Abstract
Commercial tests are often employed in clinical microbiology laboratories for antifungal susceptibility testing of filamentous fungi. Method-dependent epidemiological cutoff values (ECVs) have been defined in order to detect non-wild-type (NWT) isolates harboring resistance mechanisms. We reviewed the literature in order to find studies where commercial methods were used to evaluate for in vitro susceptibility of filamentous fungi and assess their ability to detect NWT isolates according to the available ECVs. Data were found for the gradient concentration strips Etest and MIC Test Strips (MTS), broth microdilution Sensititre YeastOne (SYO), Micronaut-AM and the agar dilution VIPcheck assays. Applying itraconazole, voriconazole and posaconazole Etest ECVs for A. fumigatus, Etest was able to detect 90.3% (84/93), 61.2% (90/147) and 86% (31/36) of isolates with known cyp51A mutations, respectively. Moreover, Etest also was able to detect 3/3 fks mutants using caspofungin ECVs and 2/3 micafungin mutant isolates. Applying the voriconazole and posaconazole SYO ECVs, 57.7% (67/116) and 100% (47/47) of mutants with known cyp51A substitutions were classified as NWT, respectively. VIPcheck detected 90.3% (159/176), 80.1% (141/176) and 66% (141/176)of mutants via itraconazole, voriconazole and posaconazole, respectively, whereas Micronaut-AM detected 88% (22/25). In conclusion, Etest posaconazole and itraconazole, as well as micafungin and caspofungin ECVs, detected A. fumigatus mutants. On the other hand, while the posaconazole SYO ECV was able to detect cyp51A mutants, similar data were not observed with the SYO voriconazole ECV.
1. Introduction
The prevalence of invasive fungal infections continues to increase due to immunocompromised individuals. In order to manage these infections three classes of antifungal agents (echinocandins, azoles and polyenes) are recommended as first-line or salvage therapy [1]. Given the emergence of isolates with intrinsic or acquired resistance associated with high mortality, the Clinical and Laboratory Standards Institute [CLSI] and the European Committee on Antifungal susceptibility Testing [EUCAST]) have developed standardized broth microdilution methods for in vitro antifungal susceptibility testing. The EUCAST method utilizes 96-microtiter flat bottom plates, RPMI medium containing 2% glucose buffered with MOPS, a 105 CFU/mL inoculum, visual and/or spectrophotometric determination of minimal inhibitory concentrations (MIC) for azoles and amphotericin B as the lowest drug concentration with >90% growth inhibition and minimal effect concentrations (MEC) for echinocandins as the lowest drug concentration with abnormal, short, and branched hyphal clusters, whereas the CLSI method utilizes 96-microtiter U-shaped plates, RPMI medium containing 0.2% glucose buffered with MOPS, a 104 CFU/mL inoculum and visual determination of MIC for azoles and amphotericin B corresponding to the lowest drug concentration with complete growth inhibition and MEC for echinocandins as the lowest drug concentration with small, rounded compact hyphal forms [2,3]. Species-specific breakpoints (BP) have also been established for interpreting MIC/MECs of some antifungal agents against the most prevalent species [4,5,6]. Results by reference methods have been correlated with in vivo outcome, as infections by azole resistant Aspergillus isolates have been associated with increased mortality [7]. Amphotericin B failure has been linked with isolates with high MICs [8,9] and micafungin therapy failed against an Aspergillus isolate with reduced susceptibility to echinocandins [10], while several preclinical models show the importance of reference MICs regarding in vivo outcome [11].
1.1. Development of Clinical Breakpoints and ECVs for Filamentous fungi
The role of antifungal susceptibility testing relies on the ability to select the most appropriate agent for the treatment of a specific fungal infection. Even though methodological differences exist between CLSI and EUCAST procedures, their results have proven to be comparable [12] and allow the categorization of the strains as susceptible or resistant by applying the established BPs. EUCAST has defined drug- and species-specific clinical BPs for Aspergillus spp. versus triazoles (itraconazole, posaconazole, voriconazole and isavuconazole) and amphotericin B [13], whereas the CLSI has recently adopted a clinical BP only for voriconazole and A. fumigatus [14]. Although BPs can predict the likelihood of clinical response to antifungal therapy, there are many species and antifungal drugs for which there are insufficient data to determine clinical BPs [4]. For those species and drugs, epidemiological cutoff values (ECOFFs for the EUCAST reference method and ECVs for the CLSI reference method) can be used in order to detect isolates with acquired resistance mechanisms [5,15].
An ECV is defined as the highest MIC of the wild-type (WT) population of a given species without a phenotypically detectable acquired resistance mechanism [4]. The main role of an ECV is to distinguish WT isolates from non-wild-type (NWT) isolates [16,17,18], i.e., isolates with MIC higher than the ECV that potentially harbors a known or unknown acquired resistance mechanism [19]. Moreover, ECVs have an important role in tracking MIC elevation and emergence of resistance. ECVs are determined based on MIC distributions integrating information from with drug resistance mechanisms whenever available, whereas BPs are based on data for ECVs, pharmacokinetic/pharmacodynamic studies and correlation of the MIC with clinical outcome [20,21,22]. Therefore, the NWT or WT is not equivalent with the terms “susceptible” or “resistant” [14] to an antifungal agent. For some species, WT isolates may naturally possess resistance mechanisms, in which case the species is considered intrinsically resistant; for example, Fusarium spp. and several drugs [14]. Thus, WT isolates may or may not respond to antifungal therapy, whereas NWT isolates are expected to be associated with clinical failure [23]. The development of ECVs is dependent on the in vitro susceptibility testing used to generate MIC values. ECVs have been published for the most common Aspergillus species [5,15], and for some Fusarium and Zygomycetes spp. [14,24] mainly for the reference methodologies (Table 1).

Table 1.
Method-dependent ECVs for clinically relevant filamentous fungi and available CLSI and EUCAST ECVs.
CLSI and EUCAST followed a strict ECV setting process in order to determine “reference” ECVs based on MIC data generated with reference methods [32]. Apart from “reference” ECVs, there are also “method-dependent” ECVs for commercial susceptibility testing methods, and particularly Etest for Aspergillus spp. [25,29]. Unfortunately, apart from the most common Aspergillus spp., “method-dependent” ECVs have not been defined for other filamentous fungi (Table 1). Considering the comparative rarity of infections caused by less prevalent molds, it may take years before sufficient reliable data will be available to establish ECVs for available commercial methods.
1.2. Commercial Methods for Antifungal Susceptibility Testing of Molds
Reference antifungal susceptibility testing methods are not widely used because they are difficult to implement and require expertise. For optimal patient management and for routine practice laboratories, antifungal susceptibility methods should be fast, accurate, user-friendly, reproducible and low-cost. Across the years, several commercially available antifungal susceptibility methods have been used in clinical and research laboratories [33]. These methods could be helpful in such limited settings for MIC/MEC determination. Commercially available susceptibility methods have been compared with reference assays in two ways: the essential agreement (EA), which is the agreement between MICs/MECs of reference and commercial methods (usually within 1–2 twofold dilutions), or the categorical agreement (CA), which is the agreement between categorization of isolates as susceptible, intermediate or resistant with commercial and reference methods [19,26]. Therefore, commercially available and ready-to-use methods could be a better alternative for the routine clinical microbiology laboratory as far as they are able to produce similar results with reference standards [26].
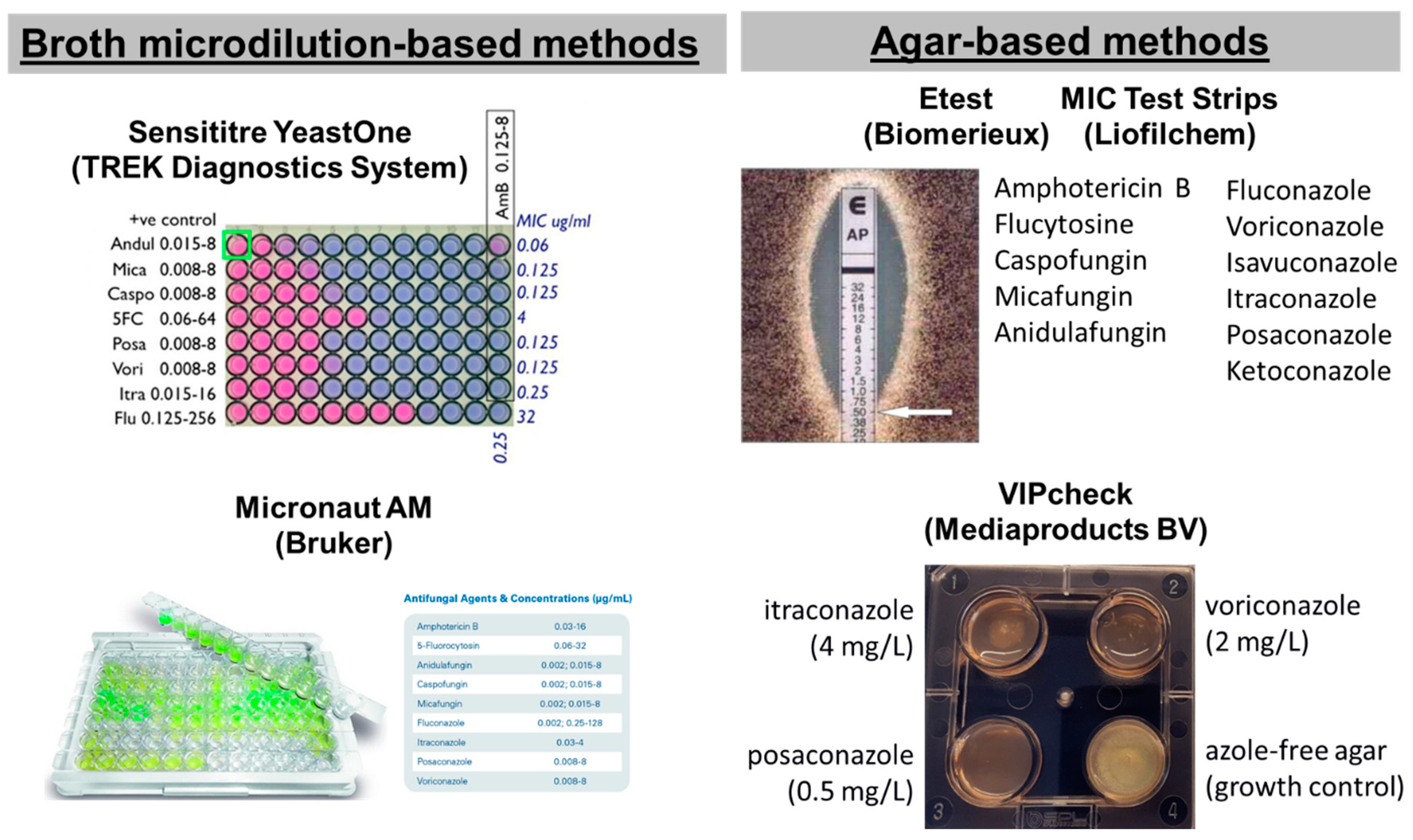
In particular, five commercial tests are available for screening antifungal resistance of moulds: two broth microdilution-based methods, the Sensititre YeastOne (SYO, TREK Diagnostics System, Cleveland, OH, USA) and the Micronaut AM (Bruker, Billerica, MA, USA) and three agar-based methods using strips with a gradient of antifungal concentrations, the Etest (Biomerieux, Tokyo, Japan), MIC Test Strips (MTS) (Liofilchem, Roseto degli Abruzzi, Italy) and the four-well plates VIPcheckTM (Mediaproducts BV, Groningen, The Netherlands) (Figure 1) [34,35]. Although they are easier compared to reference methods, they are expensive, and MIC reading can be difficult because of subtle color changes, trailing growth and isolated colonies, and may not perform equally well for all species and drugs. Most of these tests have been developed for yeasts and then applied to molds. For these reasons, commercial tests needs to be calibrated based on reference methods by using QC of reference methods, generating similar MIC distributions for WT isolates and detecting NWT isolates with different levels of resistance. The agreement between commercial and reference methodologies, together with the two-fold differences between median MICs, is summarized in Table 2. A more-than-one two-fold difference between commercial and reference methods indicates a significant difference in MIC distributions for the two methods that could lead to classification errors if reference ECV/BPs will be used.
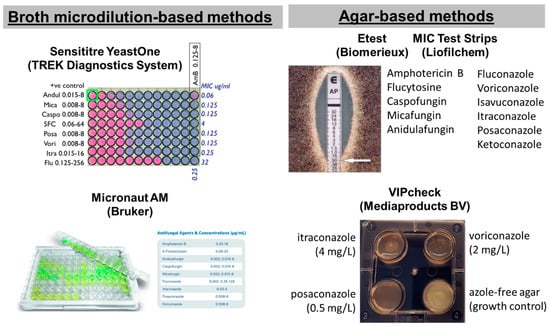
Figure 1.
Commercial tests for antifungal susceptibility testing of filamentous fungi.

Table 2.
Essential agreement ±2 two-fold dilutions (two-fold differences of median MIC) of the SYO, Etest, Micronaut-AM and VIPcheck tests compared to the CLSI method.
Concerning colorimetric methods, SYO yielded high essential agreement with the CLSI for voriconazole, posaconazole and itraconazole, except for A. fumigatus and A. nidulans isolates [37], while for the echinocandins, high essential agreement was found only for micafungin versus A. fumigatus and A. flavus isolates [36] (Table 2). Results for amphotericin B also showed high essential agreement (91.3–100%) depending on Aspergillus spp., but not for the zygomycetes. Reliable alternatives for antifungal susceptibility testing for Aspergillus and non-Aspergillus species are Etest and MTS: categorical and essential agreements of ≥90% with CLSI and EUCAST [40,45]. Briefly, concerning echinocandins for both micafungin and caspofungin, good essential agreement (>77.8%) was found for all species except A. glaucus, Scedosporium apiospermum and Scopulariopsis spp. High essential agreement was reported for the azoles and the polyenes, in particular for amphotericin B, posaconazole and voriconazole, but not itraconazole (>75%), although in some studies lower essential agreement was found. Results for Micronaut-AM in one study demonstrated good essential agreement with the CLSI reference method (>90%) for anidulafungin, voriconazole and amphotericin B, but not for itraconazole (87%) for 78 Aspergillus isolates [48]. In another study, low essential agreement (<62%) was found for all triazoles, but not for amphotericin B (100%) for 77 Aspergillus isolates with the EUCAST reference method [49]. Finally, categorical agreement was also stated for VIPcheckTM as the method can discriminate NWT isolates of A. fumigatus. Overall, good categorical agreement was found in two studies for all triazoles (80–97.8%), except in one study for posaconazole where agreement was 73.6% [50,51].
Difficulties in susceptibility testing of molds and the issue of lack of clinical data preclude the definition of BPs for commercially available methods, but method-specific ECVs have been defined for various Aspergillus spp. and antifungal drugs [4,16,29]. However, ECVs of commercial methods may differ from ECVs of reference methods when MIC distributions obtained using each method are different (Table 1). Although for most drug-species the differences are within one two-fold dilution, there are notable exceptions like Etest with caspofungin and posaconazole against A. terreus, voriconazole against A. flavus, posaconazole against A. niger, as well as amphotericin B against S. apiospermum and Scopulariopsis spp. and for SYO with caspofungin against all Aspergillus spp. and for amphotericin B against the zygomycetes. Similarly, Micronaut-AM MICs of all three triazoles were 2–3 two-fold dilutions different from EUCAST but not from CLSI MICs (except itraconazole) for Aspergillus spp. [40,45]. These results lead to susceptibility classification errors when established BPs or ECVs for reference methods are used for interpreting MICs generated with commercial methods.
2. Purpose of Review
The main purpose of the current review was to summarize the ability of commercial methods for in vitro susceptibility testing of filamentous fungi to detect NWT isolates to the triazoles, echinocandins or polyenes using available ECVs. Based on the role of the ECV to detect mutants [52], we focused on publications where data on resistance mechanisms are presented together with the MIC of the isolates for particular species and drugs [53]. For this reason, we initially describe the mechanism of actions and resistance and summarize known mutations in target and other genes that are associated with resistance. As expected, data for non-Aspergillus species were scarce and most publications reported only Etest and SYO data. The importance of validating these methods as predictors of in vitro resistance is deemed necessary as, apart from reference laboratories, the majority of clinical microbiology routine laboratories use commercial methods for in vitro susceptibility testing of molds [33]. However, as is the case for any susceptibility test, ECVs can provide incorrect classification or overlapping results between mutants and WT isolates as reported elsewhere [33]. For instance, MICs of mutants may be lower than the given ECV for particular agents and species. Although this might be true for mutations that do not elevate MICs, for mutations that are known to confer resistance, any misclassification would mean failure of the test and further optimization may be needed.
2.1. Echinocandins
Echinocandins target the fungal cell wall via non-competitive inhibition of (1,3)-β-D glucan (BDG) and—contrary to Candida spp., which is fungicidal—present fungistatic activity against Aspergillus spp. and other filamentous fungi. Despite the understanding of resistance mechanisms for Candida isolates, they are not as well documented for Aspergillus spp. There is minimal data for the treatment of invasive aspergillosis because echinocandins have been used mainly as salvage or combination therapy. Differences in innate echinocandin susceptibility show up in A. niger due to different cell wall composition [54]. fks1 gene substitutions, the effect of chitin synthesis, genetic repression in heat shock protein 90 (Hsp 90) and the reactive oxygen species (ROS) are among the molecular mechanisms of echinocandin resistance reported in Aspergillus spp. isolates [35,55].
Amino acid substitutions in the fks1 subunit of glucan synthesis lead to echinocandin resistance. Although knowledge is scarce about echinocandin resistance among Aspergillus spp., an expression analysis of the fks1 gene revealed an overexpression (three-fold higher) in comparison with a WT isolate, after a treatment failure with caspofungin in an invasive aspergillosis infection caused by A. fumigatus [56]. In order to understand echinocandin resistance, multiple in vitro models with resistance induction have been developed [57,58]. Briefly, site-directed mutation at S678Y into fks1 led to decreased susceptibility to caspofungin with MEC = 4 mg/L [57], while substitution of serine with proline in codon 678 resulted in a resistance phenotype of MEC ≥ 16 mg/L to all the three available echinocandins. This result suggests that fks1 gene modifications lead to echinocandin resistance [58]. Previously, an echinocandin-resistant A. fumigatus isolate harboring the point mutation F675S in the fks1 gene had reduced susceptibility to both caspofungin and micafungin (MEC = 2 mg/L) [10]. However, an fks1-independent mechanism of echinocandin resistance in A. fumigatus has been recently identified: an alteration of the drug–target interaction via caspofungin-induced ROS-mediated changes in the lipid composition of the glucan synthase with elevated MECs (caspofungin (4–16 mg/L) and micafungin (2–4 mg/L)) [59].
Another resistance mechanism of Aspergillus species to echinocandins is the effect of chitin synthesis. The paradoxical phenomenon or the ability of Aspergillus spp. isolates to grow in concentration above MEC is related to the increased chitin synthesis in the fungal cell wall. This phenomenon is more commonly seen with caspofungin than micafungin and anidulafungin [60]. Increased sensitivity of caspofungin mutants was in agreement with synergistic antifungal effect of a combination of chitin synthesis inhibitor and caspofungin, while Δras mutants, despite having a low level of β-glucan, are more resistant to caspofungin due to an increase in the cell wall synthesis [61]. Increased sensitivity to caspofungin was also observed among calcineurin mutants (ΔcnaA) of A. fumigatus isolates, due to low level β-glucan and chitin synthesis [62].
Hsp 90 has also been implicated in echinocandin resistance. It is involved in a wide range of signaling networks and cell processes, from control to survival of the cell cycle, as well as response to cell stress in order to maintain cell homeostasis [63]. Hsp 90 plays a key role in the evolution of azole and echinocandin resistance by activating specific cellular signaling pathways that are necessary for cell survival against membrane stress due to the antifungal agent [64]. More specifically, resistance to echinocandins is affected through calcineurin, a protein phosphatase regulator of cellular signaling. However, genetical repression of Hsp 90 leads to decreased virulence in a murine infection model of IA; the replacement of natural promoters with two artificial promoters in A. fumigatus isolates resulted in increased susceptibility to caspofungin and a canceling of the paradoxical effect [65].
Finally, a new mechanism was described via ROS production. It was found that caspofungin exposure modifies glucan synthase, rendering it insensitive to echinocandins. This mechanism of resistance involved alteration of the glucan synthase lipid microenvironment and was mediated via an off-target effect on mitochondria leading to increased ROS. Thus, it was hypothesized that caspofungin-induced ROS alters the lipid composition around glucan synthase, changing its conformation and making it insensitive to echinocandins [66]. The resistance mechanisms for other filamentous fungi have not been fully explored.
2.2. Triazoles
Azole resistance is usually associated with specific resistance mechanisms constituted by a variable number of tandem repeat (TR) integrations in cyp51A promoter and mutations in the coding gene [35]. Isolates with resistance to azoles due to TR (TR34/L98H, TR34/L98H/S297T/F495I, TR46/Y121F/T289A and TR53) have been detected throughout the world [67]. Moreover, single nucleotide polymorphisms, mainly in gene positions G54, M220 and G448 of the cyp51A gene, have been observed to be more frequent in patients with chronic pulmonary aspergillosis, long term azole therapy, and clinical treatment failures [35]. Single-point mutations in other positions have also been associated with azole resistance (G138C, F219I, P216L, G432S and G432A) [68,69,70]. Isolates with substitutions in cyp51A usually have high MIC values for voriconazole (0.5–>32 mg/L), itraconazole (0.5–32 mg/L), posaconazole (0.125–>16 mg/L) and isavuconazole (0.125–>16 mg/L) [45], depending on the mutation [35]. It is widely accepted that TR34/L98H alterations are associated with pan-azole resistance, and TR46/Y121F/T289A with a high level of voriconazole resistance but with variable posaconazole and itraconazole susceptibility. M220 alterations are associated with resistance to itraconazole and posaconazole and with variable susceptibility to voriconazole, except M220T which shows susceptibility to posaconazole and voriconazole. Similarly, G54 alterations are associated with resistance to itraconazole and posaconazole but not to voriconazole [71].
Although mutations in cyp51A have been well explored, there are also mutations or overexpression in cyp51B and cyp51C which confer resistance to the triazoles. Most Aspergillus spp., Mucorales spp. and Penicillium spp., have two paralogues (cyp51A and cyp51B), while there are few species, including A. flavus, A. oryzae and the Fusarium spp., that have three cyp51 enzymes (cyp51A, cyp51B and cyp51C) [72,73]. However, while mutations in cyp51B responsible for azole resistance in A. fumigatus have not been reported, overexpression in two clinical azole-resistant isolates suggest its possible role [74]. Interestingly, in a single study, a novel G457S mutation in cyp51B with concomitant F390L mutation in the 3-hydroxy-3-methylglutaryl-coenzyme-A-reductase-encoding gene (hmg1) showed high MICs in itraconazole, voriconazole, posaconazole and isavuconazole (>8 mg/L). The contribution of azole resistance is unclear due to both mutations [72], while reconstitution of the G457S mutation in a triazole-sensitive strain resulted in resistance to voriconazole (2 mg/l), but not to itraconazole or posaconazole [75]. Four mutations of A. flavus in cyp51C (S196F, A324P, N423D and V465M) are correlated with voriconazole resistance [76]. Moreover, it has been found that T788G missense mutation in cyp51C gene was responsible for a voriconazole-resistant A. flavus clinical isolate (MIC = 8 mg/L) [77]. Finally, in a resistant A. flavus isolate it has been reported that substitution in H349R in cyp51C showed increased gene expression but the role of azole resistance remained unclear [78].
Apart from mutations in cyp51-related genes, there are also resistance mechanisms that involve resistance to azoles among Aspergillus spp. isolates (overexpression of efflux pumps, upregulation of cyp51A, CCAAT-binding complex, upregulation of efflux pumps, hmg1 mutations, master regulators, damage resistance protein 1, mismatch repair gene (MSH2), OrmA enzyme, deletion of the CybE encoding gene (b5 CybE), oxidoreductase HorA, hapE, A. fumigatus farnesyltransferase Cox10 (Afcox10), RNA interference (RNAi)-dependent mutations, cholesterol uptake/import resistance mechanisms and cytochrome c oxidase cox7c W56* nonsense) [35,64,79]. Efflux pumps, particularly major facilitator superfamily (MFS) and ATP-binding cassette (ABC) transporters, remove toxins by coupling transport with proton gradient or adenosine triphosphate (ATP) hydrolysis [64]. In itraconazole-resistant A. fumigatus isolates, overexpression of ABC transporters (AfuMDR1, AfuMDR2 and AtrF) and upregulation of AfuMDR3 and AfuMDR4 encoding MFS-transporters have been described [80,81]. Moreover, upregulation in transporter genes (abcB/Afu1g10390, abcE, mfsA, mfsB and mfsC) [82] and cdr1B [83] were shown in response to voriconazole and itraconazole exposure in azole-susceptible and azole-resistant A. fumigatus isolates, respectively. Increased expression of cdr1B resulted in voriconazole MIC = 1 mg/L, posaconazole MIC = 0.25 mg/L and itraconazole MIC = 2 mg/L [83]. In addition, hapE is an important subunit in the CCAAT-binding complex, which plays a regulatory role of fungal phenotypes in azole resistance. It was found that six non-synonymous mutations were identified in the non-coding regions, of which resistance in the progeny was due to mutation in the hapE gene [84]. Isolates harboring these mutations in the hapE gene showed increased MICs to azoles (voriconazole, itraconazole and posaconazole MIC = 2–4 mg/L, >16 mg/L and 0.25–0.5 mg/L, respectively) [84]. Furthermore, a novel mutation (R243Q) in Afcox10 gene was shown by next-generation sequencing analysis to confer cross-resistance to itraconazole (MIC = 8 mg/L), terbinafine (MIC = 16 mg/L) and bifonazole [85]. Finally, it has been found that mutation or deletion of cox7c results explicitly in antifungal resistance to targeting enzymes, including triazoles with high MICs of voriconazole (4 mg/L), itraconazole (2 mg/L) and posaconazole (2 mg/L) in comparison with the parental strains lower MICs [79]. The resistance mechanisms for the triazoles’ other filamentous fungi have not been fully explored.
2.3. Polyenes
Amphotericin B binds to ergosterol in the fungal cell membrane, leading to pore formation in the cell membrane with ion leakage and consequently cell death [86]. The main driver of amphotericin B efficacy seems to be interference with the mitochondria from ROS/anti-ROS [87]. Although amphotericin B resistance mechanisms are not well understood, the oxidative injury by ROS has been also implicated in this resistance [88]. Resistance to amphotericin B (MIC ≥ 2 mg/L) has been reported for A. fumigatus, A. niger, A. flavus, A. lentulus, A. terreus and A. ustus [64,89]. In a study, resistance to polyenes was attributed to the depletion of ergosterol due to diminished binding to the cytoplasmic cell membrane and increased amphotericin B MICs (16 mg/L) [90]. According to a large review conducted last year, the pooled mean prevalence of amphotericin B resistance was 0.17% among 26,909 Aspergillus isolates [89]. Overall, the development of resistance to amphotericin B is rare due to its action as a rapid fungicidal agent that inhibits the fungal growth by physiochemical reaction rather than enzyme inhibition [64]. There are no studies reporting genetic mutational changes leading to increasing amphotericin B MICs. Genetic analysis of amphotericin B resistant A. fumigatus isolates identified missense variants in genes tcsB, mpkC and catA and mutations in fumarylacetoacetate hydrolase associated with amphotericin B resistance [91,92]. There are observations of resistant Aspergillus spp. isolates that failed treatment with amphotericin B, in particular an 88% mortality rate among patients treated with amphotericin B therapy for A. flavus infections [93]. Some reports indicate that alterations of the fungal cell wall show a correlation with amphotericin B resistance. In another study, an experimentally evolved A. flavus isolate was able to grow at concentrations of up to 100 μg/mL and the authors assumed that alterations in the cell wall contributed to the resistance [94]. In addition, preclinical and clinical studies showed that amphotericin B is a poor therapeutic option (96% mortality) for A. terreus isolates as it is intrinsically resistant [95], with most isolates exhibiting MIC values ≥ 2 mg/L [96]. To date, there is no A. terreus specific genomic feature that has provided an explanation for amphotericin B resistance/tolerance mechanisms [87]. In general, resistance in section Terrei is associated with modulating molecular chaperons, targeting ROS via mitochondria and shaping the cellular redox homeostasis [87]; while underlying mechanisms may be associated with the level of catalase production of this species, in comparison with A. fumigatus [97,98]. The resistance mechanisms for polyenes and other filamentous fungi need to be fully explored.
3. Gradient Concentration Strips
- Triazoles
ECVs for Etest have been determined for itraconazole, posaconazole and voriconazole and the most common Aspergillus species (A. fumigatus, A. flavus, A. terreus, A. niger, A. nidulans) in a multicenter study (Table 3) [29]. An ECV has not been determined yet for isavuconazole and therefore the upper MIC level of WT isolates (WT-UL) was used to assess A. fumigatus isolates with known cyp51A substitutions. Information about resistance mechanisms of NWT isolates exists only for A. fumigatus and mainly involves cyp51A substitutions. Applying the Etest itraconazole ECVs for A. fumigatus (2 mg/L), Etest was able to detect 78/81 isolates with known cyp51A substitutions [29]. The three strains characterized as WT were two strains with G448S and I301T and one strain with M220K substitutions in cyp51A [29]. In another study, Etest was able to detect 6/6 of mutant isolates to itraconazole and 3/6 to voriconazole [99], but unable to detect any of the three isolates tested with cyp51A substitutions for all triazoles [100]. Applying isavuconazole WT-UL (2 mg/L) determine in 40 WT isolates, 72.4% (21/29) of isolates with M220 and TR mutations were categorized as NWT (21/39), while isolates harboring G54 alterations had lower MIC values and 0/10 were detected [45]. Concerning Etest and voriconazole, results obtained from different studies were somewhat less promising in the detection of mutant isolates using an Etest ECV of 0.5 mg/L. Etest was able to detect 49/75 isolates with cyp51A substitutions [29]. Among the isolates that have been characterized as WT based on the proposed ECV (25/75), there were mutants with TR34 (0.125–0.5 mg/L, 3/38), G54E/R/W (<0.06–0.5 mg/L, 12/12), M220I/K//R/T/V (0.125–0.5 mg/L, 8/11), G138C (0.25 mg/L, 1/1) and I301T (<0.06 mg/L, 1/1) substitutions [29]. It is worth noting that, in a study including isolates with TR34, the proposed ECV was able to detect 92% (35/38) of mutant isolates [29]. Overall, Etest ECVs detected 61.2% (90/147) of all mutant isolates as NWT. More promising results were found with the Etest ECV of posaconazole (0.25 mg/L), which detected the majority of isolates (86%, 31/36) with different cyp51A substitutions including G54E/R/V/W, M220I/R/T/V/K, G448S and TR34/L98H [30,99]. Among the isolates that were not recognized were five mutants with M220I/R/T/V/K and G448S and other non-specified cyp51A substitutions with MICs ranging 0.023–0.25 mg/L. Overall, the proposed ECVs for posaconazole and itraconazole were able to detect 90.3% and 86.1% of mutant isolates with distinct mutations respectively, while results were less promising for the detection of mutant isolates for voriconazole (61.2%) (Table 3).

Table 3.
Detection of Aspergillus fumigatus isolates harboring resistance mechanisms to triazoles/echinocandins with Etest dependent ECV.
- 2.
- Echinocandins
ECVs for Etest and echinocandins, and more precisely for caspofungin and micafungin, have been determined in 2019 from three studies, including multi-laboratories with sufficient numbers of isolates tested [4,26,31]. Caspofungin ECVs have been defined for A. fumigatus (0.25 mg/L), A. flavus (0.5 mg/L), A. terreus (2 mg/L) and A. niger (0.25 mg/L), [31], while a micafungin ECV has been defined only for A. fumigatus (0.016 mg/L) [4]. There are scarce data concerning the detection of mutants using Etest ECVs as very few isolates harboring resistance mechanisms have been described in the literature and susceptibility testing is not considered as an everyday practice in routine laboratory [102]. There is only one study with micafungin and caspofungin MIC data of laboratory mutants with known fks alterations with MTS [101]. An ECV of caspofungin was able to detect 3/3 of fks mutants, while micafungin’s ECV was able to predict 2/3 fks mutants as the third isolate had an MEC of 0.004 mg/L (Table 3).
- 3.
- Polyenes
Apart from CLSI and EUCAST ECV for amphotericin B, there is also a method-dependent ECV concerning Etest and A. fumigatus (2 mg/L), A. flavus (8 mg/L), A. niger (2 mg/L) and A. terreus (16 mg/L) [25]. Method-dependent ECVs for Etest are only available for these four Aspergillus species, but not for other filamentous fungi. Etest amphotericin B ECVs were consistently higher (one or two dilutions) or the same with CLSI method. A. terreus and A. flavus have high ECVs indicating intrinsic resistance for these species [25] in agreement with clinical cases with poor outcomes after amphotericin B therapy [93,103,104]. Since there is not any known resistance mechanism for Aspergillus and amphotericin B, studies including NWT isolates are not available in the literature.
3.1. Sensititre YeastOne
- 1.
- Triazoles
SYO ECVs of triazoles were determined in two large multicentre studies [29,30]. There are only ECVs for voriconazole (1 mg/L) and A. fumigatus, A. flavus and A. terreus and for posaconazole (0.06 mg/L) and A. fumigatus. It should be noted that the ECV for posaconazole and A. fumigatus is based on the unknown mutant status of the isolates. Applying the method-specific SYO ECV of voriconazole, 21/39 of mutant isolates with known cyp51A substitutions (TR34, G54E/R/W, M220I/K//R/T/V, G138C and I301T) were detected in one study [24] and 9/10 mutant isolates with TR34/L98H and TR46/Y121F T289A in another study [37]. In a third study, the voriconazole ECV of 1 mg/L was unable to detect two mutant isolates with G54R alteration and MIC 0.125 mg/L, whereas a single isolate with MIC 8 mg/L carrying the TR34/L98H cyp51A mutation has been recognized [93]. Overall, 67/116 (57.7%) of all mutant isolates have been characterized as NWT, indicating a concern about using SYO and voriconazole in order to detect mutant strains. The posaconazole SYO ECV was able to detect all 54/54 mutant isolates with the following cyp51A substitutions: TR34/L98H, TR46/Y121F T289A and G54R [37,105,106]. Considering the WT-UL (0.5 mg/L) that has been used in order to assess findings for itraconazole and A. fumigatus, 35/37 (94.5%) isolates with TR34/L98H, TR46/Y121F T289A and G54R mutations have been detected as NWT [37] in the majority of the studies, while there was a single study that included isolates with non-specified cyp51A substitutions, in which only 9/21 (42.8%) have been characterized as NWT [37] (Table 4). In this study, isolates with itraconazole and/or voriconazole CLSI MIC > 1 mg/L were submitted to cyp51A sequence analysis for the detection of azole-resistance-associated mutations and it was found that the isolates harbored TR34/L98H and TR46/Y121F T289A mutations. Overall, 45/65 (69.2%) of all isolates with various cyp51A mutations have been recognized. These results seem promising for posaconazole but not for voriconazole.

Table 4.
Detection of Aspergillus fumigatus isolates harboring resistance mechanisms to triazoles/echinocandins with Sensititre YeastOne-dependent ECV.
- 2.
- Echinocandins
ECVs for SYO and echinocandins have not been established till now, maybe due to the difficulty of the method used for the correct estimation of MEC. Recently, it has been proposed that the optimal conditions for SYO susceptibility testing of echinocandins is the use of an inoculum of 104 CFU/mL, incubation for 20 h for A. flavus and 30 h for A. fumigatus and A. terreus and reading the first purple well. Agreement with CLSI reference method was good for micafungin (77–100%), with median (range) two-fold difference 0 (−1 to 2), −1 (−4 to 1) and −2 (−3 to −2), but poor for caspofungin (0–54%), with median (range) two-fold difference 3 (1 to 5), 3 (−4 to 4) and 2 (1 to 4) for A. fumigatus, A. flavus and A. terreus, respectively, indicating that SYO does not produce similar results as the reference method and therefore reference ECV should not be used for commercial test, while results were inconclusive for anidulafungin due to off-scale color endpoints [36]. To the best of our knowledge, there is no study where NWTs with known resistance mechanisms have been studied with SYO to echinocandins.
- 3.
- Polyenes
Regarding polyenes and especially amphotericin B, there is not a specific ECV for SYO for any filamentous fungi. In addition, there are no studies where SYO has been used for antifungal susceptibility testing of mutant isolates. Summarizing available studies, an MIC90 for 30 WT A. fumigatus isolates was 2 mg/L, while the same MIC90 was found for 24 A. fumigatus isolates harboring TR34 mutation as expected since cyp51A mutations do not affect amphotericin B susceptibility and for 10 A. niger isolates, whereas the MIC90 for 23 A. flavus strains was one dilution higher (4 mg/L) [105]. The MIC90 for 13 A. terreus which considered resistant to amphotericin was 2 mg/L indicating that SYO cannot differentiated amphotericin B susceptible from resistant species. In another study, an MIC = 1 mg/L for 2 A. fumigatus isolates harboring G54R mutations was reported, while an MIC = 2 mg/L for a single isolate with TR34/L98H mutation was also reported [106].
3.2. VIPcheck
VIPcheck is intended for the phenotypic detection of A. fumigatus resistance to itraconazole, voriconazole and posaconazole, in routine laboratories where the application of reference method is not possible [108,109]. Introduction of a 4-well plate into routine clinical laboratories has a huge impact on the early detection of azole resistance and subsequently benefits from more appropriate therapy for the patient [71]. It has been used for international A. fumigatus resistance prevalence study (SCARE), as well as other surveillance studies with overall good performance [50,51,107,110,111,112,113]. Categorical agreement with the reference method was 78.8–80% for voriconazole, 69.2–97.8% for itraconazole and 55.8–83.3% for posaconazole [50,107]. Lower categorical agreement was found for A. fumigatus cryptic species (Table 2). Moreover, the ability of VIPcheck to recognize isolates harboring TR mutations was excellent for itraconazole and voriconazole with 90.3% (159/176) and 80.1% (141/176) of mutants recognized as NWT, respectively. Results were less encouraging for posaconazole, with VIPcheck able to recognize 66% (115/176) of NWT isolates (Table 5).

Table 5.
Detection of Aspergillus fumigatus isolates harboring resistance mechanisms to triazoles/echinocandins with VIPcheckTM.
3.3. Micronaut-AM
Micronaut’s ability to detect mutants has been assessed in two studies, with one showing ability to detect 89% (8/9) of CLSI azole NWT Aspergillus isolates [48] and the other one showing ability to detect 88% (14/16) of EUCAST azole NWT isolates with 15 harboring cyp51A mutations (eight TR34/L98H, 1 TR46/Y121F/T289A, three G54E/W and three other) [49]. In the last study, 1/15 (TR46/Y121F/T289A) was resistant to voriconazole (MIC = 4 mg/L), while 10/15 and 2/15 were resistant to itraconazole (MIC = 4 mg/L) and posaconazole (MIC = 0.5–1 mg/L), respectively [49]. In contrast, according to the authors, four A. fumigatus showed major and very major discrepancies. Three isolates were classified as WT by Micronaut for voriconazole (n = 2) and for itraconazole (n = 1) contrary to CLSI (very major), while one isolate was classified as NWT with Micronaut, whereas CLSI categorized it as WT (major) [48].
4. Conclusions
In conclusion, commercial methods can easily be applied in routine laboratories which do not have access to reference methods. However, they should be carefully applied following exactly the instructions of the manufacturers, taking into account the peculiarity of MIC reading for each drug and species for each method. VIPcheck method can be used to screen Aspergillus spp. for azole resistance and gradient concentration tests can be used to test susceptibility to each drug separately, whereas Sensititre YeastOne and Micronaut-AM can be used to test all drugs simultaneously. Among the few commercially available methods for antifungal susceptibility testing of molds, there are ECVs only for Etest and SYO and for the most common Aspergillus spp. (A. fumigatus, A. flavus, A. terreus, A. niger and A. nidulans). In addition, the ability of commercial tests to detect resistance is limited to A. fumigatus and triazole where isolates with known cyp51A substitutions have been studied. Thus, the performance of commercial tests in detecting resistance for other drugs and species including A. fumigatus cryptic species is unknown. Although in some cases the number of mutant isolates used to evaluate proposed ECVs is low, some conclusions can be made: (i) for Etest method, proposed ECVs for posaconazole, itraconazole and micafungin were able to detect >86% mutants of A. fumigatus, while ECV for voriconazole and caspofungin was less able to detect NWT with known cyp51A substitutions and fks alterations (<67%); (ii) for the SYO method, ECV for posaconazole exhibit encouraging results, recognizing all mutants, whereas ECV for voriconazole detect only 57.7% (67/116) of A. fumigatus isolates with cyp51A substitutions (Table 6). This is supported by results obtained from previous reports that showed isolates with TR34/L98H were characterized by a high itraconazole MIC (>8 μg/mL), variable susceptibility to voriconazole and cross-resistance to posaconazole [105]; (iii) finally, concerning VIPcheck as a method for early and reliable detection of azole resistance in routine clinical laboratory, where the usage of reference methodologies is not available, results were encouraging, with high categorical agreement especially for A. fumigatus sensu stricto isolates [50,107]. As not all mutations result in elevated MICs and NWT isolates may harbor unknown resistance mechanisms, one should be careful when evaluating performance of an MIC test based on their ability to detect mutants. However, a well described mutation that confer resistance to a specific drug should be captured by a certain test and failure to so means poor performance. Because most commercial methods have been developed for yeasts and then applied to molds, optimal performance cannot be guaranteed. There is a need for developing commercial methods for antifungal susceptibility testing of molds, taking into account the physiological (growth rate, metabolic activity, inoculum) and pharmacological (inhibition mode, killing activity) characteristics of each drug and species. Although the development of a commercial method that would produce the same MICs as the reference method for different drugs and species may be challenging, as long as the MICs of two methods are highly correlated, method-specific ECV could be used in order to improve categorical agreement. Finally, as each species have marked physiological characteristics, optimal conditions may be different among species even of the same genus. Further efforts are needed to develop an easy and fast commercial method for detecting resistance to many drugs among molds, particularly for species other than A. fumigatus.

Table 6.
Summary of the ability of each method to detect resistance of NWT Aspergillus fumigatus isolates harboring resistance mechanisms to triazoles/echinocandins. Percent of mutant isolates classified as NWT (N mutants/total isolates) are shown for each drug and method.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ullmann, A.J.; Aguado, J.M.; Arikan-Akdagli, S.; Denning, D.W.; Groll, A.H.; Lagrou, K.; Lass-Flörl, C.; Lewis, R.E.; Munoz, P.; Verweij, P.E.; et al. Diagnosis and Management of Aspergillus Diseases: Executive Summary of the 2017 ESCMID-ECMM-ERS Guideline. Clin. Microbiol. Infect. 2018, 24, e1–e38. [Google Scholar] [CrossRef]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi M38A, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; ISBN 978-1-56238-830-0. [Google Scholar]
- Guinea, J.; Meletiadis, J.; Arikan-Akdagli, S.; Muehlethaler, K.; Kahlmeter, G.; Arendrup, M. EUCAST Definitive Document E.Def 9.4. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds. 2022. Available online: https://eucast.org/astoffungi/ (accessed on 20 January 2024).
- Salsé, M.; Gangneux, J.P.; Cassaing, S.; Delhaes, L.; Fekkar, A.; Dupont, D.; Botterel, F.; Costa, D.; Bourgeois, N.; Bouteille, B.; et al. Multicentre Study to Determine the Etest Epidemiological Cut-off Values of Antifungal Drugs in Candida spp. and Aspergillus Fumigatus Species Complex. Clin. Microbiol. Infect. 2019, 25, 1546–1552. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Overview of Antifungal ECOFFs and Clinical Breakpoints for Yeasts, Moulds and Dermatophytes Using the EUCAST E.Def 7.3, E.Def 9.3 and E.Def 11.0 Procedures, Version 3.0. 2022. Available online: https://eucast.org/astoffungi/ (accessed on 20 January 2024).
- CLSI. M60-Ed2 June 2020 Replaces M60-Ed1 Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd ed.; CLSI Supplement M60; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; ISBN 978-1-68440-083-6. [Google Scholar]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.G.; Meis, J.F. Azole Resistance in Aspergillus Fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Lionakis, M.S.; Lewis, R.E.; Chamilos, G.; Kontoyiannis, D.P. Aspergillus Susceptibility Testing in Patients with Cancer and Invasive Aspergillosis: Difficulties in Establishing Correlation between in Vitro Susceptibility Data and the Outcome of Initial Amphotericin B Therapy. Pharmacotherapy 2005, 25, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Lass-Flörl, C.; Kofler, G.; Kropshofer, G.; Hermans, J.; Kreczy, A.; Dierich, M.P.; Niederwieser, D. In-Vitro Testing of Susceptibility to Amphotericin B Is a Reliable Predictor of Clinical Outcome in Invasive Aspergillosis. J. Antimicrob. Chemother. 1998, 42, 497–502. [Google Scholar] [CrossRef]
- Jiménez-Ortigosa, C.; Moore, C.; Denning, D.W.; Perlin, D.S. Emergence of Echinocandin Resistance Due to a Point Mutation in the Fks1 Gene of Aspergillus Fumigatus in a Patient with Chronic Pulmonary Aspergillosis. Antimicrob. Agents Chemother. 2017, 61, e01277-17. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Lewis, R.E.; Kontoyiannis, D.P. Role and Interpretation of Antifungal Susceptibility Testing for the Management of Invasive Fungal Infections. J. Fungi 2021, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Cuenca-Estrella, M.; Cantón, E. EUCAST and CLSI: Working Together Towards a Harmonized Method for Antifungal Susceptibility Testing. Curr. Fungal Infect. Rep. 2013, 7, 59–67. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing, Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 20 January 2024).
- Espinel-Ingroff, A.; Turnidge, J. The Role of Epidemiological Cutoff Values (ECVs/ECOFFs) in Antifungal Susceptibility Testing and Interpretation for Uncommon Yeasts and Moulds. Rev. Iberoam. Micol. 2016, 33, 63–75. [Google Scholar] [CrossRef]
- CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 4th ed.; CLSI Supplement M57S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; ISBN 978-1-68440-159-8. [Google Scholar]
- Espinel-Ingroff, A.; Alvarez-Fernandez, M.; Cantón, E.; Carver, P.L.; Chen, S.C.A.; Eschenauer, G.; Getsinger, D.L.; Gonzalez, G.M.; Govender, N.P.; Grancini, A.; et al. Multicenter Study of Epidemiological Cutoff Values and Detection of Resistance in Candida spp. to Anidulafungin, Caspofungin, and Micafungin Using the Sensititre YeastOne Colorimetric Method. Antimicrob. Agents Chemother. 2015, 59, 6725–6732. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Pfaller, M.A.; Bustamante, B.; Canton, E.; Fothergill, A.; Fuller, J.; Gonzalez, G.M.; Lass-Flörl, C.; Lockhart, S.R.; Martin-Mazuelos, E.; et al. Multilaboratory Study of Epidemiological Cutoff Values for Detection of Resistance in Eight Candida Species to Fluconazole, Posaconazole, and Voriconazole. Antimicrob. Agents Chemother. 2014, 58, 2006. [Google Scholar] [CrossRef]
- Turnidge, J.; Kahlmeter, G.; Kronvall, G. Statistical Characterisation of Bacterial Wild-Type MIC Value Distributions and the Determination of Epidemiological Cut-off Values. Clin. Microbiol. Infect. 2006, 12, 418–425. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Dannaoui, E. Should Etest MICs for Yeasts Be Categorized by Reference (BPs/ECVs) or by Etest (ECVs) Cutoffs as Determinants of Emerging Resistance? Curr. Fungal Infect. Rep. 2020, 14, 120–129. [Google Scholar] [CrossRef]
- Kahlmeter, G. The 2014 Garrod Lecture: EUCAST—Are We Heading towards International Agreement? J. Antimicrob. Chemother. 2015, 70, 2427–2439. [Google Scholar] [CrossRef]
- Dalhoff, A.; Ambrose, P.G.; Mouton, J.W. A Long Journey from Minimum Inhibitory Concentration Testing to Clinically Predictive Breakpoints: Deterministic and Probabilistic Approaches in Deriving Breakpoints. Infection 2009, 37, 296–305. [Google Scholar] [CrossRef]
- Turnidge, J.; Paterson, D.L. Setting and Revising Antibacterial Susceptibility Breakpoints. Clin. Microbiol. Rev. 2007, 20, 391–408. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Ghannoum, M.A.; Alexander, B.D. Establishment and Use of Epidemiological Cutoff Values for Molds and Yeasts by Use of the Clinical and Laboratory Standards Institute M57 Standard. J. Clin. Microbiol. 2017, 55, 1262. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Colombo, A.L.; Cordoba, S.; Dufresne, P.J.; Fuller, J.; Ghannoum, M.; Gonzalez, G.M.; Guarro, J.; Kidd, S.E.; Meis, J.F.; et al. International Evaluation of MIC Distributions and Epidemiological Cutoff Value (ECV) Definitions for Fusarium Species Identified by Molecular Methods for the CLSI Broth Microdilution Method. Antimicrob. Agents Chemother. 2016, 60, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Arendrup, M.; Canton, E.; Cordob, S.; Dannaoui, E.; Garcia-Rodriguez, J.; Gonzalez, G.M.; Govender, N.P.; Martin-Mazuelos, E.; Lackner, M.; et al. Multicenter Study of Method-Dependent Epidemiological Cutoff Values for Detection of Resistance in Candida spp. and Aspergillus spp. to Amphotericin B and Echinocandins for the Etest Agar Diffusion Method. Antimicrob. Agents Chemother. 2016, 61, 1792–1808. [Google Scholar] [CrossRef] [PubMed]
- Dannaoui, E.; Espinel-Ingroff, A. Antifungal Susceptibly Testing by Concentration Gradient Strip Etest Method for Fungal Isolates: A Review. J. Fungi 2019, 5, 108. [Google Scholar] [CrossRef]
- CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 3rd ed.; CLSI Supplement M59; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Data from the EUCAST MIC Distribution Website. Available online: https://eucast.org/astoffungi/ (accessed on 20 January 2024).
- Espinel-Ingroff, A.; Turnidge, J.; Alastruey-Izquierdo, A.; Botterel, F.; Canton, E.; Castro, C.; Chen, Y.C.; Chen, Y.; Chryssanthou, E.; Dannaoui, E.; et al. Method-Dependent Epidemiological Cutoff Values for Detection of Triazole Resistance in Candida and Aspergillus Species for the Sensititre Yeastone Colorimetric Broth and Etest Agar Diffusion Methods. Antimicrob. Agents Chemother. 2019, 63, e01651-18. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Turnidge, J.; Alastruey-Izquierdo, A.; Dannaoui, E.; Garcia-Effron, G.; Guinea, J.; Kidd, S.; Pelaez, T.; Sanguinetti, M.; Meletiadis, J.; et al. Posaconazole MIC Distributions for Aspergillus Fumigatus Species Complex by Four Methods: Impact of Cyp51a Mutations on Estimation of Epidemiological Cutoff Values. Antimicrob. Agents Chemother. 2018, 62, e01916-17. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Sasso, M.; Turnidge, J.; Arendrup, M.; Botterel, F.; Bourgeois, N.; Bouteille, B.; Canton, E.; Cassaing, S.; Dannaoui, E.; et al. Etest ECVs/ECOFFs for Detection of Resistance in Prevalent and Three Nonprevalent Candida spp. to Triazoles and Amphotericin B and Aspergillus spp. to Caspofungin: Further Assessment of Modal Variability. Antimicrob. Agents Chemother. 2021, 65, e0109321. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. MIC Distributions and Epidemiological Cut-Off Value (ECOFF) Setting, EUCAST SOP 10.2. 2021. Available online: https://eucast.org/astoffungi/ (accessed on 20 January 2024).
- Borman, M.; Espinel-Ingroff, A. Commercial Methods for Antifungal Susceptibility Testing of Yeasts: Strengths and Limitations as Predictors of Resistance. J. Fungi 2022, 8, 309. [Google Scholar]
- Berkow, E.L.; Lockhart, S.R.; Ostrosky-Zeichner, L. Antifungal Susceptibility Testing: Current Approaches. Clin. Microbiol. Rev. 2020, 33, e00069-19. [Google Scholar] [CrossRef]
- Arastehfar, A.; Carvalho, A.; Houbraken, J.; Lombardi, L.; Garcia-Rubio, R.; Jenks, J.D.; Rivero-Menendez, O.; Aljohani, R.; Jacobsen, I.D.; Berman, J.; et al. Aspergillus Fumigatus and Aspergillosis: From Basics to Clinics. Stud. Mycol. 2021, 100, 100115. [Google Scholar] [CrossRef] [PubMed]
- Siopi, M.; Pournaras, S.; Meletiadis, J. Comparative Evaluation of Sensititre YeastOne and CLSI M38-A2 Reference Method for Antifungal Susceptibility Testing of Aspergillus spp. against Echinocandins. J. Clin. Microbiol. 2017, 55, 1714–1719. [Google Scholar] [CrossRef]
- Mello, E.; Posteraro, B.; Vella, A.; De Carolis, E.; Torelli, R.; D’Inzeo, T.; Verweij, P.E.; Sanguinetti, M. Susceptibility Testing of Common and Uncommon Aspergillus Species against Posaconazole and Other Mold-Active Antifungal Azoles Using the Sensititre Method. Antimicrob. Agents Chemother. 2017, 61, e00168-17. [Google Scholar] [CrossRef]
- Martín-Mazuelos, E.; Pemán, J.; Valverde, A.; Chaves, M.; Serrano, M.C.; Cantón, E. Comparison of the Sensititre YeastOne Colorimetric Antifungal Panel and Etest with the NCCLS M38-A Method to Determine the Activity of Amphotericin B and Itraconazole against Clinical Isolates of Aspergillus spp. J. Antimicrob. Chemother. 2003, 52, 365–370. [Google Scholar] [CrossRef]
- Torres-Narbona, M.; Guinea, J.; Martínez-Alarcón, J.; Peláez, T.; Bouza, E. In Vitro Activities of Amphotericin B, Caspofungin, Itraconazole, Posaconazole, and Voriconazole against 45 Clinical Isolates of Zygomycetes: Comparison of CLSI M38-A, Sensititre YeastOne, and the Etest. Antimicrob. Agents Chemother. 2007, 51, 1126. [Google Scholar] [CrossRef][Green Version]
- Lamoth, F.; Alexander, B.D. Comparing Etest and Broth Microdilution for Antifungal Susceptibility Testing of the Most-Relevant Pathogenic Molds. J. Clin. Microbiol. 2015, 53, 3176–3181. [Google Scholar] [CrossRef] [PubMed]
- Pinto, E.; Lago, M.; Branco, L.; Vale-Silva, L.A.; Pinheiro, M.D. Evaluation of Etest Performed in Mueller-Hinton Agar Supplemented with Glucose for Antifungal Susceptibility Testing of Clinical Isolates of Filamentous Fungi. Mycopathologia 2014, 177, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.C.; Morilla, D.; Valverde, A.; Chávez, M.; Espinel-Ingroff, A.; Claro, R.; Ramírez, M.; Martín Mazuelos, E. Comparison of Etest with Modified Broth Microdilution Method for Testing Susceptibility of Aspergillus spp. to Voriconazole. J. Clin. Microbiol. 2003, 41, 5270. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Boyken, L.; Hollis, R.J.; Diekema, D.J. In Vitro Susceptibility Testing of Filamentous Fungi: Comparison of Etest and Reference M38-A Microdilution Methods for Determining Posaconazole MICs. Diagn. Microbiol. Infect. Dis. 2003, 45, 241–244. [Google Scholar] [CrossRef]
- Martos, A.I.; Romero, A.; González, M.T.; González, A.; Serrano, C.; Castro, C.; Pemán, J.; Cantón, E.; Martn-Mazuelos, E. Evaluation of the Etest Method for Susceptibility Testing of Aspergillus spp. and Fusarium spp. to Three Echinocandins. Med. Mycol. 2010, 48, 858–861. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Verweij, P.; Nielsen, H.V. Evaluation of MIC Strip Isavuconazole Test for Susceptibility Testing of Wild-Type and Non-Wild-Type Aspergillus Fumigatus Isolates. Antimicrob. Agents Chemother. 2017, 61, e01659-16. [Google Scholar] [CrossRef] [PubMed]
- Ozkutuk, A.; Ergon, C.; Metin, D.Y.; Yucesoy, M.; Polat, S.H. Comparison of Disk Diffusion, E-Test and Broth Microdilution Test in Determination of Susceptibility of Aspergillus Species to Amphotericin B, Itraconazole and Voriconazole. J. Chemother. 2008, 20, 87–92. [Google Scholar] [CrossRef]
- Al-Hatmi, A.M.S.; Normand, A.C.; Ranque, S.; Piarroux, R.; De Hoog, G.S.; Meletiadis, J.; Meis, J.F. Comparative Evaluation of Etest, EUCAST, and CLSI Methods for Amphotericin B, Voriconazole, and Posaconazole against Clinically Relevant Fusarium Species. Antimicrob. Agents Chemother. 2016, 61, e01671-16. [Google Scholar] [CrossRef]
- Nuh, A.; Ramadan, N.; Schelenz, S.; Armstrong-James, D. Comparative Evaluation of MIRONAUT-AM and CLSI Broth Microdilution Method for Antifungal Susceptibility Testing of Aspergillus Species against Four Commonly Used Antifungals. Med. Mycol. 2020, 58, 1085–1090. [Google Scholar] [CrossRef]
- Gyurtane Szabo, N.; Joste, V.; Houzé, S.; Dannaoui, E.; Bonnal, C. Comparison of the Micronaut-AM System and the EUCAST Broth Microdilution Reference Method for MIC Determination of Four Antifungals against Aspergillus Fumigatus. J. Fungi 2023, 9, 721. [Google Scholar] [CrossRef]
- Tsitsopoulou, A.; Posso, R.; Vale, L.; Bebb, S.; Johnson, E.; White, P.L. Determination of the Prevalence of Triazole Resistance in Environmental Aspergillus Fumigatus Strains Isolated in South Wales, UK. Front. Microbiol. 2018, 9, 386222. [Google Scholar] [CrossRef]
- Serrano-Lobo, J.; Gómez, A.; Rodríguez-Sánchez, B.; Muñoz, P.; Escribano, P.; Guinea, J. Azole-Resistant Aspergillus Fumigatus Clinical Isolate Screening in Azole-Containing Agar Plates (EUCAST E.Def 10.1): Low Impact of Plastic Trays Used and Poor Performance in Cryptic Species. Antimicrob. Agents Chemother. 2021, 65, e0048221. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Principles and Procedures for the Development of Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 1st ed.; CLSI Document M57; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Arendrup, M.C.; Garcia-Effron, G.; Lass-Flörl, C.; Lopez, A.G.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M.; Perlin, D.S. Echinocandin Susceptibility Testing of Candida Species: Comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, Disk Diffusion, and Agar Dilution Methods with RPMI and IsoSensitest Media. Antimicrob. Agents Chemother. 2010, 54, 426. [Google Scholar] [CrossRef] [PubMed]
- Imhof, A.; Balajee, S.A.; Marr, K.A. New Methods to Assess Susceptibilities of Aspergillus Isolates to Caspofungin. J. Clin. Microbiol. 2003, 41, 5683–5688. [Google Scholar] [CrossRef]
- e Silva, A.P.; Miranda, I.M.; Branco, J.; Oliveira, P.; Faria-Ramos, I.; Silva, R.M.; Rodrigues, A.G.; Costa-de-Oliveira, S. FKS1 Mutation Associated with Decreased Echinocandin Susceptibility of Aspergillus Fumigatus Following Anidulafungin Exposure. Sci. Rep. 2020, 10, 1–6. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Garcia-Effron, G.; Buzina, W.; Mortensen, K.L.; Reiter, N.; Lundin, C.; Jensen, H.E.; Lass-Florl, C.; Perlin, D.S.; Bruun, B. Breakthrough Aspergillus Fumigatus and Candida Albicans Double Infection during Caspofungin Treatment: Laboratory Characteristics and Implication for Susceptibility Testing. Antimicrob. Agents Chemother. 2009, 53, 1185–1193. [Google Scholar] [CrossRef]
- Gardiner, R.E.; Souteropoulos, P.; Park, S.; Perlin, D.S. Characterization of Aspergillus Fumigatus Mutants with Reduced Susceptibility to Caspofungin. Med. Mycol. 2005, 43, S299–S305. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.M.F.; Garcia-Effron, G.; Park, S.; Perlin, D.S. A Ser678Pro Substitution in Fks1p Confers Resistance to Echinocandin Drugs in Aspergillus Fumigatus. Antimicrob. Agents Chemother. 2007, 51, 4174. [Google Scholar] [CrossRef]
- Satish, S.; Jiménez-Ortigosa, C.; Zhao, Y.; Lee, M.H.; Dolgov, E.; Krüger, T.; Park, S.; Denning, D.W.; Kniemeyer, O.; Brakhage, A.A.; et al. Stress-Induced Changes in the Lipid Microenvironment of β-(1,3)-d-Glucan Synthase Cause Clinically Important Echinocandin Resistance in Aspergillus Fumigatus. mBio 2019, 10, e00779-19. [Google Scholar] [CrossRef]
- Antachopoulos, C.; Meletiadis, J.; Sein, T.; Roilides, E.; Walsh, T.J. Concentration-Dependent Effects of Caspofungin on the Metabolic Activity of Aspergillus Species. Antimicrob. Agents Chemother. 2007, 51, 881–887. [Google Scholar] [CrossRef]
- Jimeńez-Ortigosa, C.; Aimanianda, V.; Muszkieta, L.; Mouyna, I.; Alsteens, D.; Pire, S.; Beau, R.; Krappmann, S.; Beauvais, A.; Dufrêne, Y.F.; et al. Chitin Synthases with a Myosin Motor-Like Domain Control the Resistance of Aspergillus Fumigatus to Echinocandins. Antimicrob. Agents Chemother. 2012, 56, 6121. [Google Scholar] [CrossRef] [PubMed]
- Fortwendel, J.R.; Juvvadi, P.R.; Perfect, B.Z.; Rogg, L.E.; Perfect, J.R.; Steinbach, W.J. Transcriptional Regulation of Chitin Synthases by Calcineurin Controls Paradoxical Growth of Aspergillus Fumigatus in Response to Caspofungin. Antimicrob. Agents Chemother. 2010, 54, 1555. [Google Scholar] [CrossRef] [PubMed]
- Ancuceanu, R.; Hovaneț, M.V.; Cojocaru-Toma, M.; Anghel, A.I.; Dinu, M. Potential Antifungal Targets for Aspergillus sp. from the Calcineurin and Heat Shock Protein Pathways. Int. J. Mol. Sci. 2022, 23, 12543. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Chowdhary, A. Molecular Bases of Antifungal Resistance in Filamentous Fungi. Int. J. Antimicrob. Agents 2017, 50, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Juvvadi, P.R.; Gehrke, C.; Asfaw, Y.G.; Steinbach, W.J. Transcriptional Activation of Heat Shock Protein 90 Mediated via a Proximal Promoter Region as Trigger of Caspofungin Resistance in Aspergillus Fumigatus. J. Infect. Dis. 2014, 209, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Satish, S.; Perlin, D.S. Echinocandin Resistance in Aspergillus Fumigatus Has Broad Implications for Membrane Lipid Perturbations That Influence Drug-Target Interactions. Microbiol. Insights 2019, 12, 117863611989703. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; Cuenca-Estrella, M.; Mellado, E. Triazole Resistance in Aspergillus Species: An Emerging Problem. Drugs 2017, 77, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Dabas, Y.; Xess, I.; Bakshi, S.; Mahapatra, M.; Seth, R. Emergence of Azole-Resistant Aspergillus Fumigatus from Immunocompromised Hosts in India. Antimicrob. Agents Chemother. 2018, 62, e02264-17. [Google Scholar] [CrossRef]
- Berkow, E.L.; Nunnally, N.S.; Bandea, A.; Kuykendall, R.; Beer, K.; Lockhart, S.R. Detection of TR34/L98H CYP51A Mutation through Passive Surveillance for Azole-Resistant Aspergillus Fumigatus in the United States from 2015 to 2017. Antimicrob Agents Chemother 2018, 62, e02240-17. [Google Scholar] [CrossRef]
- Wiederhold, N.P.; Gil, V.G.; Gutierrez, F.; Lindner, J.R.; Albataineh, M.T.; McCarthy, D.I.; Sanders, C.; Fan, H.; Fothergill, A.W.; Sutton, D.A. First Detection of TR34 L98H and TR46 Y121F T289A Cyp51 Mutations in Aspergillus Fumigatus Isolates in the United States. J. Clin. Microbiol. 2016, 54, 168–171. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Verweij, P.E.; Mouton, J.W.; Lagrou, K.; Meletiadis, J. Multicentre Validation of 4-Well Azole Agar Plates as a Screening Method for Detection of Clinically Relevant Azole-Resistant Aspergillus Fumigatus. J. Antimicrob. Chemother. 2017, 72, 3325–3333. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, I.; Lucio, J.; Amich, J.; Cuesta, I.; Arroyo, R.S.; Alcazar-Fuoli, L.; Mellado, E. A Cyp51B Mutation Contributes to Azole Resistance in Aspergillus Fumigatus. J. Fungi 2020, 6, 315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L.; Lv, Q.; Yan, L.; Wang, Y.; Jiang, Y. The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef]
- Buied, A.; Moore, C.B.; Denning, D.W.; Bowyer, P. High-Level Expression of Cyp51B in Azole-Resistant Clinical Aspergillus Fumigatus Isolates. J. Antimicrob. Chemother. 2013, 68, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Handelman, M.; Meir, Z.; Scott, J.; Shadkchan, Y.; Liu, W.; Ben-Ami, R.; Amich, J.; Osherov, N. Point Mutation or Overexpression of Aspergillus Fumigatus, Encoding Lanosterol 14α-Sterol Demethylase, Leads to Triazole Resistance. Antimicrob. Agents Chemother. 2021, 65, AAC0125221. [Google Scholar] [CrossRef]
- Sharma, C.; Kumar, R.; Kumar, N.; Masih, A.; Gupta, D.; Chowdhary, A. Investigation of Multiple Resistance Mechanisms in Voriconazole-Resistant Aspergillus Flavus Clinical Isolates from a Chest Hospital Surveillance in Delhi, India. Antimicrob. Agents Chemother. 2018, 62, e01928-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sun, Y.; Chen, W.; Liu, W.; Wan, Z.; Bu, D.; Li, R. The T788G Mutation in the Cyp51C Gene Confers Voriconazole Resistance in Aspergillus Flavus Causing Aspergillosis. Antimicrob. Agents Chemother. 2012, 56, 2598. [Google Scholar] [CrossRef]
- Lucio, J.; Gonzalez-Jimenez, I.; Rivero-Menendez, O.; Alastruey-Izquierdo, A.; Pelaez, T.; Alcazar-Fuoli, L.; Mellado, E. Point Mutations in the 14-α Sterol Demethylase Cyp51A or Cyp51C Could Contribute to Azole Resistance in Aspergillus Flavus. Genes 2020, 11, 1217. [Google Scholar] [CrossRef]
- Chen, M.; Zhong, G.; Wang, S.; Chen, P.; Li, L. Deletion of Cox7c Results in Pan-Azole Resistance in Aspergillus Fumigatus. Antimicrob. Agents Chemother. 2022, 66, e0015122. [Google Scholar] [CrossRef]
- Tobin, M.B.; Peery, R.B.; Skatrud, P.L. Genes Encoding Multiple Drug Resistance-like Proteins in Aspergillus Fumigatus and Aspergillus Flavus. Gene 1997, 200, 11–23. [Google Scholar] [CrossRef]
- Chowdhary, A.; Sharma, C.; Hagen, F.; Meis, J.F. Exploring Azole Antifungal Drug Resistance in Aspergillus Fumigatus with Special Reference to Resistance Mechanisms. Future Med. 2014, 9, 697–711. [Google Scholar] [CrossRef] [PubMed]
- da Silva Ferreira, M.E.; Malavazi, I.; Savoldi, M.; Brakhage, A.A.; Goldman, M.H.S.; Kim, H.S.; Nierman, W.C.; Goldman, G.H. Transcriptome Analysis of Aspergillus Fumigatus Exposed to Voriconazole. Curr. Genet. 2006, 50, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, M.G.; Bromley, M.; Buied, A.; Moore, C.B.; Rajendran, R.; Rautemaa, R.; Ramage, G.; Denning, D.W.; Bowyer, P. The Cdr1B Efflux Transporter Is Associated with Non-Cyp51a-Mediated Itraconazole Resistance in Aspergillus Fumigatus. J. Antimicrob. Chemother. 2013, 68, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Camps, S.M.T.; Dutilh, B.E.; Arendrup, M.C.; Rijs, A.J.M.M.; Snelders, E.; Huynen, M.A.; Verweij, P.E.; Melchers, W.J.G. Discovery of a HapE Mutation That Causes Azole Resistance in Aspergillus Fumigatus through Whole Genome Sequencing and Sexual Crossing. PLoS ONE 2012, 7, e50034. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, P.; Gao, R.; Li, Y.; Zhang, A.; Liu, F.; Lu, L. Screening and Characterization of a Non-Cyp51A Mutation in an Aspergillus Fumigatus Cox10 Strain Conferring Azole Resistance. Antimicrob. Agents Chemother. 2017, 61, e02101-16. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.R.H.; Bicanic, T.; Salim, R.; Hope, W. Liposomal Amphotericin B (AmBisome®): A Review of the Pharmacokinetics, Pharmacodynamics, Clinical Experience and Future Directions. Drugs 2016, 76, 485. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Dietl, A.M.; Kontoyiannis, D.P.; Brock, M. Aspergillus Terreus Species Complex. Clin. Microbiol. Rev. 2021, 34, e0031120. [Google Scholar] [CrossRef]
- Blum, G.; Hörtnagl, C.; Jukic, E.; Erbeznik, T.; Pümpel, T.; Dietrich, H.; Nagl, M.; Speth, C.; Rambach, G.; Lass-Flörl, C. New Insight into Amphotericin B Resistance in Aspergillus Terreus. Antimicrob. Agents Chemother. 2013, 57, 1583. [Google Scholar] [CrossRef]
- Fakhim, H.; Badali, H.; Dannaoui, E.; Nasirian, M.; Jahangiri, F.; Raei, M.; Vaseghi, N.; Ahmadikia, K.; Vaezi, A. Trends in the Prevalence of Amphotericin B-Resistance (AmBR) among Clinical Isolates of Aspergillus Species. J. Mycol. Med. 2022, 32, 101310. [Google Scholar] [CrossRef]
- Walsh, T.J.; Petraitis, V.; Petraitiene, R.; Field-Ridley, A.; Sutton, D.; Ghannoum, M.; Sein, T.; Schaufele, R.; Peter, J.; Bacher, J.; et al. Experimental Pulmonary Aspergillosis Due to Aspergillus Terreus: Pathogenesis and Treatment of an Emerging Fungal Pathogen Resistant to Amphotericin B. J. Infect. Dis. 2003, 188, 305–319. [Google Scholar] [CrossRef]
- Fan, Y.; Korfanty, G.A.; Xu, J. Genetic Analyses of Amphotericin b Susceptibility in Aspergillus Fumigatus. J. Fungi 2021, 7, 860. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.M.; Shi, G.H.; Dai, Y.; Fang, W.X.; Wu, Q. Identifying Genetic Variants Associated with Amphotericin B (AMB) Resistance in Aspergillus Fumigatus via k-Mer—Based GWAS. Front. Genet. 2023, 14, 1133593. [Google Scholar] [CrossRef]
- Hadrich, I.; Makni, F.; Neji, S.; Cheikhrouhou, F.; Bellaaj, H.; Elloumi, M.; Ayadi, A.; Ranque, S. Amphotericin B in Vitro Resistance Is Associated with Fatal Aspergillus Flavus Infection. Med. Mycol. 2012, 50, 829–834. [Google Scholar] [CrossRef]
- Seo, K.; Akiyoshi, H.; Ohnishi, Y. Alteration of Cell Wall Composition Leads to Amphotericin B Resistance in Aspergillus Flavus. Microbiol. Immunol. 1999, 43, 1017–1025. [Google Scholar] [CrossRef]
- Chowdhary, A.; Masih, A.; Sharma, C. Azole Resistance in Moulds—Approach to Detection in a Clinical Laboratory. Curr. Fungal Infect. Rep. 2016, 10, 96–106. [Google Scholar] [CrossRef]
- Kathuria, S.; Sharma, C.; Singh, P.K.; Agarwal, P.; Agarwal, K.; Hagen, F.; Meis, J.F.; Chowdhary, A. Molecular Epidemiology and In-Vitro Antifungal Susceptibility of Aspergillus Terreus Species Complex Isolates in Delhi, India: Evidence of Genetic Diversity by Amplified Fragment Length Polymorphism and Microsatellite Typing. PLoS ONE 2015, 10, e0118997. [Google Scholar] [CrossRef]
- Jukic, E.; Blatzer, M.; Posch, W.; Steger, M.; Binder, U.; Lass-Flörl, C.; Wilflingseder, D. Oxidative Stress Response Tips the Balance in Aspergillus Terreus Amphotericin B Resistance. Antimicrob. Agents Chemother. 2017, 61, e00670-17. [Google Scholar] [CrossRef]
- Blum, G.; Perkhofer, S.; Haas, H.; Schrettl, M.; Würzner, R.; Dierich, M.P.; Lass-Flörl, C. Potential Basis for Amphotericin B Resistance in Aspergillus Terreus. Antimicrob. Agents Chemother. 2008, 52, 1553–1555. [Google Scholar] [CrossRef]
- Burgel, P.R.; Baixench, M.T.; Amsellem, M.; Audureau, E.; Chapron, J.; Kanaan, R.; Honoré, I.; Dupouy-Camet, J.; Dusser, D.; Klaassen, C.H.; et al. High Prevalence of Azole-Resistant Aspergillus Fumigatus in Adults with Cystic Fibrosis Exposed to Itraconazole. Antimicrob. Agents Chemother. 2012, 56, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Kano, R.; Sobukawa, H.; Murayama, S.Y.; Hirose, D.; Tanaka, Y.; Kosuge, Y.; Hasegawa, A.; Kamata, H. In Vitro Resistance of Aspergillus Fumigatus to Azole Farm Fungicide. J. Infect. Chemother. 2016, 22, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Siopi, M.; Perlin, D.S.; Arendrup, M.C.; Pournaras, S.; Meletiadis, J. Comparative Pharmacodynamics of Echinocandins against Aspergillus Fumigatus Using an In Vitro Pharmacokinetic/Pharmacodynamic Model That Correlates with Clinical Response to Caspofungin Therapy: Is There a Place for Dose Optimization? Antimicrob. Agents Chemother. 2021, 65, e01618-20. [Google Scholar] [CrossRef] [PubMed]
- Aruanno, M.; Glampedakis, E.; Lamoth, F. Echinocandins for the Treatment of Invasive Aspergillosis: From Laboratory to Bedside. Antimicrob. Agents Chemother. 2019, 63, e00399-1. [Google Scholar] [CrossRef] [PubMed]
- Lass-Flörl, C.; Arendrup, M.C.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M.; Donnelly, P.; Hope, W. EUCAST Technical Note on Amphotericin B. Clin. Microbiol. Infect. 2011, 17, E27–E29. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Cuenca-Estrella, M.; Fothergill, A.; Fuller, J.; Ghannoum, M.; Johnson, E.; Pelaez, T.; Pfaller, M.A.; Turnidge, J. Wild-Type MIC Distributions and Epidemiological Cutoff Values for Amphotericin B and Aspergillus spp. for the CLSI Broth Microdilution Method (M38-A2 Document). Antimicrob. Agents Chemother. 2011, 55, 5150–5154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Hsieh, M.I.; Choi, P.C.; Wu, C.J. Comparison of the Sensititre YeastOne and CLSI M38-A2 Microdilution Methods in Determining the Activity of Amphotericin B, Itraconazole, Voriconazole, and Posaconazole against Aspergillus Species. J. Clin. Microbiol. 2018, 56, e00780-18. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.J.; Subedi, S.; Halliday, C.L.; Hibbs, D.E.; Lai, F.; Lopez-Ruiz, F.J.; Harper, L.; Park, R.F.; Cuddy, W.S.; Biswas, C.; et al. Surveillance for Azole Resistance in Clinical and Environmental Isolates of Aspergillus Fumigatus in Australia and Cyp51A Homology Modelling of Azole-Resistant Isolates. J. Antimicrob. Chemother. 2018, 73, 2347–2351. [Google Scholar] [CrossRef]
- Chen, Y.C.; Kuo, S.F.; Wang, H.C.; Wu, C.J.; Lin, Y.S.; Li, W.S.; Lee, C.H. Azole Resistance in Aspergillus Species in Southern Taiwan: An Epidemiological Surveillance Study. Mycoses 2019, 62, 1174–1181. [Google Scholar] [CrossRef]
- Guinea, J.; Verweij, P.E.; Meletiadis, J.; Mouton, J.W.; Barchiesi, F.; Arendrup, M.C.; Arikan-Akdagli, S.; Castanheira, M.; Chryssanthou, E.; Friberg, N.; et al. How to: EUCAST Recommendations on the Screening Procedure E.Def 10.1 for the Detection of Azole Resistance in Aspergillus Fumigatus Isolates Using Four-Well Azole-Containing Agar Plates. Clin. Microbiol. Infect. 2019, 25, 681–687. [Google Scholar] [CrossRef]
- Buil, J.B.; Van Der Lee, H.A.L.; Rijs, A.J.M.M.; Zoll, J.; Hovestadt, J.A.M.F.; Melchers, W.J.G.; Verweij, P.E. Single-Center Evaluation of an Agar-Based Screening for Azole Resistance in Aspergillus Fumigatus by Using VIPcheck. Antimicrob. Agents Chemother. 2017, 61, e01250-17. [Google Scholar] [CrossRef]
- van der Linden, J.W.M.; Arendrup, M.C.; Warris, A.; Lagrou, K.; Pelloux, H.; Hauser, P.M.; Chryssanthou, E.; Mellado, E.; Kidd, S.E.; Tortorano, A.M.; et al. Prospective Multicenter International Surveillance of Azole Resistance in Aspergillus Fumigatus. Emerg. Infect. Dis. 2015, 21, 1041. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, K.L.; Mellado, E.; Lass-Flörl, C.; Rodriguez-Tudela, J.L.; Johansen, H.K.; Arendrup, M.C. Environmental Study of Azole-Resistant Aspergillus Fumigatus and Other Aspergilli in Austria, Denmark, and Spain. Antimicrob. Agents Chemother. 2010, 54, 4545–4549. [Google Scholar] [CrossRef] [PubMed]
- Abdolrasouli, A.; Scourfield, A.; Rhodes, J.; Shah, A.; Elborn, J.S.; Fisher, M.C.; Schelenz, S.; Armstrong-James, D. High Prevalence of Triazole Resistance in Clinical Aspergillus Fumigatus Isolates in a Specialist Cardiothoracic Centre. Int. J. Antimicrob. Agents 2018, 52, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Lestrade, P.P.; van der Velden, W.J.F.M.; Bouwman, F.; Stoop, F.J.; Blijlevens, N.M.A.; Melchers, W.J.G.; Verweij, P.E.; Donnelly, J.P. Epidemiology of Invasive Aspergillosis and Triazole-Resistant Aspergillus Fumigatus in Patients with Haematological Malignancies: A Single-Centre Retrospective Cohort Study. J. Antimicrob. Chemother. 2018, 73, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).