.; dePamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA Viruses in Hymenopteran Pollinators: Evidence of Inter-Taxa Virus Transmission via Pollen and Potential Impact on Non-Apis Hymenopteran Species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef] [PubMed]
- Sachman-Ruiz, B.; Narváez-Padilla, V.; Reynaud, E. Commercial Bombus Impatiens as Reservoirs of Emerging Infectious Diseases in Central México. Biol. Invasions 2015, 17, 2043–2053. [Google Scholar] [CrossRef] [Green Version]
- Zioni, N.; Soroker, V.; Chejanovsky, N. Replication of Varroa destructor Virus 1 (VDV-1) and a Varroa destructor Virus 1–Deformed Wing Virus Recombinant (VDV-1–DWV) in the Head of the Honey Bee. Virology 2011, 417, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed Wing Virus and Varroa destructor Virus-1 May Prevail in Varroa destructor-Infested Honeybee Colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef]
- Evans, J.D.; Hung, A.C. Molecular Phylogenetics and the Classification of Honey Bee Viruses. Arch. Virol. 2000, 145, 2015–2026. [Google Scholar] [CrossRef]
- Grabensteiner, E.; Ritter, W.; Carter, M.J.; Davison, S.; Pechhacker, H.; Kolodziejek, J.; Boecking, O.; Derakhshifar, I.; Moosbeckhofer, R.; Licek, E.; et al. Sacbrood Virus of the Honeybee (Apis mellifera): Rapid Identification and Phylogenetic Analysis Using Reverse Transcription-PCR. Clin. Diagn. Lab. Immunol. 2001, 8, 93–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, C.; Schröder, M.; Bienefeld, K.; Genersch, E. Detection of Viral Sequences in Semen of Honeybees (Apis mellifera): Evidence for Vertical Transmission of Viruses through Drones. J. Invert. Pathol. 2006, 92, 105–108. [Google Scholar] [CrossRef] [PubMed]
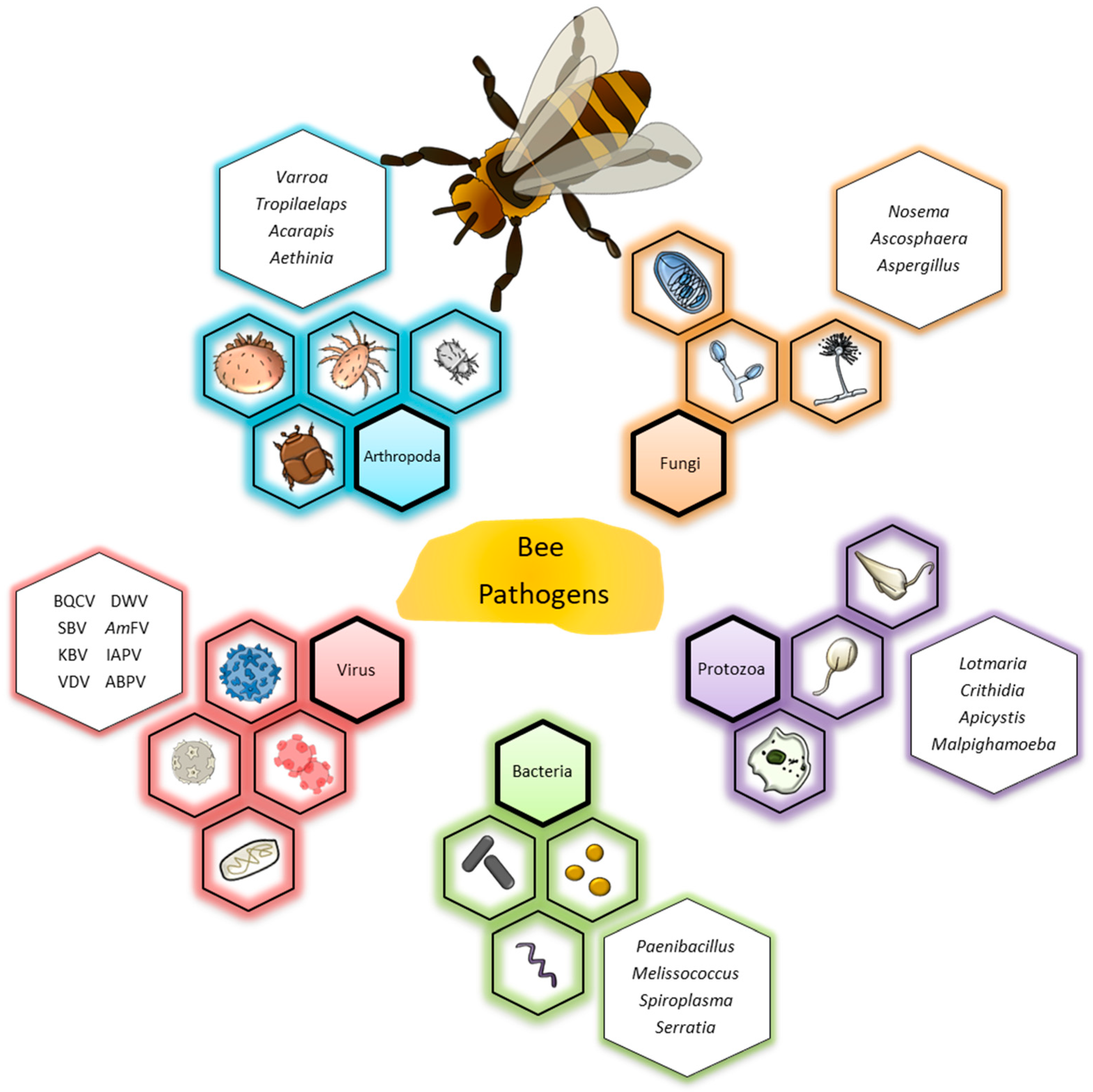

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lannutti, L.; Gonzales, F.N.; Dus Santos, M.J.; Florin-Christensen, M.; Schnittger, L. Molecular Detection and Differentiation of Arthropod, Fungal, Protozoan, Bacterial and Viral Pathogens of Honeybees. Vet. Sci. 2022, 9, 221. https://doi.org/10.3390/vetsci9050221
Lannutti L, Gonzales FN, Dus Santos MJ, Florin-Christensen M, Schnittger L. Molecular Detection and Differentiation of Arthropod, Fungal, Protozoan, Bacterial and Viral Pathogens of Honeybees. Veterinary Sciences. 2022; 9(5):221. https://doi.org/10.3390/vetsci9050221
Chicago/Turabian StyleLannutti, Lucas, Fernanda Noemi Gonzales, Maria José Dus Santos, Mónica Florin-Christensen, and Leonhard Schnittger. 2022. "Molecular Detection and Differentiation of Arthropod, Fungal, Protozoan, Bacterial and Viral Pathogens of Honeybees" Veterinary Sciences 9, no. 5: 221. https://doi.org/10.3390/vetsci9050221
APA StyleLannutti, L., Gonzales, F. N., Dus Santos, M. J., Florin-Christensen, M., & Schnittger, L. (2022). Molecular Detection and Differentiation of Arthropod, Fungal, Protozoan, Bacterial and Viral Pathogens of Honeybees. Veterinary Sciences, 9(5), 221. https://doi.org/10.3390/vetsci9050221









