Identification of Five Serotypes of Enteropathogenic Escherichia coli from Diarrheic Calves and Healthy Cattle in Belgium and Comparative Genomics with Shigatoxigenic E. coli
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Previous Studies
2.2. Escherichia coli Collections
2.3. Genomic O Serotyping
2.4. Whole Genome Sequencing
2.5. Phylogenetic Tree
3. Results
3.1. Genomic O Serotyping
3.2. Identification of O:H Serotypes, MLSTypes, and Eae Gene Subtypes by WGS
3.3. Identification of Virulotypes by WGS
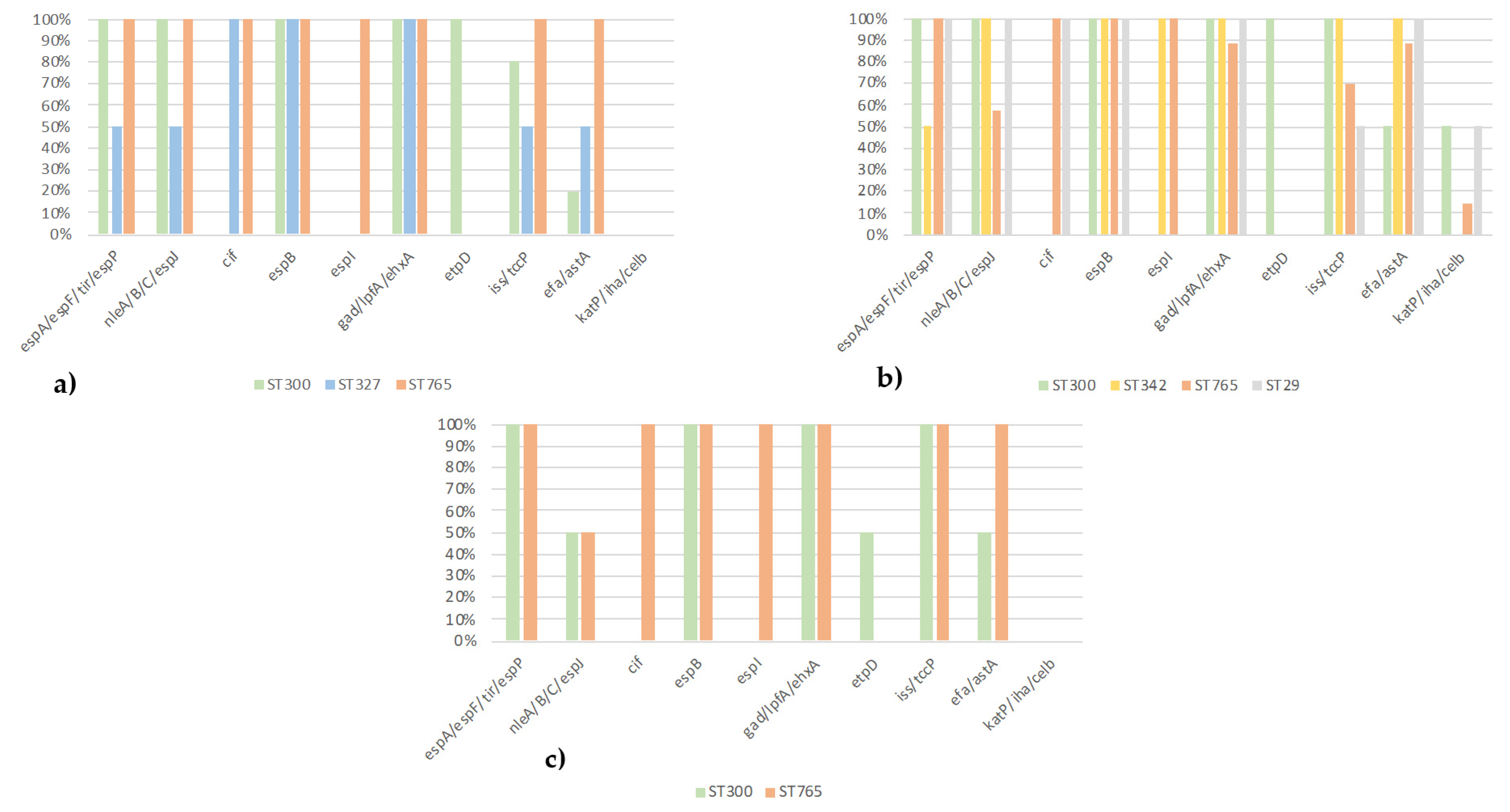
3.4. Comparison of Serotypes, ST, and Virulotypes of EPEC from Diarrheic Calves, Healthy Cattle, and Healthy Calves
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mainil, J.G.; Fairbrother, J.M. Pathogenic Escherichia coli in domestic mammals and birds. In Pathogenic Escherichia coli: Molecular and Cellular Microbiology; Morabito, S., Ed.; Horizon Scientific Press and Caister Academic Press: Norwich, UK, 2014; pp. 19–43. [Google Scholar]
- Tozzoli, R.; Scheutz, F. Diarrhoeagenic Escherichia coli infections in humans. In Pathogenic Escherichia coli: Molecular and Cellular Microbiology; Morabito, S., Ed.; Horizon Scientific Press and Caister Academic Press: Norwich, UK, 2014; pp. 1–18. [Google Scholar]
- Piérard, D.; De Greve, H.; Haesebrouck, F.; Mainil, J.G. O157:H7 and O104:H4 Vero/Shiga toxin-producing Escherichia coli: Respective role of cattle and humans. Vet. Res. 2012, 43, 13. [Google Scholar] [CrossRef] [PubMed]
- Fakih, I.; Thiry, D.; Duprez, J.N.; Saulmont, M.; Iguchi, A.; Piérard, D.; Jouant, L.; Daube, G.; Ogura, Y.; Hayashi, T.; et al. Identification of Shiga toxin-producing (STEC) and enteropathogenic (EPEC) Escherichia coli in diarrhoeic calves and comparative genomics of O5 bovine and human STEC. Vet. Microbiol. 2017, 202, 16–22. [Google Scholar] [CrossRef]
- Habets, A.; Crombé, F.; Nakamura, K.; Guérin, V.; De Rauw, K.; Piérard, D.; Saulmont, M.; Hayashi, T.; Mainil, J.G.; Thiry, D. Genetic characterization of Shigatoxigenic and enteropathogenic Escherichia coli O80:H2 from diarrheic and septicemic calves and relatedness to human Shigatoxigenic E. coli O80:H2. J. Appl. Microbiol. 2020, 130, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Thiry, D.; De Rauw, K.; Takaki, S.; Duprez, J.N.; Iguchi, A.; Piérard, D.; Korsak, N.; Mainil, J.G. Low prevalence of the “gang of seven” and absence of the O80:H2 serotypes among Shigatoxigenic and enteropathogenic Escherichia coli (STEC and EPEC) in intestinal contents of healthy cattle at two slaughterhouses in Belgium in 2014. J. Appl. Microbiol. 2018, 124, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Habets, A.; Engelen, F.; Duprez, J.N.; Devleesschauwer, B.; Heyndrickx, M.; De Zutter, L.; Thiry, D.; Cox, E.; Mainil, J.G. Identification of Shigatoxigenic and enteropathogenic Escherichia coli serotypes in healthy young dairy calves in Belgium by recto-anal mucosal swabbing. J. Vet. Sci. 2020, 7, 167. [Google Scholar] [CrossRef]
- Iguchi, A.; Iyoda, S.; Seto, K.; Morita-Ishihara, T.; Scheutz, F.; Ohnishi, M. The pathogenic E. coli working group in Japan. Escherichia coli O-genotyping PCR: A comprehensive and practical platform for molecular O serogrouping. J. Clin. Microbiol. 2015, 53, 2427–2432. [Google Scholar] [CrossRef] [PubMed]
- Mekata, H.; Iguchi, A.; Kawano, K.; Kirino, Y.; Kobayashi, I.; Misawa, N. Identification of O serotypes, genotypes, and virulotypes of Shiga toxin-producing Escherichia coli isolates, including non-O157 from beef cattle in Japan. J. Food Prot. 2014, 77, 1269–1274. [Google Scholar] [CrossRef]
- Engelen, F.; Thiry, D.; Devleesschauwer, B.; Heyndrickx, M.; Mainil, J.; De Zutter, L.; Cox, E. Pathogenic potential of Escherichia coli O157 and O26 isolated from young Belgian dairy calves by recto-anal mucosal swab culturing. J. Appl. Microbiol. 2021, 131, 964–972. [Google Scholar] [CrossRef]
- Engelen, F.; Thiry, D.; Devleesschauwer, B.; Mainil, J.; De Zutter, L.; Cox, E. Occurrence of ‘gang of five’ Shiga toxin-producing Escherichia coli serogroups on Belgian dairy cattle farms by overshoe sampling. Lett. Appl. Microbiol. 2021, 72, 415–419. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its application to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, M. Real-time whole genome sequencing for routine typing, surveillance and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- De Rauw, K.; Thiry, D.; Caljon, B.; Saulmont, M.; Mainil, J.G.; Piérard, D. Characteristics of Shigatoxin producing and enteropathogenic Escherichia coli of the emerging serotype O80:H2 isolated from humans and diarrhoeic calves in Belgium. Clin. Microbiol. Infect. 2018, 25, 111.e5–111.e8. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, L.; Jelsbak, L.; Sicheritz-pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 52, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.9. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Lukjancenko, O.; Wassenaar, T.M.; Ussery, D.W. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 2010, 60, 708–720. [Google Scholar] [CrossRef] [PubMed]
- De Rauw, K.; Jacobs, S.; Piérard, D. Twenty-seven years of screening for Shiga toxin-producing Escherichia coli in a university hospital. Brussels, Belgium, 1987–2014. PLoS ONE 2018, 13, e0199968. [Google Scholar] [CrossRef]
- Gilmour, M.W.; Tabor, H.; Wang, G.; Clark, C.G.; Tracz, D.M.; Olson, A.B.; Mascarenhas, M.; Karmali, M.A.; Mailman, T.; Ng, L. Isolation and genetic characterization of a coinfection of non-O157 Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 2007, 45, 3771–3773. [Google Scholar] [CrossRef]
- Sheng, H.; Duan, M.; Hunter, S.S.; Minnich, S.A.; Settles, M.L.; New, D.D.; Chase, J.R.; Fagnan, M.W.; Hovde, J. High-quality complete genome sequences of three bovine Shiga toxin-producing Escherichia coli O177:H- (fliCH25) isolates harboring virulent stx2 and multiple plasmids. Genome. Announc. 2018, 6, e01592-17. [Google Scholar] [CrossRef] [Green Version]
- Bielaszewska, M.; Middendorf, B.; Ko, R.; Friedrich, A.W.; Fruth, A.; Karch, H.; Schmidt, M.A.; Mellmann, A. Shigatoxin-negative attaching and affacing Escherichia coli: Distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 2008, 47, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.A.; Menge, C.; Eichhorn, I.; Semmler, T.; Wieler, L.H.; Pickard, D.; Belka, A. The accessory genome of Shiga toxin-producing Escherichia coli defines a persistent colonization type in cattle. Appl. Environ. Microbiol. 2016, 82, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Chen, X.; Zheng, S.; Han, D.; Wang, Y.; Wang, R.; Wang, B.; Chen, Y. Prevalence and genetic diversity of human diarrheagic Escherichia coli isolates by multilocus sequence typing. Int. J. Infect. Dis. 2018, 67, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Bibbal, D.; Loukiadis, E.; Kérourédan, M.; Peytavin de Garam, C.; Ferré, F.; Cartier, P.; Gay, M.; Oswald, E.; Auvray, F.; Brugère, H. Intimin gene (eae) subtype-based real-time PCR strategy for specific detection of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 in cattle feces. Appl. Environ. Microbiol. 2014, 80, 1177–1184. [Google Scholar] [CrossRef]
- Matussek, A.; Jernber, C.; Einemo, I.M.; Monecke, S.; Ehricht, R.; Engelmann, I.; Löfgren, S.; Mernelius, S. Genetic makeup of Shiga toxin-producing Escherichia coli in relation to clinical symptoms and duration of shedding: A microarray analysis of isolates from Swedish children. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1433–1441. [Google Scholar] [CrossRef]
- Peigne, C.; Bidet, P.; Mahjoub-Messai, F.; Plainvert, C.; Barbe, V.; Médigue, C.; Frapy, E.; Nassif, X.; Denamur, E.; Bingen, E.; et al. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 2009, 77, 2272–2284. [Google Scholar] [CrossRef]
- Leung, P.H.; Peiris, J.S.; Ng, W.W.; Robins-Browne, R.M.; Bettelheim, K.A.; Yam, W.C. A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 7549–7553. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Vo, T.T.; Vu-Khac, H. Virulence factors in Escherichia coli isolated from calves with diarrhea in Vietnam. J. Vet. Sci. 2011, 12, 159–164. [Google Scholar] [CrossRef]
- Dong, H.-J.; Lee, S.; Kim, W.; An, J.-U.; Kim, J.; Kim, D.; Cho, S. Prevalence, virulence potential, and pulsed-field gel electrophoresis profiling of Shiga toxin-producing Escherichia coli strains from cattle. Gut. Pathog. 2017, 9, 22. [Google Scholar] [CrossRef]
- Okuno, K.; Awasthi, S.P.; Kopprio, G.A.; Iguchi, A.; Hatanaka, N.; Hinenoya, A.; Lara, R.J.; Yamasaki, S. Prevalence, O-genotype and Shiga toxin (Stx) 2 subtype of Stx-producing Escherichia coli strains isolated from Argentinean beef cattle. J. Vet. Med. Sci. 2021, 83, 630–636. [Google Scholar] [CrossRef]
- Navarro-Garcia, F. Escherichia coli O104:H4 Pathogenesis: An enteroaggregative E. coli/Shiga Toxin- Producing E. coli explosive cocktail of high virulence. Microbiol Spectr. 2014, 6, 505–532. [Google Scholar] [CrossRef] [PubMed]
- Ingelbeen, B.; Bruyand, M.; Mariani-Kurkjian, P.; Le Hello, S.; Danis, K.; Sommen, C.; Bonacorsi, S.; de Valk, H. Emerging Shiga-toxin-producing Escherichia coli serogroup O80 associated hemolytic and uremic syndrome in France, 2013–2016: Differences with other serogroups. PLoS ONE 2018, 12, e0207492. [Google Scholar] [CrossRef] [PubMed]
 healthy cattle,
healthy cattle,  diarrheic calves,
diarrheic calves,  dairy calves, and
dairy calves, and  humans whose sequences of 3424 were compared to other strains based on [4,19]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura–Nei model and 1000 bootstrap replicates [18]. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
humans whose sequences of 3424 were compared to other strains based on [4,19]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura–Nei model and 1000 bootstrap replicates [18]. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
 healthy cattle,
healthy cattle,  diarrheic calves,
diarrheic calves,  dairy calves, and
dairy calves, and  humans whose sequences of 3424 were compared to other strains based on [4,19]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura–Nei model and 1000 bootstrap replicates [18]. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
humans whose sequences of 3424 were compared to other strains based on [4,19]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura–Nei model and 1000 bootstrap replicates [18]. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
| Gene | Primer | Sequence 5′-3′ | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| O123/186 | O123/186-F | TTTCAACAGGTTCGAATGCC | 362 | [8] |
| O123/186-R | CCCACCAATACCACTGGAATA | |||
| O156 | O156-F | GGAAAATGGAACATTTAGCGG | 236 | [8] |
| O156-R | TCGGAGTGCCAACCAAAATA | |||
| O177 | O177-F | CCGATACACCGGATGGATTAT | 427 | [8] |
| O177-R | AAGCCAGTACCCAGAACAGGA | |||
| O182 | O182-F | CGGTGATGGTTCTATTCTTGG | 502 | [8] |
| O182-R | TGCTTGCACCAACTGTGTTA | |||
| O183 | O183-F | CGTGGTAACCAATTTCGCAA | 666 | [8] |
| O183-R | GGGAATAACGAACGGTTTACA |
| O:H Serotype (Nr. Isolates) | MLST (Nr. Isolates) | Host (Nr. Isolates) | Virulotype (Nr. Isolates) | LEE-Located T3SS- and T3SE-Encoding Genes: Nr. +Ve Isolates | Non-LEE-Encoded T3SE-Encoding Genes: Nr. +Ve Isolates | Other Virulence Genes: Nr. +Ve Isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| espA/espF/tir/espP | espB | nleA/B/C/espJ | cif | espI | gad/lpfA/ehxA | etpD | iss/tccP | efa1/astA | katP/iha/celB | ||||
| O156:H8 (3) | ST327 (3) | HC (3) | eaeθ (3) b | 0 to 3 | 3° | 0 to 3 | 3 | 0 | 3 | 0 | 0 to 3 | 0–1 | 0 |
| O156:H25 (12) | ST300 (11) | HC (11) | eaeζ (10) c,d | 10 | 10° | 10 | 0 | 0 | 10 | 10 | 6 | 0 | 0 |
| eaeζ, stx2g (1) c | 1 | 1° | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | |||
| ST4942 (1) | HU (1) | eaeζ, stx1a (1) | 1 | 1° | 1 | 0 | 0 | 1 | 1 | 1 | 0–1 | 0 | |
| O177:H11 (17) | ST765 (15) | DC (8) | eaeβ (8) | 8 | 8 | 2 to 8 | 8 | 8 | 7 | 0 | 4 to 7 | 7 | 0–1 |
| HC (1) | eaeβ (1) | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| HCS (6) | eaeβ, stx1a (1) | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| eaeβ, stx2c (1), | 1 | 0 | 1 | 1 | 0 | ||||||||
| stx2c (1) | 1 | 1 | 1 | 1 | 1 | 4 | 0 | 4 | 4 | 0 | |||
| ST29 (2) | DC (1) | eaeβ (4) e | 4 | 4 | 2 to 4 | 4 | 4 | 1 | 0 | 0–1 | 1 | 0–1 | |
| HU (1) | eaeβ (1) | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0–1 | ||
| eaeβ, stx1a (1) | 1 | 1 | 1 | 0 | 0 | ||||||||
| O177:H25 (2) | ST342 (2) | DC (2) | eaeβ (1) | 0–1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| eaeβ, stx2c (1) | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |||
| 0O182:H25 (10) | ST300 (10) | DC (4) | eaeζ (2) | 2 | 2° | 2 | 0 | 0 | 2 | 2 | 2 | 1 | 0 |
| eaeζ, stx1a (2) | 2 | 2° | 2 | 0 | 0 | 2 | 2 | 2 | 0–2 | 0–1 | |||
| HC (2) | eaeζ (2) | 2 | 2° | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | ||
| HCS (2) | eaeζ (2) f | 2 | 2° | 0–2 | 0 | 0 | 2 | 1–2 | 2 | 1–2 | 0 | ||
| HU (2) | eaeζ, stx1a (2) | 2 | 2° | 1–2 | 0 | 0 | 2 | 2 | 0–2 | 0 | 0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habets, A.; Touzain, F.; Lucas, P.; Huong, N.T.T.; Iguchi, A.; Crombé, F.; Korsak, N.; Piérard, D.; Saulmont, M.; Cox, E.; et al. Identification of Five Serotypes of Enteropathogenic Escherichia coli from Diarrheic Calves and Healthy Cattle in Belgium and Comparative Genomics with Shigatoxigenic E. coli. Vet. Sci. 2022, 9, 492. https://doi.org/10.3390/vetsci9090492
Habets A, Touzain F, Lucas P, Huong NTT, Iguchi A, Crombé F, Korsak N, Piérard D, Saulmont M, Cox E, et al. Identification of Five Serotypes of Enteropathogenic Escherichia coli from Diarrheic Calves and Healthy Cattle in Belgium and Comparative Genomics with Shigatoxigenic E. coli. Veterinary Sciences. 2022; 9(9):492. https://doi.org/10.3390/vetsci9090492
Chicago/Turabian StyleHabets, Audrey, Fabrice Touzain, Pierrick Lucas, Nguyen Thi Thu Huong, Atsushi Iguchi, Florence Crombé, Nicolas Korsak, Denis Piérard, Marc Saulmont, Eric Cox, and et al. 2022. "Identification of Five Serotypes of Enteropathogenic Escherichia coli from Diarrheic Calves and Healthy Cattle in Belgium and Comparative Genomics with Shigatoxigenic E. coli" Veterinary Sciences 9, no. 9: 492. https://doi.org/10.3390/vetsci9090492





