Abstract
As environmental and health concerns increase, the trend toward sustainable agriculture is moving toward using biological agents. About 60% of all biological fungicides have Trichoderma species as the active ingredient, with T. harzianum as the most common species in these products. However, the name T. harzianum has often been used incorrectly in culture collections, databases, and scientific literature due to the division of the Harzianum clade (HC) into more than 95 cryptic species, with only one being named T. harzianum. In this study, 49 strains previously identified as T. harzianum in three surveys of Trichoderma species from soils in South and Central America were re-identified using phylogenetic analyses based on tef1α, rpb2, and ITS sequences obtained from GenBank. These were combined with the HC species from two other studies, which were identified based on the current taxonomy. Based on the results of the five surveys of the total 148 strains in HC, 11 species were identified. T. afroharzianum, T. lentiforme, and T. endophyticum, followed by T. azevedoi and T. harzianum, were the dominant species of the HC in South and Central America. This is the first report to identify dominant Trichoderma species within the HC in South and Central American soil based on multiple studies. These results will be useful in selecting strains within the clade for the formulation of biocontrol and biofertilizer products on the continent.
1. Introduction
A significant number of investigations have documented the use of beneficial microbes for disease suppression and plant growth enhancement. Species in the genus Trichoderma stand out for these plant-beneficial activities. Trichoderma as a genus was introduced in 1794 by Persoon []. The importance of Trichoderma in agriculture, specifically as a biocontrol agent (BCA) against fungal plant diseases, has been known since the 1930s []. Then, in the 1980s, studies showed growth promotion of various crops by application of Trichoderma species [,]. More recently, Trichoderma is being employed also in environmental remediation processes [,].
However, only in the 1990s did commercial products with Trichoderma as an active ingredient become commercially available with reasonable success []. One of the most common species in those products is Trichoderma harzianum. Samuels and Hebbar [] assembled a list of commercial Trichoderma biocontrol products that have T. harzianum as the active ingredient in 21 out of 55 products, which was higher than any other Trichoderma species, reflecting the importance of the species for biocontrol. Also, in a compiled list of publications, Zin and Badaluddin [] showed investigations involving the effectiveness of Trichoderma species against fungal crop pathogens. Within the list, T. harzianum was the most studied species (11 out of 18) and showed high effectiveness against various crop diseases. Taxonomically, T. harzianum was only one of the nine aggregate species described by Rifai []. Aggregate species, per Rifai, means a group of more than one species that are morphologically identical but biologically different. Taxonomy based on DNA sequencing of specific markers started in the late 1990s, which resulted in an exponential expansion of the number of species in the genus Trichoderma. The species, morphologically identified as T. harzianum, appeared to split into different clades. Those clades, in some cases, were marked by Roman numerals or Arabic numbers [,], without any coordination in numbering. These studies clearly showed that T. harzianum could represent several species that are morphologically indistinguishable. Therefore, the concept Harzianum clade (HC) started to replace T. harzianum. In 2015, Chaverri et al. [] accepted 14 species within the HC, including a few that were already described. The number of species in the HC continued to expand [,,,,,]. Unfortunately, the split of HC did not resolve the confusion about the name completely. There are many sequences for strains deposited in databases including, GenBank as T. harzianum, even though T. harzianum is only one of the uncommon species among more than 95 described species within the clade, limiting the full value of the databases. There is another problem with the HC species. HC boundaries are not clearly identified, and mistakes happen when other species are included in the HC even though they are phylogenetically positioned outside the clade. For example, Chaverri et al. [] did not include T. tawa, T. tomentosum, and T. velutinum within the HC. However, Zheng et al. [] included all three species within the clade.
There are several surveys exploring Trichoderma in the soil in different parts of the world that have reported species in HC as T. harzianum. In this study, strains identified as T. harzianum in three survey studies for the isolation of Trichoderma strains from the soil of South and Central America were re-analyzed phylogenetically based on the available sequencing data of three loci: translation elongation factor 1α (tef1α), RNA polymerase subunit II (rpb2), and the internal transcribed spacers (ITS) obtained from GenBank. These three loci have been recommended for the identification of Trichoderma species []. After re-identification, the number of strains for each species was added to the numbers of respective species in another two studies where species in the HC were identified according to the current taxonomy. This was carried out to determine the dominant soil species of the HC on the continent. Knowing the exact dominant Trichoderma species in the soil from a given geographical region could help biocontrol investigations. It would also facilitate the identification of indigenous species that compete well in the soil and may have the ability to establish endophytic relationships with plants, resulting in better exploitation of plant beneficial activities by the Trichoderma species.
2. Materials and Methods
2.1. Evaluating the Accuracy of Trichoderma harzianum Strains Deposited in GenBank
To evaluate the accuracy of the identification of T. harzianum strains deposited at the National Center for Biotechnology Information (NCBI) GenBank (https://www.ncbi.nlm.nih.gov/genbank/), a search was carried out on 10 June 2024 for “Trichoderma harzianum tef1α” in the GenBank. The initial 100 sequence hits were downloaded in the FASTA file format. Sequences of sixteen ex-type strains in HC retrieved from GenBank were then added to this file as references. The FASTA file was aligned using Clustal Omega, version 1.2.4 (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 10 June 2024) and adjusted manually using the software Mesquite version 3.81 []. Then after, the file was used to construct phylogenetic trees using two methods: (1) A parsimony tree was obtained using PAUP version 4.0a (https://phylosolutions.com/paup-test/, accessed on 10 June 2024). The tree was produced using a heuristic search with a starting tree obtained by 1000 random stepwise additions of sequences, tree-bisection-reconnection (TBR) as the branch-swapping algorithm with MULTREES in effect. Gaps were treated as missing characters. Supports for branches were assessed with 1000 replicates of bootstrap. (2) A maximum likelihood tree was obtained using MEGA X, version 11.0.10 with the substitution model predetermined using MEGA X []. Support for the clades was assessed with 1000 bootstrap replicates.
2.2. Evaluation of the Dominant Species of the Harzianum Clade from South and Central America
To determine the dominant species of the HC in soils in South and Central America, strains identified based on tef1α in the studies of Hoyos-Carvajal et al., Smith et al., and Druzhinina et al. [,,] as T. harzianum (49 strains) were re-analyzed phylogenetically based on the DNA sequencing data of three loci, tef1α, rpb2, and ITS. The sequences for each locus were downloaded from the GenBank and aligned with reference sequences, particularly for the ex-type specimens of known species in the clade described in Chaverri et al. [] and del Carmen et al. [] using the Clustal Omega, version 1.2.4 (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 10 June 2024). All the strains used in the phylogenetic analysis are listed in Table 1. The alignment files for the three genes were concatenated and adjusted visually using the software Mesquite version 3.81 []. The alignment file was used to construct phylogenetic trees as described above. The trees obtained by both methods were essentially identical in topology, and thus only the parsimony tree constructed by PAUP is presented.

Table 1.
Trichoderma species with their origin, strain number, GenBank accession number, and the number of strains with identical tef1α GenBank accession numbers included in Figure 1.
2.3. Tabulation of HC Species from Soil in South and Central America
After re-identification of the strains of HC from the three studies, as detailed in Section 2.2, the number of strains of each species was added to the number of corresponding species from two other studies by Inglis et al. and Barrera et al. [,], which had identified nearly 100 isolates in the HC according to the correct taxonomy. The results were tabulated to determine the most prevalent species within the HC in soils of South and Central America, based on the total 148 strains of HC.
3. Results
3.1. Re-Identification of the Trichoderma harzianum Strains Deposited in GenBank
Initially, we searched GenBank for “Trichoderma harzianum tef1α” and phylogenetically analyzed the first 100 sequences. Based on the phylogenetic tree (Figure 1), only 22 out of 100 sequences clustered in fully supported clade (C1) with the ex-type strain of T. harzianum (CBS 226.95, AF348101); therefore, these strains are being identified as T. harzianum. Among the remaining 78 strains, many of them were recognized as other species in the HC as they fit into clades C2–C7 which include the type strains of the following species: T. rifaii, T. xixiacum, T. guizhouense, T. afroharzianum, T. atrobrunneum, and T. rugulosum, respectively. There were other strains that match into the HC but could not be identified as any known Trichoderma species of that clade. Two strains with accession numbers OQ200374 and OQ200375 did not belong to the HC and were identified as T. virens.
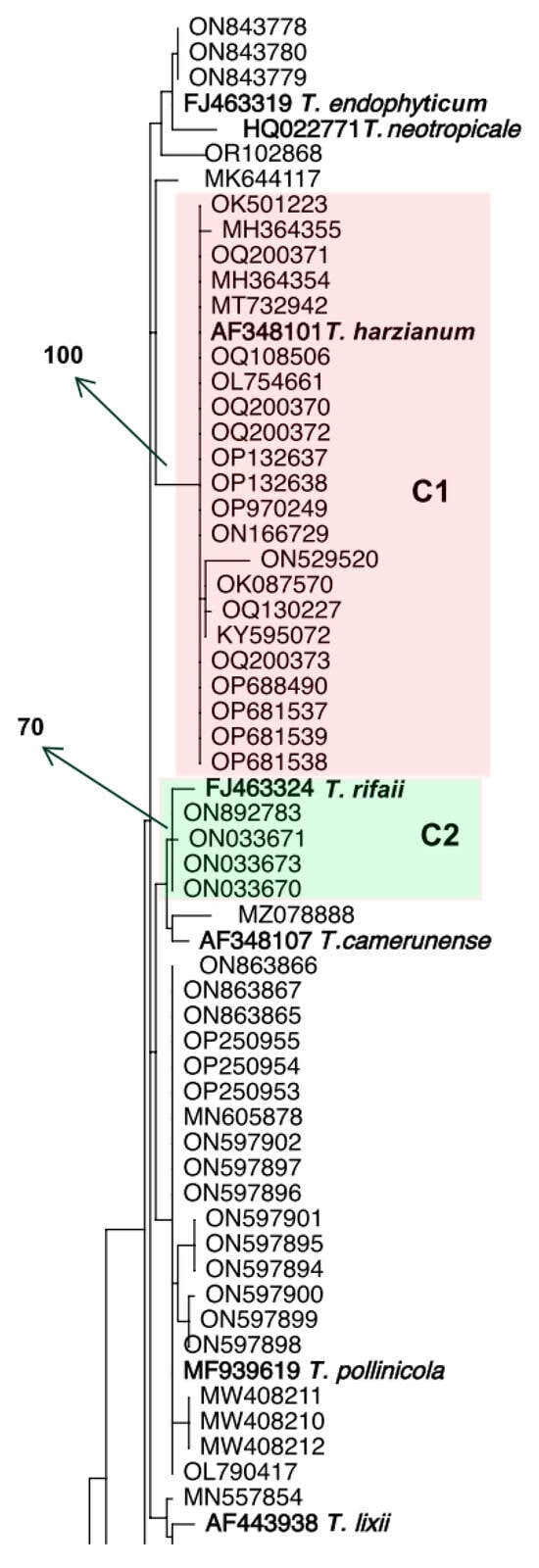
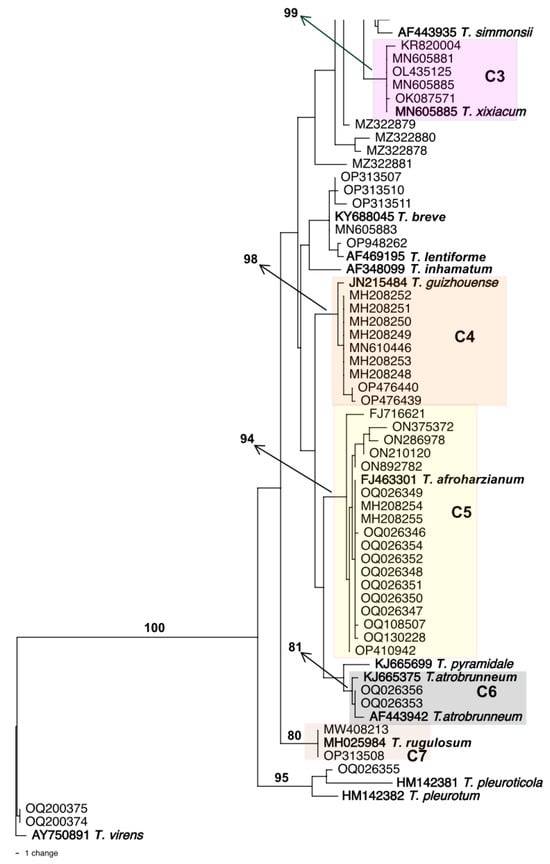
Figure 1.
Phylogenetic tree based on tef1α sequence data for 100 strains retrieved from GenBank deposited as Trichoderma harzianum. The leaves are identified by GenBank accession numbers. Numbers given above the branches indicate bootstrap values of 70% obtained via 1000 replications. The boldface indicates type strains included to identify the clades. Colored clades C1–C7 indicate strains clustered with a type strain of an HC species with a bootstrap value of 70%. The tree was rooted to an ex-type specimen of T. virens.
3.2. Evaluation of the Dominant Trichoderma Species of the Harzianum Clade from South and Central America
In the three survey studies by Hoyos-Carvajal et al., Smith et al., and Druzhinina et al. [,,] for isolation of Trichoderma species in the soil of Central and South America, all the strains in the HC were reported as T. harzianum, which does not reflect the split of the clade into more than 95 species. Phylogenetic re-analyses of sequencing data (tef1α, rpb2, and ITS) of the 49 strains (Figure 2) showed that 25 strains clustered with the type strain of T. lentiforme in clade C1 with a high bootstrap value of 79 and are recognized here as belonging to that species.
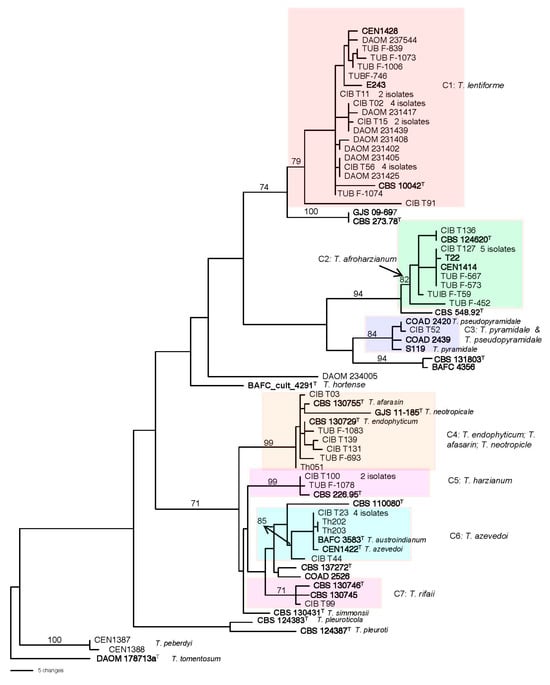
Figure 2.
One of the most parsimonious trees generated by phylogenetic analysis of combined DNA sequences of tef1α, rpb2, and ITS. Numbers given above the branches indicate bootstrap values of 70% obtained via 1000 replications. Leaves are identified by strain numbers. Boldface indicates reference strains included to identify the clades. T After strain numbers indicate the ex-type strain. Clades (C1–C7) refer to lineages that have strains identified in this study. Tree was rooted to the type strain of T. tomentosum.
In clade C2, 10 strains from the three studies formed a highly supported clade with the ex-type strain of T. afroharzianum and two other reference sequences of strains previously identified as T. afroharzianum supporting the identification of these strains as T. afroharzianum.
In clade C3, one isolate from Peru, CIB T52 clustered with three reference strains identified as T. pyramidale and T. pseudopyramidale. However, the CIB T52 is closer to T. pseudopyramidale than to T. pyramidale. These two species are closely related, but T. pseudopyramidale has been found in Africa and the other in South America [,].
The clade C4 included six strains together with the type strains of T. endophyticum, T. neotropicale, and T. afarasin. The type strain of T. neotropicle seems to be distantly related to the clade through a long branch, and T. afarasin is known to be an African species not found outside that continent. Therefore, it is most likely that the six isolates in the clade were T. endophytcum.
In clade C5, two strains CIB T100 and TUB-F1078 from two different studies clustered with the type strain of T. harzianum (CBS 226.95) with a high bootstrap value (BS = 99) and were recognized as T. harzianum.
In clade C6, seven strains nested with the ex-type strain of T. azevedoi (CEN1422); in most cases the sequences were identical. In the study of Barrera et al. [], five strains from Argentina were identified as T. austroindianum. Four of the five strains have identical tef1α sequences. The other strain has 1 gap difference with the other four isolates. The type strain of T. austroindianum had sequences highly homologous to those of T. azevedoi strains and fell in clade 6 with T. azevedoi, and thus, we believe that the two species represent only one species. Accordingly, the strains of T. austroindianum were tabulated with the T. azevedoi as, on a priority basis, the latter species was described before T. austroindianum [].
In clade C7, one isolate from Colombia, CIB T99, formed a clade with the type strain of T. rifaii and another reference strain of T. rifaii with a BS value of 71, suggesting the correct identification of the strain as T. rifaii.
The strain DAOM 234005 did not fit into any clade representatives of the known species of the Harzianum clade. Moreover, based on BLAST search, the tef1α sequence of this strain had no close homologous sequence to it in the GenBank; the nearest sequence to it was accession number KU238051 with a genetic distance of 0.0578 as determined by PAUP. This accession number belongs to a strain (TS187) from Malaysia deposited in GenBank as Trichoderma sp. []. Thus, we consider this strain as a possible new species in the HC.
A previous study [] described 12 isolates from Brazil as T. peberdyi belonging to HC. However, the phylogenetic tree (Figure 2) showed that T. peberdyi is positioned between three species: T. tomentosum (outgroup), T. pleuroti, and T. pleuroticola, all of which were considered outside of HC [].
The results of our re-identification of species of HC combined with the previously identified species in two other studies [,] are presented in Table 2 and summarized in Figure 3. Overall, strains of 11 species of HC were isolated in soils of South and Central America. T. afroharzianum was the most common species in the region, with 44 strains found in four out of five studies. This is a well-known biocontrol species, and the strain T22 is an example []. T. lentiforme was the second most common species, with 39 strains from four out of five studies. The strains of both species were distributed in all the regions from north to the south of the continent.

Table 2.
Trichoderma species in the Harzianum clade in soils of South and Central America based on data of five survey studies.
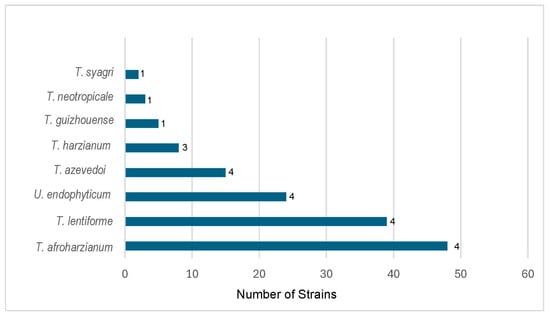
Figure 3.
Dominant species of Trichoderma in the Harazianum clade in the soil in Central and South America. The numbers at the end of the bars represent the frequency of presence of a species in five studies.
4. Discussion
Biocontrol programs are in place in South and Central America, notably in nations like Brazil, Argentina, Colombia, and Mexico, to manage plant diseases and encourage crop development with biological agents []. One of the most important fungal names in the field of biocontrol products for plant diseases and/or plant health promotion are species in the genus Trichoderma and, in particular, T. harzianum [,,]. However, confusion exists about the name T. harzianum being used for all the species within the Harzianum clade, despite the revision of the phylogeny of the clade and the description of more than 95 species that started in 2015 []. In fact, the problem is widespread and persistent in scientific publications, databases, and commercial products. As an example, after searching the GenBank for “Trichoderma harzianum tef1α” and identifying the first 100 sequences, this study revealed that only 22 clustered with the ex-type of the T. harzianum strain (GenBank accession number AF348101) and could be identified as T. harzianum, indicating 78% misidentification in the name of strains deposited as T. harzianum. What is disturbing about this is the fact that 100% of the sequences used in Figure 1 were deposited between 2018 and 2023, at least three years after the major revision of the HC []. Precise naming of species of HC is critical as beneficial properties of biocontrol and plant growth promotion are species-specific or even strain-specific. Due to improper usage of the name, there is a lack of studies exploring which species in the clade are dominant species in soil, despite the HC species being commonly isolated in many survey investigations. Dominance in the soil is an important criterion for selecting any biocontrol strain, as it reflects the fungus’s high ability to compete, outgrow, and suppress other species, leading to a higher potential for the fungus to establish endophytic relationships with plants and induce systemic resistance to pathogens. Through inducing systemic resistance, the Trichoderma species provides benefits other than disease resistance, such as growth promotion, resistance to abiotic stress, and high efficiency in using nitrogen [].
In this study, we re-analyzed the data from three survey studies published in 2005–2013 using multi-locus phylogeny. The number of re-identified species (49) was added to their respective species from the two other studies where HC species were identified, taking into account the split of the clade [,,,,]. T. afroharzianum and T. lentiforme, respectively, were the top two dominant species on the continent and were obtained from north to south. T. afroharzianum is cosmopolitan and a well-known species of biocontrol agents, and the T. afroharzianum strain T22 is well known as a biocontrol and biofertilizer strain []. T. lentiforme was reported to be mainly an endophytic fungus []. The dominance of T. lentiforme in soil is a new report of our findings. Having this ability in soil and the ability to establish endophytic relations with plants are top criteria for selection with biocontrol and biofertilizer properties. Recently, T. lentiforme was found to have biocontrol activity against Sclerotinia sclerotiorum and had growth stimulant properties in cotton [].
T. endophyticum and T. azevedoi are the third and fourth most common species, respectively, on the continent. T. endophyticum was shown to be exclusively endophytic based on the strains available in a previous study []. However, this study showed that this species is a soil fungus as well (Table 2). T. azevedoi was described in the study of Inglis et al. []. However, the strains are not geographically restricted to the continent of South America, as strains of this species have also been found in Australia, e.g., strain number BRIP 74284 with tef1α GenBank accession number of OR802290. In this context, we corrected the name of a species described by Barrera et al. [] as T. austroindianum. Five strains of T. austroindianum appear to have identical or highly homologous tef1α sequences to those of T. azevedoi, and both clustered in one highly supported clade. Thus, we renamed all five strains of T. austroindianum as T. azevedoi in Table 1. Here we stress the importance of (1) BLAST search of the sequences of tef1α or rpb2 loci of unknown Trichoderma strains before describing them as new species to avoid duplication of species naming and (2) to include the most homologous species to it in the phylogenic analyses. The phylogenetic trees in Barrera et al. [] did not include T. azevedoi in the analyses, which is probably the reason that the authors overlooked this error.
The boundaries of the species within the HC are equally unclear. For example, Chaverri et al. [] did not include T. tawa, T. tomentosum, and T. velutinum within the clade of Harzianum. However, Zheng et al. [] included all the above three species within the clade. In another example, Chaverri et al. [] excluded T. amazonicum and T. pleuroticola from HC. Yet Chen and Zhuang [] placed both species inside the HC. In this regard, Inglis et al. [] described a new species named T. peberdyi as part of the HC. Based on BLAST search at NCBI GenBank and our phylogenetic tree (Figure 2), this species is closely related to T. tomentosum. Based on Chaverri et al. [], T. tomentosum is not part of the clade. Therefore, T. perbedyi was not considered part of the HC clade and was excluded from our list of HC species (Table 2, Figure 3). Setting a genetic distance limit within HC species could be a way to solve the boundary issue.
We also attempted to compare the dominant species in Central and South America with those of other continents. However, there is currently insufficient data that quantitatively show the number of HC strains within the total number of isolated Trichoderma strains. Thus, comparing South and Central America with other continents was not feasible. Nevertheless, we found reports that allowed us to make a comparison with our results. For example, a survey of Trichoderma isolates from the soil of India [] showed that among 15 strains in HC identified, 11 were T. afroharzianum. These data corroborate the high prevalence of T. afroharzianum in the soil in South and Central America. On the other hand, a survey study from Iran showed that T. afroharzianum was not a dominant HC species from soil in the western region of the country []. In fact, T. harzianum was the most frequently isolated species. In another study, Tang et al. [] showed that species of HC were the most prevalent Trichoderma species from soil in the region of Zoige Alpine of China. Among species of the HC, T. harzianum was the most prevalent, representing about 72% of the species in the HC (37/51). This finding differs from what we obtained in South and Central America. T. harzianum is known to be a species of cold temperature region, and the temperature range of the soil in the region where samples were obtained ranges from −10 °C in winter to 15 °C in summer []. This may have caused a bias in species dominance. In South Africa, du Plessis et al. [] showed that species of T. afroharzianum, T. atrobrunneum, and T. camerunense were the only species within HC obtained from the soil in South Africa; however, the authors did not report the frequency of these of each species to know the most prevalent species in that region.
5. Conclusions
Within the genus Trichoderma, T. harzianum is one of the most well-known species for biocontrol and plant growth promotion. The Harzianum clade’s (HC) split has led to the improper usage of this species’ name in scientific publications, databases, and commercial products. This study used data from five investigations to identify Trichoderma species within the HC in the soil of South and Central America. The species T. afroharzianum, T. lentiforme, T. endophyticum, T. azevedoi, and T. harzianum were found to be the dominant species of the HC in South and Central America. However, the soil sampling in those five studies was not obtained in a statistical manner and order. Thus, we consider the findings from this study to provide a rough estimate of the dominant species of HC on the continent. Selecting strains or species for biocontrol and growth promotion from them could be an expensive and arduous undertaking. Identifying the dominant species within HC and selecting strains from them can expedite the commercialization process by reducing the time and expense associated with strain selection. Moreover, these strains may have better fitness traits than the rest of the species in HC. We further emphasize the need for additional research to be carried out in the future from different continents, as we were unable to locate any data with which to compare our findings. Moreover, we emphasize the significance of accurately identifying Trichoderma species prior to depositing their sequences into databases such as GenBank or culture collection institutes such as the American Type Culture Collection (ATCC).
Author Contributions
A.I. conceptualization, data generation, analysis, interpretation, writing—original draft preparation and editing; D.K.L. conceptualization, analysis, interpretation, writing, and editing; P.P.J. data collection and analyses, and P.S. feedback, and formatting of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USDA-ARS in-house projects 8042-21220-259-000D and 8042-22000-320-000D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Samuels, G.J.; Hebbar, P.K. Trichoderma: Identification and Agricultural Applications; APS Press: Eagan, MN, USA, 2015. [Google Scholar]
- Weindling, R. Trichoderma lignorum as a Parasite of Other Soil Fungi. Phytopathology 1932, 22, 837–845. [Google Scholar]
- Lindsey, D.L.; Baker, R. Effect of Certain Fungi on Dwarf Tomatoes Grown under Gnotobiotic Conditions. Phytopathology 1967, 57, 1262–1263. [Google Scholar]
- Chang, Y.-C.; Chang, Y.-C.; Baker, R.; Kleifeld, O.; Chet, I. Increased Growth of Plants in the Presence of the Biological Control Agent Trichoderma harzianum. Plant Dis. 1986, 70, 145–148. [Google Scholar] [CrossRef]
- Raspanti, E.; Cacciola, S.O.; Gotor, C.; Romero, L.C.; García, I. Implications of cysteine metabolism in the heavy metal response in Trichoderma harzianum and in three Fusarium species. Chemosphere 2009, 76, 48–54. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A potential bioremediator for environmental clean up. Clean Techn. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Harman, G.E. Trichoderma—Not Just for Biocontrol Anymore. Phytoparasitica 2011, 39, 103–108. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological Functions of Trichoderma spp. for Agriculture Applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Rifai, M.A. A Revision of the Genus Trichoderma. Mycol Pap. 1969, 116, 1–56. [Google Scholar]
- Chaverri, P.; Castlebury, L.A.; Samuels, G.J.; Geiser, D.M. Multilocus Phylogenetic Structure within the Trichoderma harzianum/Hypocrea lixii Complex. Mol. Phylogenet. Evol. 2003, 27, 302–313. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Bissett, J.; Druzhinina, I.; Kullnig-Gradinger, C.; Szakacs, G. Genetic and Metabolic Diversity of Trichoderma: A Case Study on South-East Asian Isolates. Fungal Genet. Biol. 2003, 38, 310–319. [Google Scholar] [CrossRef]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum Species Complex and the Re-Identification of Commercial Biocontrol Strains. Mycologia 2015, 107, 558–590. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, R.; Sun, Q.; Wu, B.; Sun, J.-Z. Four New Species of Trichoderma in the Harzianum Clade from Northern China. MycoKeys 2020, 73, 109–132. [Google Scholar] [CrossRef] [PubMed]
- del Carmen, H.; Rodríguez, M.; Evans, H.C.; de Abreu, L.M.; de Macedo, D.M.; Ndacnou, M.K.; Bekele, K.B.; Barreto, R.W. New Species and Records of Trichoderma Isolated as Mycoparasites and Endophytes from Cultivated and Wild Coffee in Africa. Sci. Rep. 2021, 11, 5671. [Google Scholar] [CrossRef]
- Zheng, H.; Qiao, M.; Lv, Y.; Du, X.; Zhang, K.-Q.; Yu, Z. New Species of Trichoderma Isolated as Endophytes and Saprobes from Southwest China. J. Fungi 2021, 7, 467. [Google Scholar] [CrossRef]
- Qin, W.-T.; Zhuang, W.-Y. Seven New Species of Trichoderma Hypocreales in the Harzianum and Strictipile Clades. Phytotax 2017, 305, 121–139. [Google Scholar] [CrossRef]
- Zhang, Y.-B.; Zhuang, W.-Y. New Species of Trichoderma in the Harzianum, Longibrachiatum and Viride Clades. Phytotaxa 2018, 379, 131–142. [Google Scholar] [CrossRef]
- Cao, Z.-J.; Qin, W.-T.; Zhao, J.; Liu, Y.; Wang, S.-X.; Zheng, S.-Y. Three New Trichoderma Species in Harzianum Clade Associated with the Contaminated Substrates of Edible Fungi. J. Fungi 2022, 8, 1154. [Google Scholar] [CrossRef]
- Cai, F.; Druzhinina, I.S. In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 2021, 107, 1–69. [Google Scholar] [CrossRef]
- Maddison, W.P. Mesquite: A Modular System for Evolutionary Analysis. Version 3.18. 2007. Available online: http://www.mesquiteproject.org (accessed on 15 June 2024).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hoyos-Carvajal, L.; Orduz, S.; Bissett, J. Genetic and Metabolic Biodiversity of Trichoderma from Colombia and Adjacent Neotropic Regions. Fungal Genet. Biol. 2009, 46, 615–631. [Google Scholar] [CrossRef]
- Smith, A.; Beltrán, C.A.; Kusunoki, M.; Cotes, A.M.; Motohashi, K.; Kondo, T.; Deguchi, M. Diversity of Soil-Dwelling Trichoderma in Colombia and Their Potential as Biocontrol Agents against the Phytopathogenic Fungus Sclerotinia sclerotiorum (Lib.) de Bary. J. Gen. Plant Pathol. 2013, 79, 74–85. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Komoń, M.; Bissett, J.; Szakacs, G.; Kubicek, C.P. An Oligonucleotide Barcode for Species Identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Inglis, P.W.; Mello, S.C.; Martins, I.; Silva, J.B.; Macêdo, K.; Sifuentes, D.N.; Valadares-Inglis, M.C. Trichoderma from Brazilian Garlic and Onion Crop Soils and Description of Two New Species: Trichoderma azevedoi and Trichoderma peberdyi. PLoS ONE 2020, 15, e0228485. [Google Scholar] [CrossRef] [PubMed]
- Barrera, V.A.; Iannone, L.; Romero, A.I.; Chaverri, P. Expanding the Trichoderma harzianum Species Complex: Three New Species from Argentine Natural and Cultivated Ecosystems. Mycologia 2021, 113, 1136–1155. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Crous, P.W.; Redhead, S.A.; Reynolds, D.R.; Samson, R.A.; Seifert, K.A.; Taylor, J.W.; Wingfield, M.J.; Abaci, Ö.; Aime, C.; et al. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2011, 2, 105–111. [Google Scholar] [CrossRef]
- Cummings, N.J.; Ambrose, A.; Braithwaite, M.; Bissett, J.; Roslan, H.A.; Abdullah, J.; Stewart, A.; Agbayani, F.V.; Steyaert, J.; Hill, R.A. Diversity of root-endophytic Trichoderma from Malaysian Borneo. Mycol. Prog. 2016, 15, 50. [Google Scholar] [CrossRef]
- Van Lenteren, J.C.; Bueno, V.H.P.; Luna, M.G. Biological control in Latin America and the Caribbean: Information sources, organizations, types and approaches in biological control. In Biological Control in Latin America and the Caribbean: Its Rich History and Bright Future; Lenteren, J.C.V., Bueno, V.H.P., Luna, M.G., Colmeneraz, Y.C., Eds.; CAB International: Wallingford, UK, 2020; pp. 1–20. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhao, Q.; Li, W.; Gao, L.; Liu, G. Strain Improvement of Trichoderma harzianum for Enhanced Biocontrol Capacity: Strategies and Prospects. Front. Microbiol. 2023, 14, 1146210. [Google Scholar] [CrossRef]
- Han, W.; Wu, Z.; Zhong, Z.; Williams, J.; Jacobsen, S.E.; Sun, Z.; Tang, Y. Assessing the Biosynthetic Inventory of the Biocontrol Fungus Trichoderma afroharzianum T22. J. Agric. Food Chem. 2023, 71, 11502–11519. [Google Scholar] [CrossRef]
- Silva, L.G.; Camargo, R.C.; Mascarin, G.M.; Nunes, P.S.d.O.; Dunlap, C.; Bettiol, W. Dual Functionality of Trichoderma: Biocontrol of Sclerotinia sclerotiorum and Biostimulant of Cotton Plants. Front. Plant Sci. 2022, 13, 983127. [Google Scholar] [CrossRef]
- Chen, K.; Zhuang, W.Y. Discovery from a large-scaled survey of Trichoderma in soil of China. Sci Rep. 2017, 7, 9090. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jambhulkar, P.P.; Singh, B.; Raja, M.; Ismaiel, A.; Lakshman, D.K.; Tomar, M.; Sharma, P. Genetic diversity and antagonistic properties of Trichoderma strains from the crop rhizospheres in southern Rajasthan, India. Sci Rep. 2024, 14, 8610. [Google Scholar] [CrossRef] [PubMed]
- Mirzaeipour, Z.; Bazgir, E.; Zafari, D.; Darvishnia, M. Isolation and identification of Harzianum clade species of Trichoderma from Khorramabad County. Mycol. Iran. 2023, 10, 67–78. [Google Scholar] [CrossRef]
- Tang, G.T.; Li, Y.; Zhou, Y.; Zhou, Y.; Zhu, Y.H.; Zheng, X.J.; Chang, X.L.; Zhang, S.R.; Gong, G.S. Diversity of Trichoderma species associated with soil in the Zoige alpine wetland of Southwest China. Sci. Rep. 2022, 12, 21709. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Lu, Q.; Zhao, Q.; Wang, J.; Ouyang, H. Effects of alpine wetland landscapes on regional climate on the Zoige Plateau of China. Adv. Meteorol. 2013, 2013, 972430. [Google Scholar] [CrossRef]
- du Plessis, I.L.; Druzhinina, I.S.; Atanasova, L.; Yarden, O.; Jacobs, K. The diversity of Trichoderma species from soil in South Africa, with five new additions. Mycologia 2018, 110, 559–583. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).