Genetic Diversity and Structure of the Main Danubian Horse Paternal Genealogical Lineages Based on Microsatellite Genotyping
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Welfare and Ethical Statement
2.2. Sample Collection
2.3. DNA Extraction
2.4. Microsatellite Markers
2.5. PCR Amplification and Fragment Analysis
2.6. Statistical Analysis
3. Results
3.1. Polymorphism of Microsatellite Markers
3.2. Genetic Diversity within and among the Genealogical Lineages
3.3. Genetic Structure and Principal Coordinate Analysis
4. Discussion
4.1. Population Genetic Diversity of Paternal Lineages in the Danubian Horse
4.2. Genetic Differentiation within and between Paternal Lineages in the Danubian Horse
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaidarska, V.M.; Ignatova, M.M.; Lytskanov, P.I. Genetic resources of farm animals in Bulgaria—Conservation and management. Anim. Breed. Genet. 2017, 53, 35–43. [Google Scholar] [CrossRef]
- Tanchev, S. Conservation of genetic resources of autochthonous domestic livestock breeds in Bulgaria. A review. Bulg. J. Agric. Sci. 2015, 21, 1262–1271. [Google Scholar]
- Thornton, P.K. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2853–2867. [Google Scholar] [CrossRef] [Green Version]
- Adelman, M.; Thompson, K. Introduction to equestrian cultures in global and local contexts. In Equestrian Cultures in Global and Local Contexts; Springer: Cham, Switzerland, 2017; pp. 1–14. [Google Scholar]
- Prescott, W.H. Revival: History of the Conquest of Mexico: With a Preliminary View of the Ancient Mexican Civilisation and the Life of the Conqueror, Hernando Cortes, 2nd ed.; Routledge: London, UK, 2018; p. 782. [Google Scholar]
- Klecel, W.; Martyniuk, E. From the Eurasian steppes to the Roman circuses: A review of early development of horse breeding and management. Animals 2021, 11, 1859. [Google Scholar] [CrossRef]
- Pinheiro, M.; Kjöllerström, H.J.; Oom, M.M. Genetic diversity and demographic structure of the endangered Sorraia horse breed assessed through pedigree analysis. Livest. Sci. 2013, 152, 1–10. [Google Scholar] [CrossRef]
- Librado, P.; Fages, A.; Gaunitz, C.; Leonardi, M.; Wagner, S.; Khan, N.; Hanghøj, K.; Alquraishi, S.A.; Alfarhan, A.H.; Al-Rasheid, K.A.; et al. The evolutionary origin and genetic makeup of domestic horses. Genetics 2016, 204, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cressent, M.; Jez, C. The French horse industry at present. Adv. Anim. BioSci. 2013, 4, 54–65. [Google Scholar] [CrossRef]
- Leinonen, R.M. From Servant to Therapist: The Changing Meanings of Horses in Finland, 1st ed.; Routledge: London, UK, 2016; pp. 68–82. [Google Scholar]
- Marshall, F.B.; Dobney, K.; Denham, T.; Capriles, J.M. Evaluating the roles of directed breeding and gene flow in animal domestication. Proc. Natl. Acad. Sci. USA 2014, 111, 6153–6158. [Google Scholar] [CrossRef] [Green Version]
- Druml, T.; Neuditschko, M.; Grilz-Seger, G.; Horna, M.; Ricard, A.; Mesarič, M.; Cotman, M.; Pausch, H.; Brem, G. Population networks associated with runs of homozygosity reveal new insights into the breeding history of the Haflinger horse. J. Hered. 2018, 109, 384–392. [Google Scholar] [CrossRef]
- Barzev, G.; Yordanov, G.; Yuseinov, Y. The Bulgarian primitive horse in the area of Stara Planina mountain. Trakia J. Sci. 2005, 3, 74–76. [Google Scholar]
- Karaivanov, R.; Petrov, A.; Dobrev, D.; Tsankov, T. Development, structure, exterior characteristics and standardization of the Danubian horse breed. In Experian for Standardize of Horse Breeds in Bulgaria; Agropres: Stara Zagora, Bulgaria, 1971; pp. 82–96. [Google Scholar]
- Karaivanov, R.; Barzev, G.; Karadzhov, T. Development and status of families in the Danubian horse breed. In International Symposium on Half Breed Equine Breeding; Agropres: Stara Zagora, Bulgaria, 1989; pp. 82–96. [Google Scholar]
- ISAG/FAO Standing Committee. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans. Measurement of Domestic Animal Diversity (MoDAD): Recommended Microsatellite Markers. Available online: http://dad.fao.org/cgi-bin/getblob.cgi?sid=ca53b91a6f7c80be8e7066f4a50 (accessed on 13 June 2022).
- Binns, M.M.; Holmes, N.G.; Rolliman, A.; Scott, A.M. The identification of polymorphic microsatellite loci in the horse and their use in thoroughbred parentage testing. Br. Vet. J. 1995, 151, 9–15. [Google Scholar] [CrossRef]
- Bowling, A.T.; Eggleston-Stott, M.L.; Byrns, G.; Clark, R.S.; Dileanis, S.; Wictum, E. Validation of microsatellite markers for routine horse parentage testing. Anim. Genet. 1997, 28, 247–252. [Google Scholar] [CrossRef]
- Irvin, Z.; Giffard, J.; Brandon, R.; Breen, M.; Bell, K. Equine dinucleotide repeat polymorphisms at loci ASB 21, 23, 25 and 37–43. Anim. Genet. 1998, 29, 67. [Google Scholar] [PubMed]
- Guérin, G.; Bertaud, M.; Amigues, Y. Characterization of seven new horse microsatellites: HMS1, HMS2, HMS3, HMS5, HMS6, HMS7 and HMS8. Anim. Genet. 1994, 25, 62. [Google Scholar] [PubMed]
- Ellegren, H.; Johansson, M.; Sandberg, K.; Andersson, L. Cloning of highly polymorphic microsatellites in the horse. Anim. Genet. 1992, 23, 133–142. [Google Scholar] [CrossRef]
- Marklund, S.; Ellegren, H.; Eriksson, S.; Sandberg, K.; Andersson, L. Parentage testing and linkage analysis in the horse using a set of highly polymorphic microsatellites. Anim. Genet. 1994, 25, 19–23. [Google Scholar] [CrossRef]
- Haeringen, H.; Bowling, A.T.; Stott, M.L.; Lenstra, J.A.; Zwaagstra, K.A. A highly polymorphic horse microsatellite locus: VHL20. Anim. Genet. 1994, 25, 207. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Yeh, F.C.; Yang, R.-C.; Boyle, T.B.J.; Ye, Z.-H.; Mao, J.X. PopGene, the user-friendly shareware for population genetic analysis, molecular biology and biotechnology center. POPGENE User-Friendly Shareware Popul. Genet. Anal. 1997, 10, 295–301. [Google Scholar]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Karaivanov, R. Development, structure, exterior features and standardization of the Danube breed. An attempt to standardize horses in Bulgaria. Zhivotnovudstvo 1971, 21, 16–22. [Google Scholar]
- Karaivanov, R. Origin, Genealogical Structure and Development of the Danube Horse Breed. Ph.D. Thesis, Higher Institute of Zootechnics andVeterinary Medicine, Stara Zagora, Bulgaria, 1975; pp. 1–269. [Google Scholar]
- Moravcikova, N.; Kasarda, R.; Kukuckova, V.; Vostry, L.; Kadlecík, O. Genetic diversity of Old Kladruber and Nonius horse populations through microsatellite variation analysis. Acta. Agric. Slov. 2016, 107, 45–49. [Google Scholar]
- Lukanova, N.; Vlaeva, R.; Hristova, D.; Georgieva, S.; Barzev, G. Study on the genetic diversity of Trotter horses populations in Bulgaria. Agricul. Sci. 2015, 7, 159–165. [Google Scholar]
- Barzev, G.; Zhelyazkov, E.; Barzeva, V.; Hristova, D.; Sabev, Z. Genetic diversity in Bulgarian Thoroughbred using microsatellite DNA markers. Agric. Sci. Technol. 2010, 2, 116–120. [Google Scholar]
- Zeng, L.; Chen, N.; Yao, Y.; Dang, R.; Chen, H.; Lei, C. Analysis of genetic diversity and structure of Guanzhong horse using microsatellite markers. Anim. Biotechnol. 2019, 30, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, M.; La Rosa, V.; Rosati, R.; Chiofalo, V. Genetic diversity of the Italian Thoroughbred horse population. Ital. J. Anim. Sci. 2019, 18, 538–545. [Google Scholar] [CrossRef]
- Fornal, A.; Kowalska, K.; Zabek, T.; Piestrzynska-Kajtoch, A.; Musiał, A.D.; Ropka-Molik, K. Genetic diversity and population structure of Polish Konik horse based on individuals from all the male founder lines and microsatellite markers. Animals 2020, 10, 1569. [Google Scholar] [CrossRef]
- Seyedabadi, H.R.; Sofla, S.S. Microsatellite analysis for parentage verification and genetic characterization of the Turkmen horse population. Kafkas Univ. Vet. Fak. Derg. 2017, 23, 467–471. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Vostry, L.; Vostra Vydrova, H.; Hofmanova, B.; Vesela, Z.; Majzlik, I. Genetic diversity in Czech Haflinger horses. Poljoprivreda 2015, 21, 163–165. [Google Scholar] [CrossRef]
- Grilz-Seger, G.; Druml, T.; Neuditschko, M.; Dobretsberger, M.; Horna, M.; Brem, G. High-resolution population structure and runs of homozygosity reveal the genetic architecture of complex traits in the Lipizzan horse. BMC Genom. 2019, 20, 174. [Google Scholar] [CrossRef] [Green Version]
- Funk, S.M.; Guedaoura, S.; Juras, R.; Raziq, A.; Landolsi, F.; Luís, C.; Martínez, A.M.; Musa Mayaki, A.; Mujica, F.; Oom, M. do M.; et al. Major inconsistencies of inferred population genetic structure estimated in a large set of domestic horse breeds using microsatellites. Ecol. Evol. 2020, 10, 4261–4279. [Google Scholar] [CrossRef] [Green Version]
- Fornal, A.; Kowalska, K.; Zabek, T.; Piestrzynska-Kajtoch, A.; Musiał, A.D.; Ropka-Molik, K. Genetic variability and population structure of Polish Konik horse maternal lines based on microsatellite markers. Genes 2021, 12, 546. [Google Scholar] [CrossRef]
- Popova, M.; Lukanova, N.; Vlaeva, R. Monitoring of the sire lines of the Danube horse breed. Anim. Sci. 2020, 57, 11–18. [Google Scholar]
- Karadzhov, T. Influence of Some Genetic and Non-Genetic Factors on Reproductive Performance and Exterior Measurements in Pleven and Danube Horses. Ph.D. Thesis, Thracian University, Stara Zagora, Bulgaria, 1997. [Google Scholar]

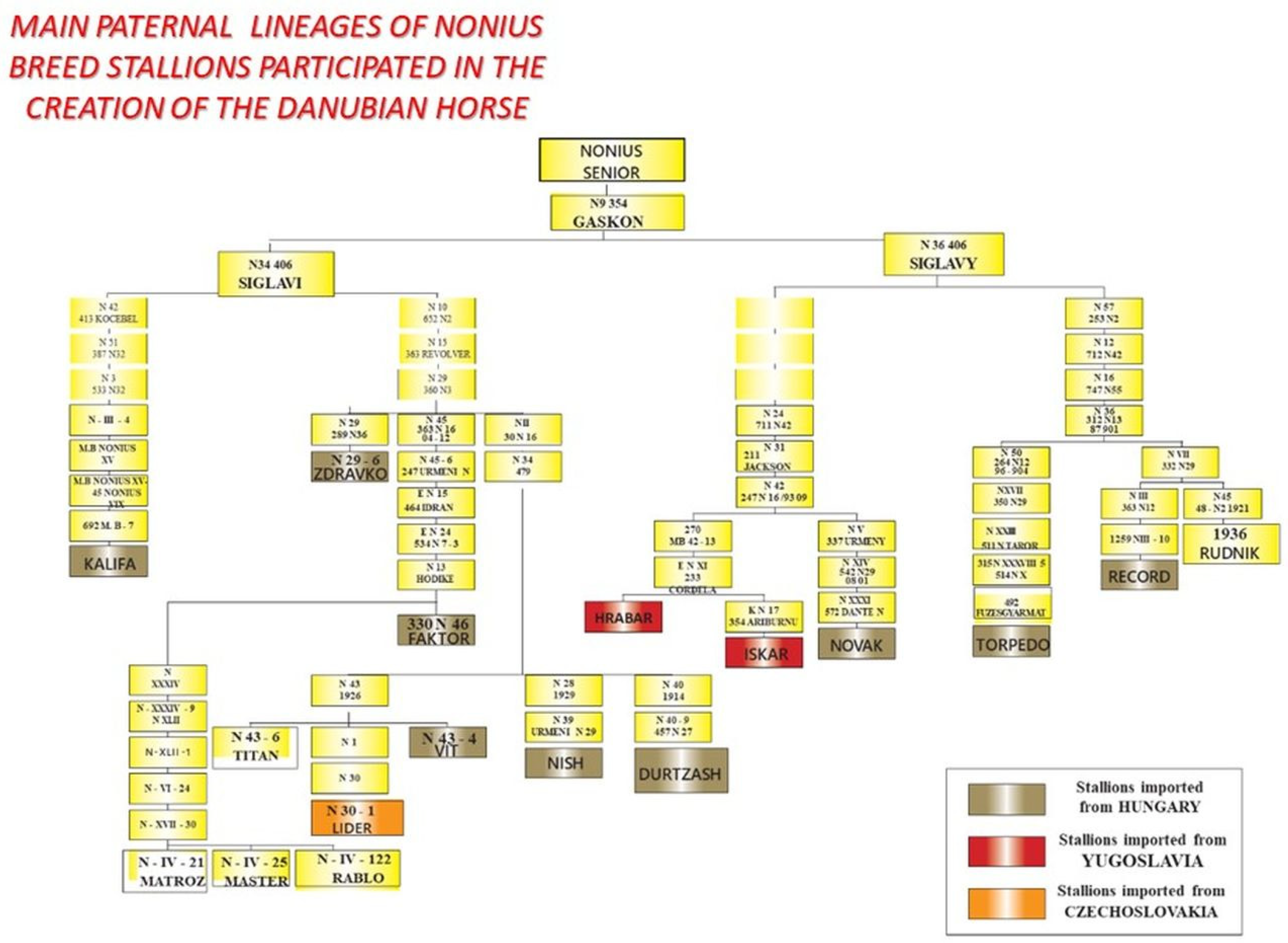
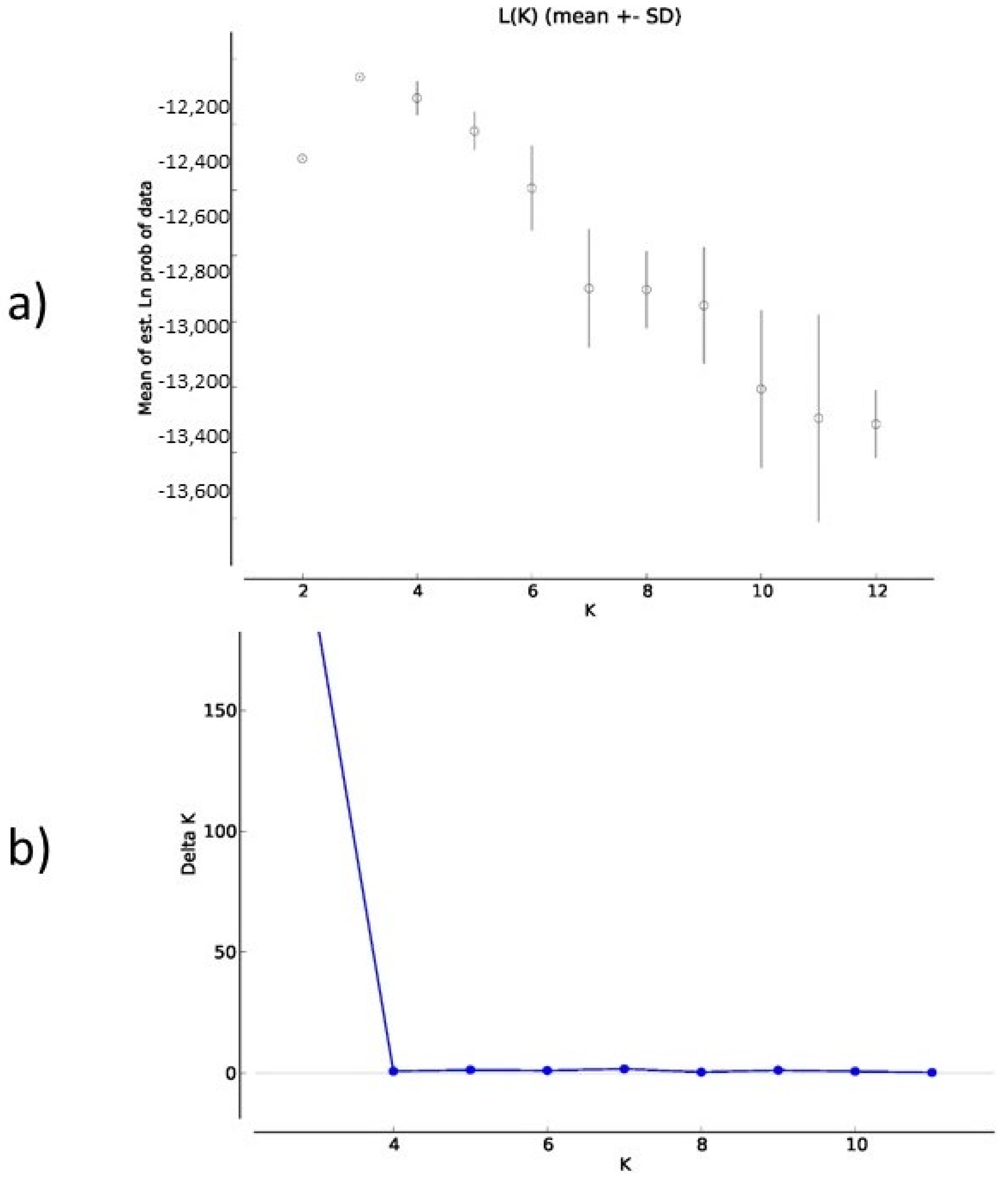
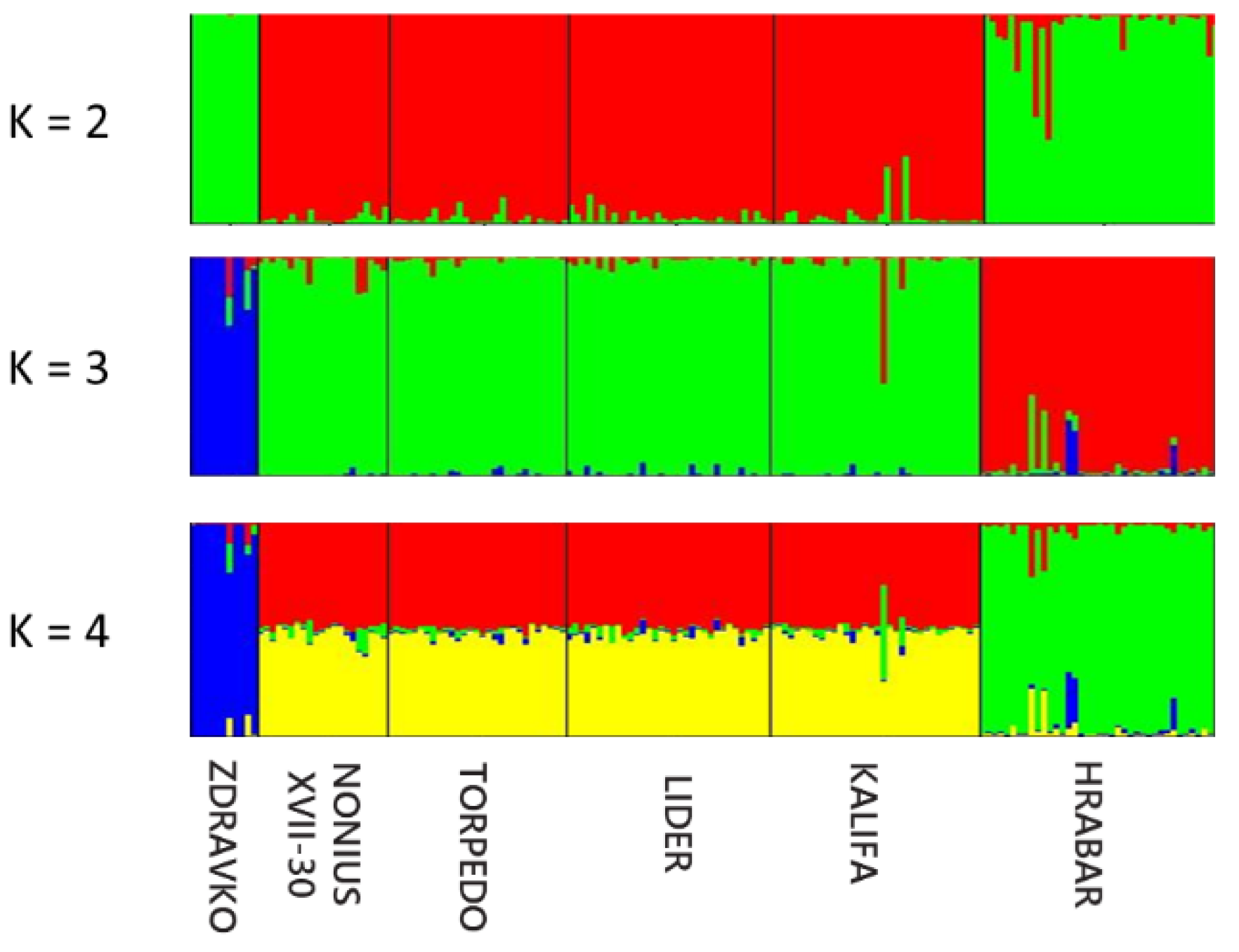

| Locus | Chrom. Location | Motif | Reference | Primer Sequences 5′–3′ | Annealing T (°C) | Amplicon Length (bp) |
|---|---|---|---|---|---|---|
| AHT4 | 24q14 | (AC)nAT(AC)n | Binns et al. [17] | F: AACCGCCTGAGCAAGGAAGT R: CCCAGAGAGTTTACCCT | 60 | 144–164 |
| AHT5 | 8 | (GT)n | Binns et al. [17] | F: ACGGACACATCCCTGCCTGC R: GCAGGCTAAGGAGGCTCAGC | 60 | 126–144 |
| ASB2 | 15q21.3-q23 | (GT)n | Bowling et al. [18] | F: CCACTAAGTGTCGTTTCAGAAGG R: CACAACTGAGTTCTCTGATAGG | 55 | 216–250 |
| ASB17 | 2p14-p15 | (AC)n | Bowling et al. [18] | F: ACCATTCAGGATCTCCACCG R: GAGGGCGGTACCTTTGTACC | 60 | 87–129 |
| ASB23 | 3q22 | (TG)n | Irvin et al. [19] | F: GCAAGGATGAAGAGGGCAGC R: CTGGTGGGTTAGATGAGAAGTC | 58 | |
| HMS1 | 15 | (TG)n | Guerin et al. [20] | F: CATCACTCTTCATGTCTGCTTGG R: TTGACATAAATGCTTATCCTATGGC | 58 | 170–186 |
| HMS2 | 10 | (CA)n(TC)2 | Guerin et al. [20] | F: CTTGCAGTCGAATGTGTATTAAATG R: ACGGTGGCAACTGCCAAGGAAG | 58 | 222–248 |
| HMS3 | 9 | (TG)2(CA)2TC(CA)n and (TG)2(CA)2TC(CA)nGA(CA)5 | Guerin et al. [20] | F: CCATCCTCACTTTTTCACTTTGTT R: CCAACTCTTTGTCACATAACAAGA | 60 | 148–170 |
| HMS6 | 4 | (GT)n | Guerin et al. [20] | F: GAAGCTGCCAGTATTCAACCATTG R: CTCCATCTTGTGAAGTGTAACTCA | 60 | 151–169 |
| HMS7 | 1q25 | (AC)2(CA)n | Guerin et al. [20] | F: TGTTGTTGAAACATACCTTGACTGT R: CAGGAAACTCATGTTGATACCATC | 60 | 165–185 |
| HTG4 | 9 | (TG)nAT(AG)5AAG (GA)5 ACAG(AGGG)3 | Ellegren et al. [21] | F: CTATCTCAGTCTTGATTGCAGGAC R: CTCCCTCCCTCCCTCTGTTCTC | 55 | 127–139 |
| HTG6 | 15q26-q27 | (TG)n | Ellegren et al. [21] | F: GTTCACTGAATGTCAAATTCTGCT R: CCTGCTTGGAGGCTGTGATAAGAT | 58 | 84–102 |
| HTG7 | 4 | (GT)n | Marklund et al. [22] | F: CCTGAAGCAGAACATCCCTCCTTG R: ATAAAGTGTCTGGGCAGAGCTGCT | 58 | 118–128 |
| HTG10 | 21 | (TG)n and TATC(TG)n | Marklund et al. [22] | F: TTTTTATTCTGATCTGTCACATTT R: CAATTCCCGCCCCACCCCCGGCA | 55 | 95–115 |
| VHL20 | 30 | (TG)n | Van Haeringen et al. [23] | F: CAAGTCCTCTTACTTGAAGACTAG R: AACTCAGGGAGAATCTTCCTCAG | 60 | 87–105 |
| Locus | Na | Ne | PIC | Ho | He | I | FIT a | FIS | FST a | DST | HT | GST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHT4 | 11.17 | 8.62 | 0.81 | 0.75 | 0.74 | 1.98 | 0.158 | 0.012 | 0.148 | 0.611 | 0.885 | 0.131 |
| AHT5 | 11.50 | 9.28 | 0.78 | 0.88 | 0.84 | 2.16 | 0.040 | −0.046 | 0.082 | 0.509 | 0.915 | 0.066 |
| ASB2 | 11.33 | 9.20 | 0.77 | 0.91 | 0.85 | 2.18 | −0.008 | −0.073 | 0.061 | 0.363 | 0.904 | 0.044 |
| ASB17 | 10.00 | 4.80 | 0.69 | 0.94 | 0.79 | 1.85 | −0.146 | −0.201 | 0.047 | 0.158 | 0.823 | 0.032 |
| ASB23 | 11.83 | 6.77 | 0.74 | 0.96 | 0.85 | 2.13 | −0.077 | −0.132 | 0.049 | 0.260 | 0.890 | 0.033 |
| HMS1 | 12.67 | 9.96 | 0.65 | 0.80 | 0.79 | 2.19 | 0.119 | −0.016 | 0.132 | 0.657 | 0.910 | 0.116 |
| HMS2 | 12.50 | 10.26 | 0.69 | 0.86 | 0.83 | 2.24 | 0.066 | −0.033 | 0.096 | 0.575 | 0.918 | 0.079 |
| HMS3 | 13.33 | 10.74 | 0.73 | 0.92 | 0.86 | 2.36 | 0.009 | −0.059 | 0.065 | 0.452 | 0.923 | 0.048 |
| HMS6 | 12.50 | 9.59 | 0.67 | 0.83 | 0.81 | 2.19 | 0.060 | −0.022 | 0.119 | 0.661 | 0.920 | 0.103 |
| HMS7 | 13.33 | 11.28 | 0.75 | 0.86 | 0.89 | 2.42 | 0.068 | 0.028 | 0.041 | 0.277 | 0.927 | 0.023 |
| HTG4 | 12.67 | 9.80 | 0.71 | 0.89 | 0.85 | 2.26 | 0.022 | −0.051 | 0.069 | 0.438 | 0.913 | 0.053 |
| HTG6 | 12.83 | 9.86 | 0.68 | 0.77 | 0.82 | 2.23 | 0.151 | 0.058 | 0.099 | 0.559 | 0.912 | 0.081 |
| HTG7 | 12.67 | 10.55 | 0.72 | 0.92 | 0.86 | 2.31 | 0.002 | −0.064 | 0.062 | 0.423 | 0.919 | 0.046 |
| HTG10 | 12.83 | 10.63 | 0.75 | 0.90 | 0.89 | 2.38 | 0.025 | −0.015 | 0.040 | 0.277 | 0.927 | 0.022 |
| VHL20 | 13.17 | 10.90 | 0.73 | 0.91 | 0.87 | 2.36 | 0.025 | −0.032 | 0.055 | 0.385 | 0.924 | 0.037 |
| Mean (SE) | 12.29 (0.44) | 9.48 (0.42) | 0.73 (0.15) | 0.87 (0.02) | 0.84 (0.02) | 2.22 (0.07) | 0.037 (0.021) | −0.043 (0.016) | 0.078 (0.009) | 0.459 (0.459) | 0.907 0.007 | 0.061 0.009 |
| Lineage | Locus | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHT4 | AHT5 | ASB2 | ASB17 | ASB23 | HMS1 | HMS2 | HMS3 | HMS6 | HMS7 | HTG4 | HTG6 | HTG7 | HTG10 | VHL20 | |
| Zdravko | 0.803 | 0.892 | 0.785 | 0.046 * | 0.075 | 0.991 | 0.214 | 0.977 | 0.909 | 0.695 | 0.509 | 0.062 | 0.370 | 0.222 | 0.386 |
| NONIUS XVII-30 | 0.148 | 0.932 | 0.141 | 0.987 | 0.064 | 0.028 * | 0.797 | 0.074 | 0.842 | 0.658 | 0.699 | 0.587 | 0.874 | 0.193 | 0.109 |
| Torpedo | 0.455 | 0.495 | 0.762 | 0.979 | 0.357 | 0.162 | 0.849 | 0.377 | 0.553 | 0.209 | 0.446 | 0.774 | 0.249 | 0.877 | 0.803 |
| Lider | 0.375 | 0.578 | 0.823 | 0.959 | 0.559 | 0.654 | 0.720 | 0.320 | 0.169 | 0.475 | 0.159 | 0.887 | 0.382 | 0.047 * | 0.429 |
| Kalifa | 0.561 | 0.915 | 0.629 | 0.713 | 0.996 | 0.002 ** | 0.796 | 0.305 | 0.645 | 0.374 | 0.467 | 0.011 * | 0.393 | 0.575 | 0.788 |
| Hrabar | 0.874 | 0.270 | 0.835 | 0.000 *** | 0.006 ** | 0.370 | 0.292 | 0.121 | 0.921 | 0.325 | 0.471 | 0.052 | 0.275 | 0.837 | 0.792 |
| Lineage | Na | Ne | I | Ho | He | NPA | No. Different Alleles (Freq ≥ 5%) |
|---|---|---|---|---|---|---|---|
| Zdravko | 4.20 | 2.73 | 1.07 | 0.65 | 0.57 | - | 3.06 |
| NONIUS XVII-30 | 13.93 | 10.72 | 2.45 | 0.91 | 0.89 | - | 7.47 |
| Torpedo | 14.33 | 11.19 | 2.50 | 0.94 | 0.90 | - | 10.73 |
| Lider | 14.60 | 11.57 | 2.54 | 0.92 | 0.91 | - | 9.13 |
| Kalifa | 14.60 | 11.40 | 2.53 | 0.93 | 0.91 | 1 | 9.13 |
| Hrabar | 12.07 | 9.29 | 2.19 | 0.88 | 0.83 | - | 8.733 |
| Mean | 27.67 | 12.29 | 2.22 | 2.22 | 0.87 | - | 8.04 |
| Zdravko | NONIUS XVII-30 | Torpedo | Lider | Kalifa | Hrabar | |
|---|---|---|---|---|---|---|
| Zdravko | 0.000 | 1.142 | 1.055 | 0.960 | 1.023 | 0.660 |
| NONIUS XVII-30 | 0.122 | 0.000 | 0.206 | 0.259 | 0.247 | 0.533 |
| Torpedo | 0.118 | 0.010 | 0.000 | 0.188 | 0.214 | 0.541 |
| Lider | 0.115 | 0.012 | 0.009 | 0.000 | 0.171 | 0.538 |
| Kalifa | 0.117 | 0.012 | 0.010 | 0.008 | 0.000 | 0.542 |
| Hrabar | 0.103 | 0.034 | 0.034 | 0.034 | 0.034 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yordanov, G.; Mehandjyiski, I.; Palova, N.; Atsenova, N.; Neov, B.; Radoslavov, G.; Hristov, P. Genetic Diversity and Structure of the Main Danubian Horse Paternal Genealogical Lineages Based on Microsatellite Genotyping. Vet. Sci. 2022, 9, 333. https://doi.org/10.3390/vetsci9070333
Yordanov G, Mehandjyiski I, Palova N, Atsenova N, Neov B, Radoslavov G, Hristov P. Genetic Diversity and Structure of the Main Danubian Horse Paternal Genealogical Lineages Based on Microsatellite Genotyping. Veterinary Sciences. 2022; 9(7):333. https://doi.org/10.3390/vetsci9070333
Chicago/Turabian StyleYordanov, Georgi, Ivan Mehandjyiski, Nadezhda Palova, Nedyalka Atsenova, Boyko Neov, Georgi Radoslavov, and Peter Hristov. 2022. "Genetic Diversity and Structure of the Main Danubian Horse Paternal Genealogical Lineages Based on Microsatellite Genotyping" Veterinary Sciences 9, no. 7: 333. https://doi.org/10.3390/vetsci9070333
APA StyleYordanov, G., Mehandjyiski, I., Palova, N., Atsenova, N., Neov, B., Radoslavov, G., & Hristov, P. (2022). Genetic Diversity and Structure of the Main Danubian Horse Paternal Genealogical Lineages Based on Microsatellite Genotyping. Veterinary Sciences, 9(7), 333. https://doi.org/10.3390/vetsci9070333






