Role of Flavohemoglobins in the Development and Aflatoxin Biosynthesis of Aspergillus flavus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. FHb Sequence Analysis
2.3. FHb Deletion and Overexpression
2.4. qRT-PCR and Gene Expression Analysis
2.5. Phenotypic Characterization
2.6. Abiotic Stress Assay
2.7. Pathogenicity Assay
2.8. Aflatoxin Extraction
2.9. UPLC-MS/MS Assay
2.10. NO Detection Assay
2.11. Statistical Analysis
3. Results
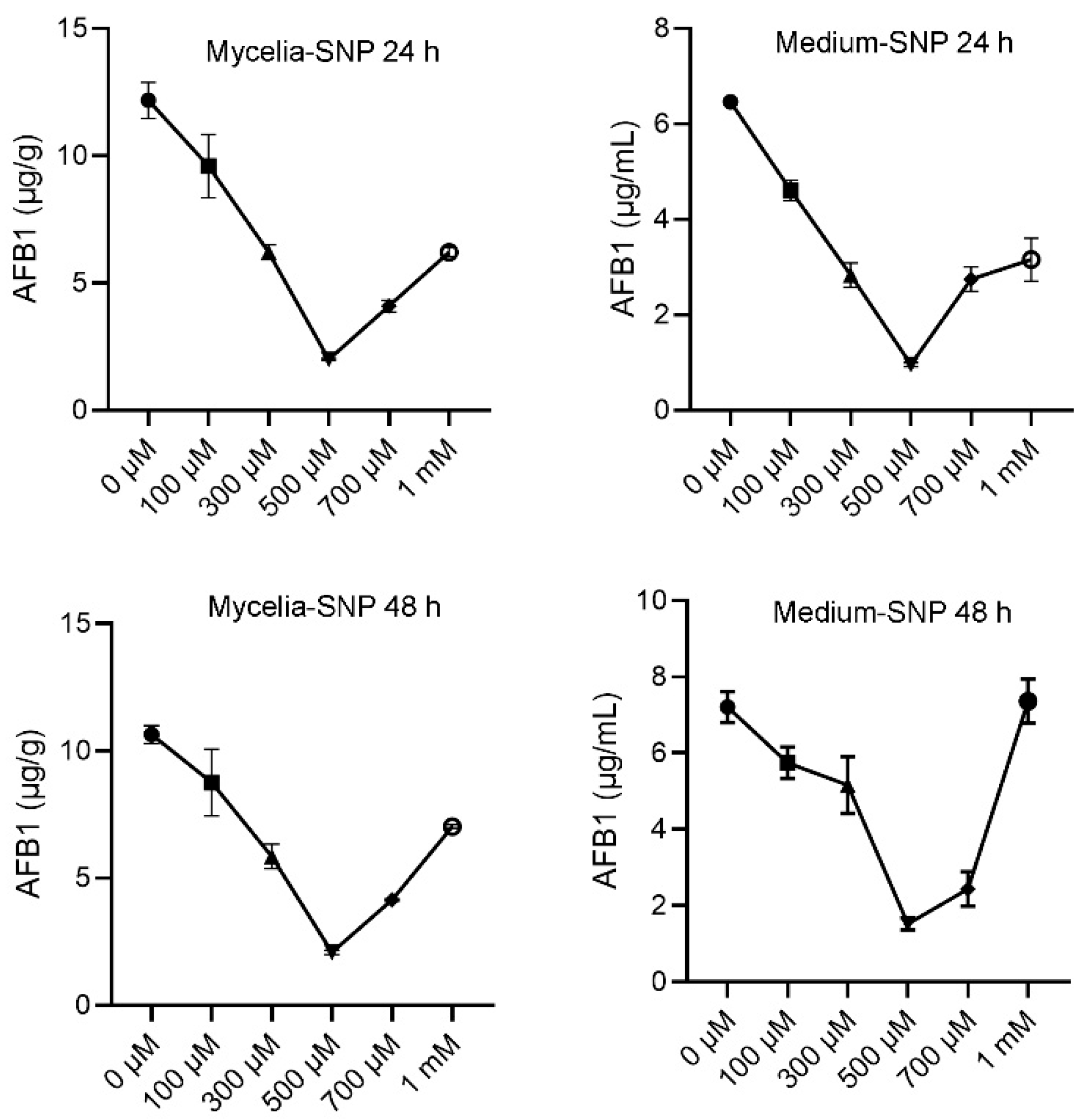
3.1. Exogenous NO Affects Aflatoxins Production in A. flavus
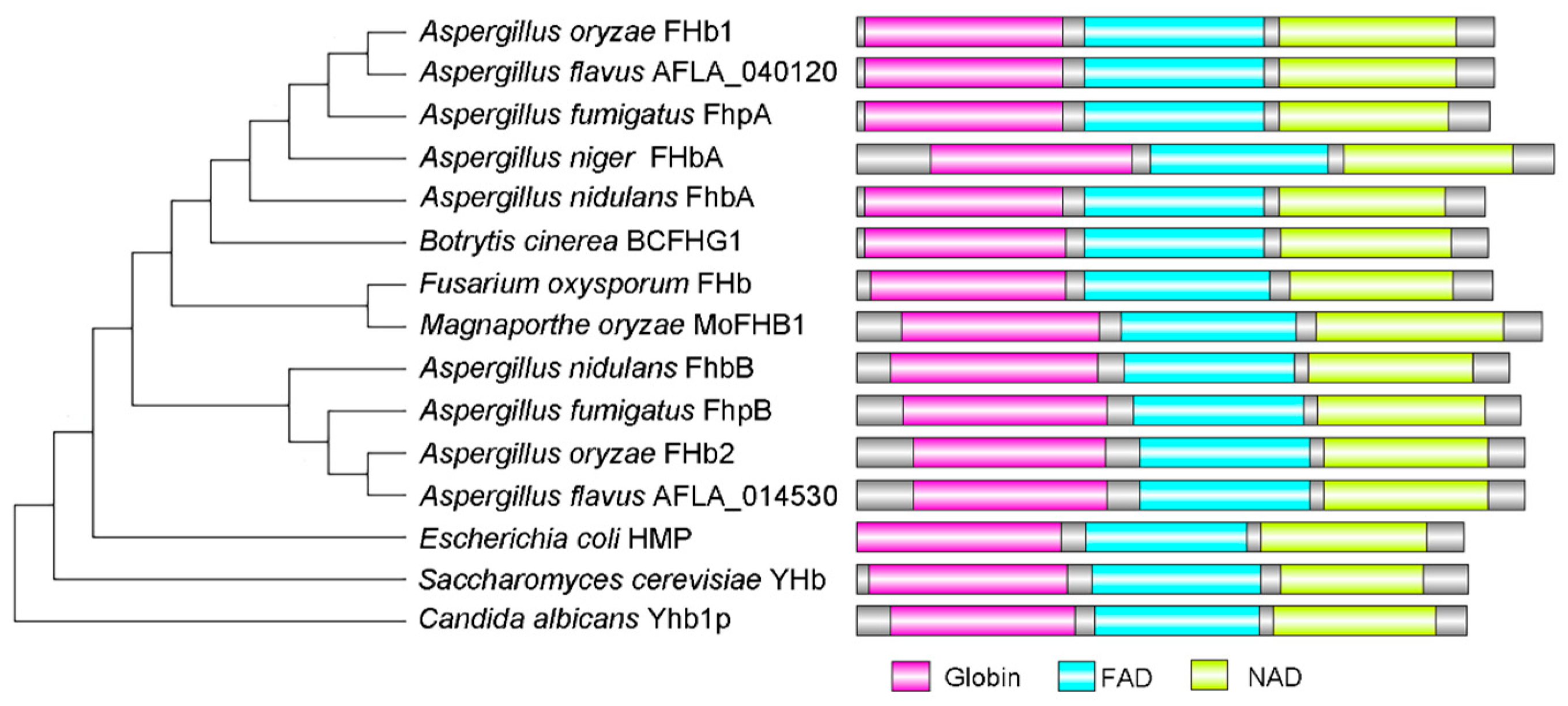
3.2. Characterization of Flavohemoglobins in A. flavus
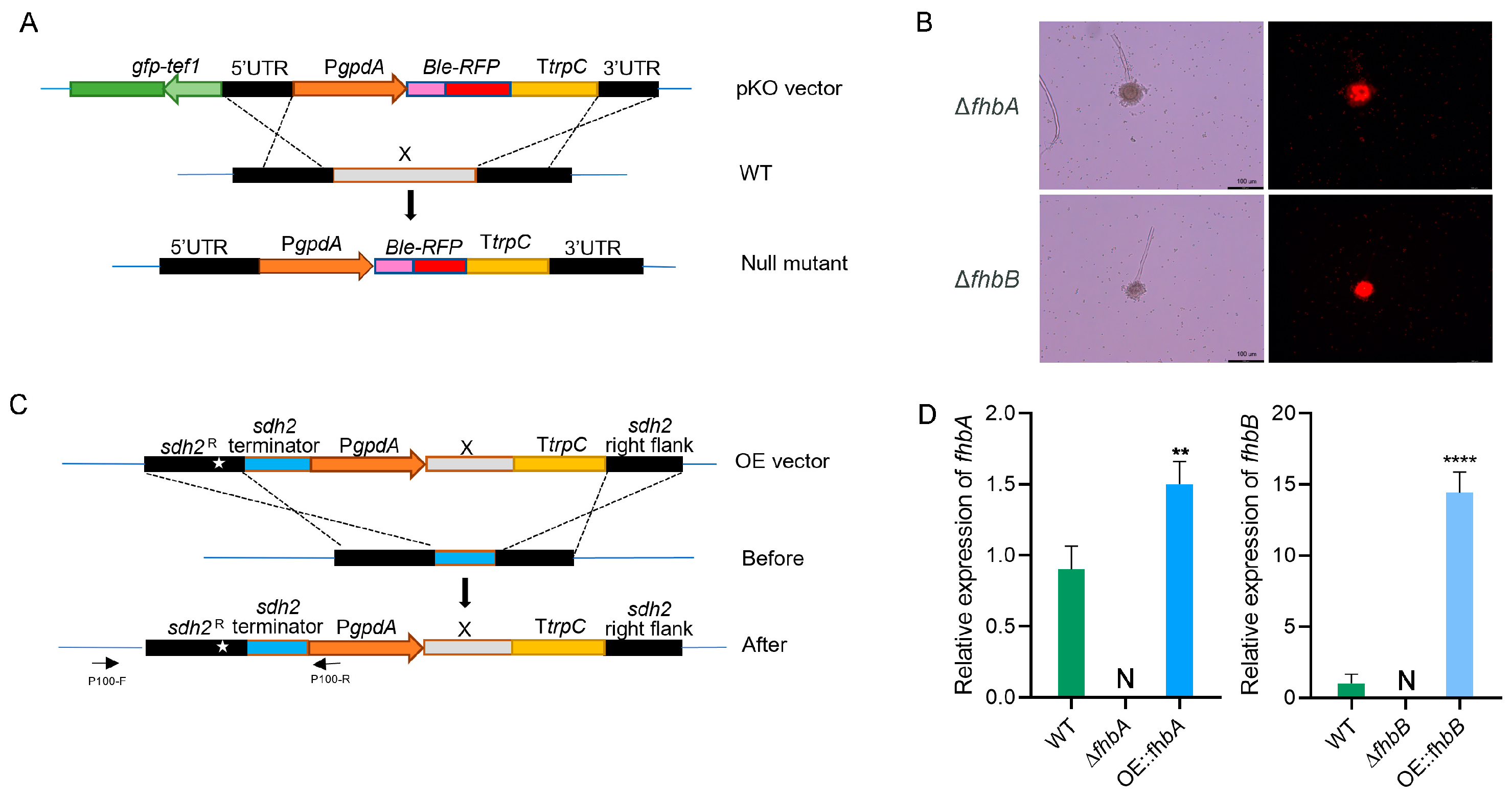
3.3. Construction of fhbA and fhbB Deletion and Overexpression Mutants
3.4. fhbA Plays a Key Role in Sustaining NO Homeostasis in A. flavus
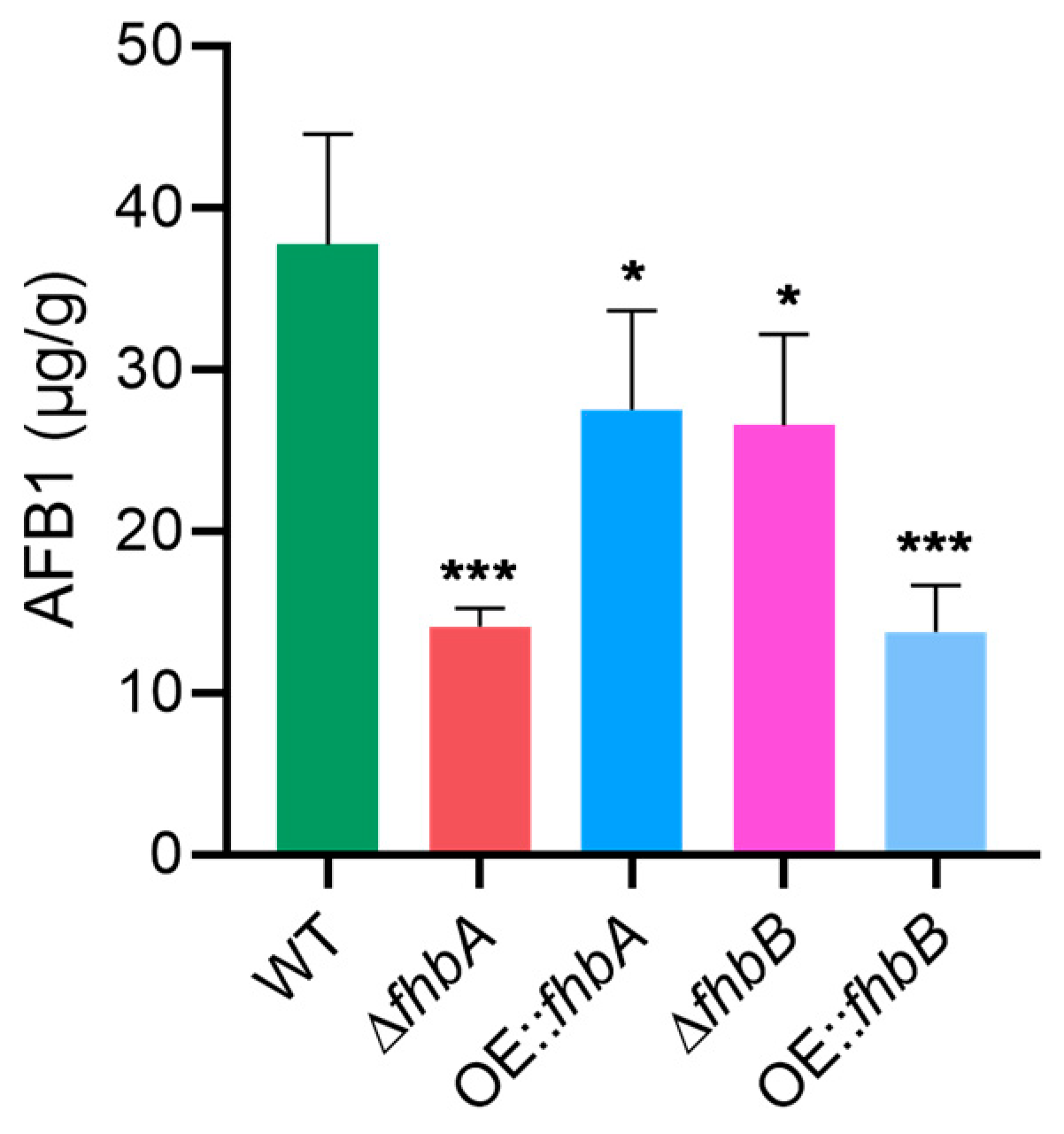
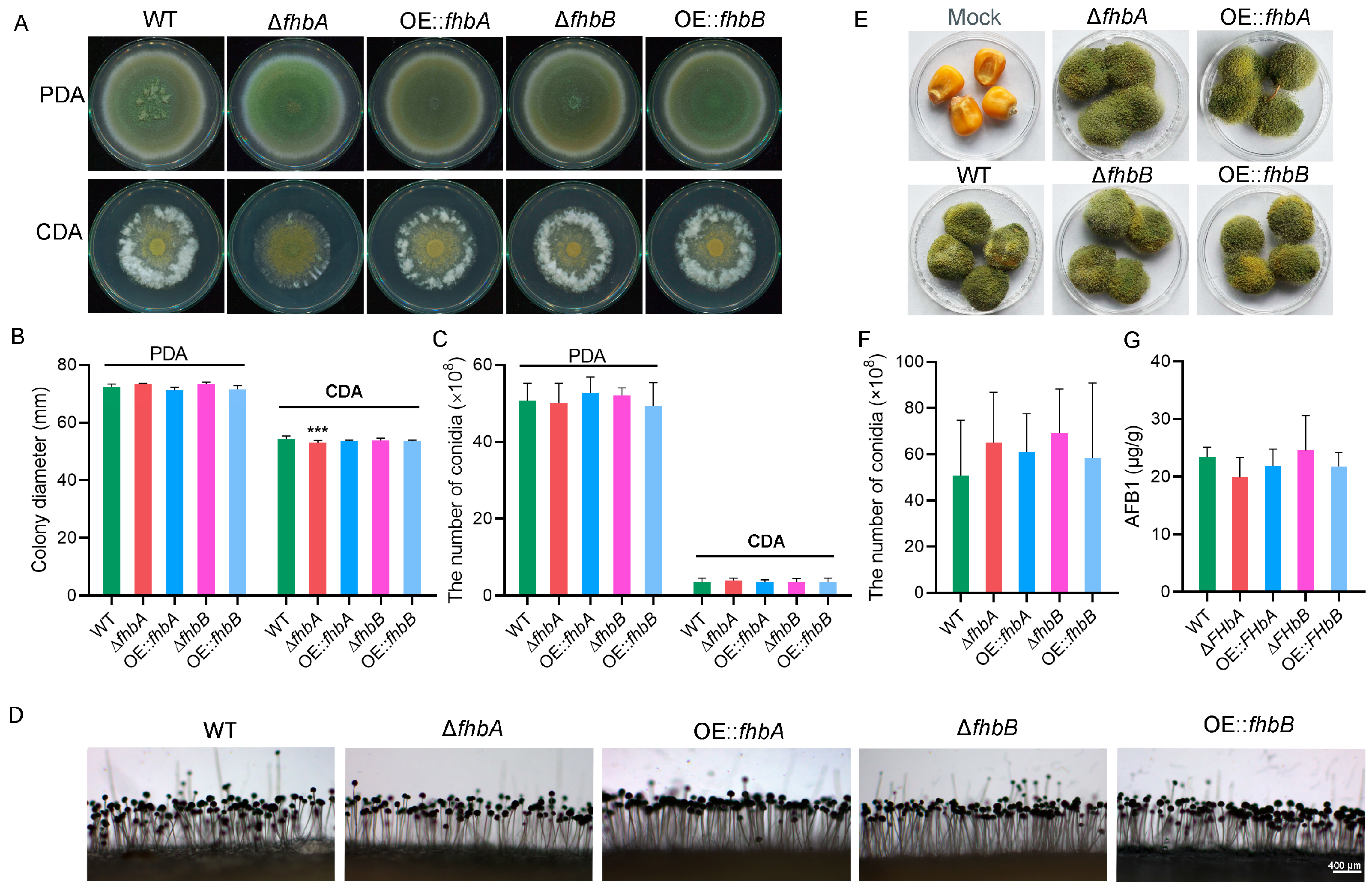
3.5. Different Role of fhbA and fhbB in the Biosynthesis of AFB1
3.6. fhbA Affects Mycelial Growth
3.7. fhbA Promotes Conidial Germination
3.8. fhbs Involved in Sclerotial Development
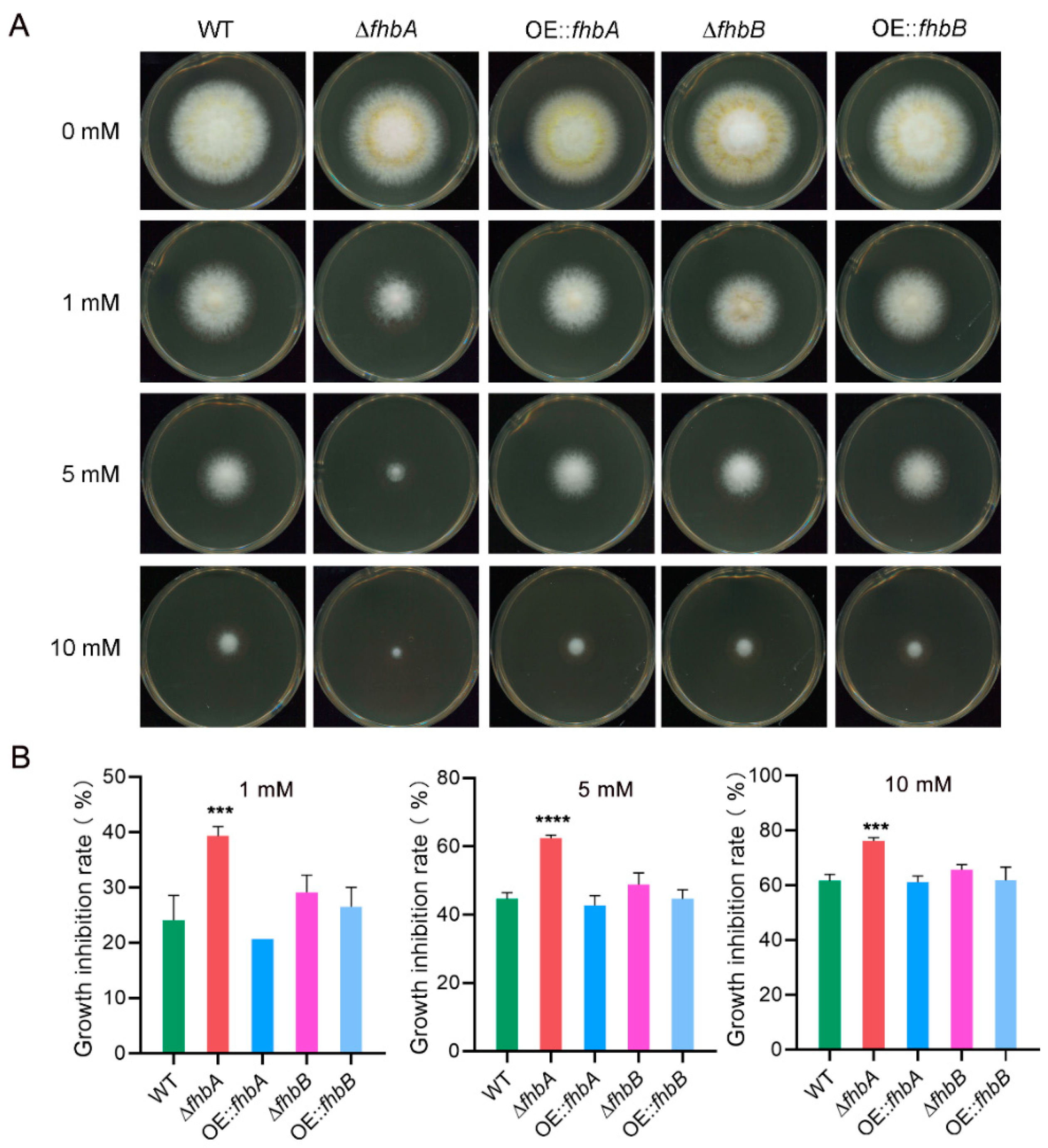
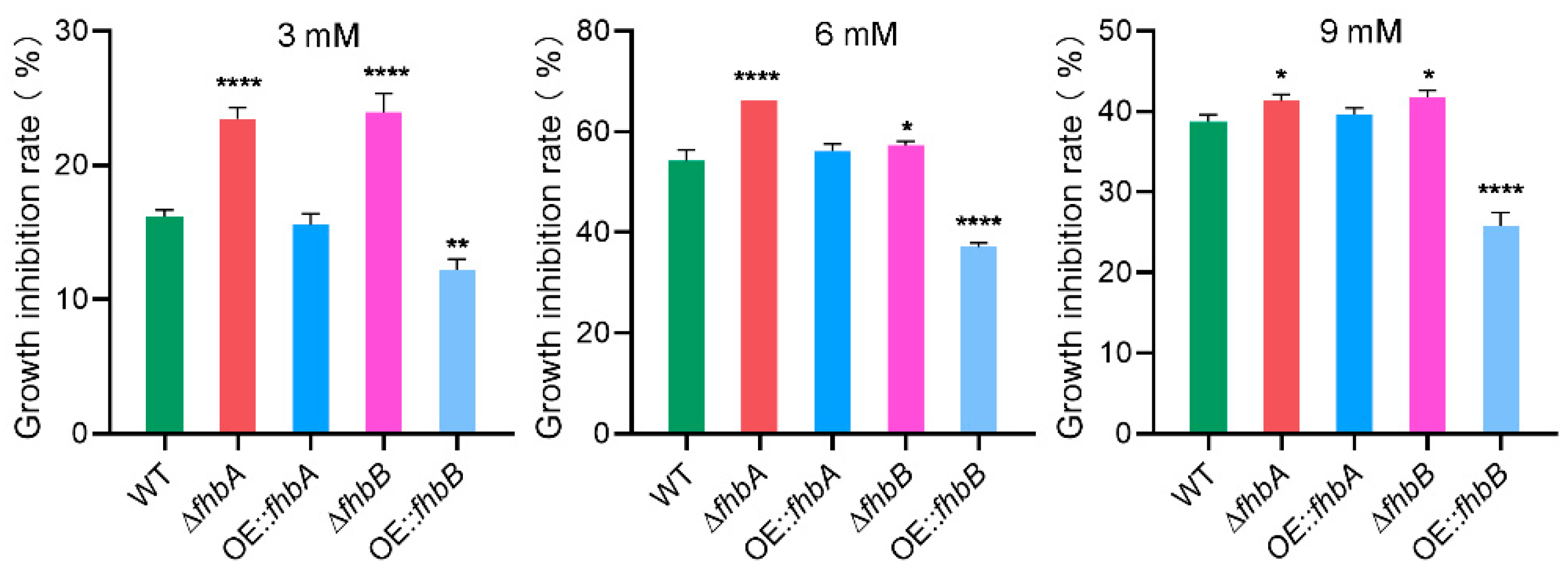
3.9. fhbA and fhbB Response to Oxidative Stress
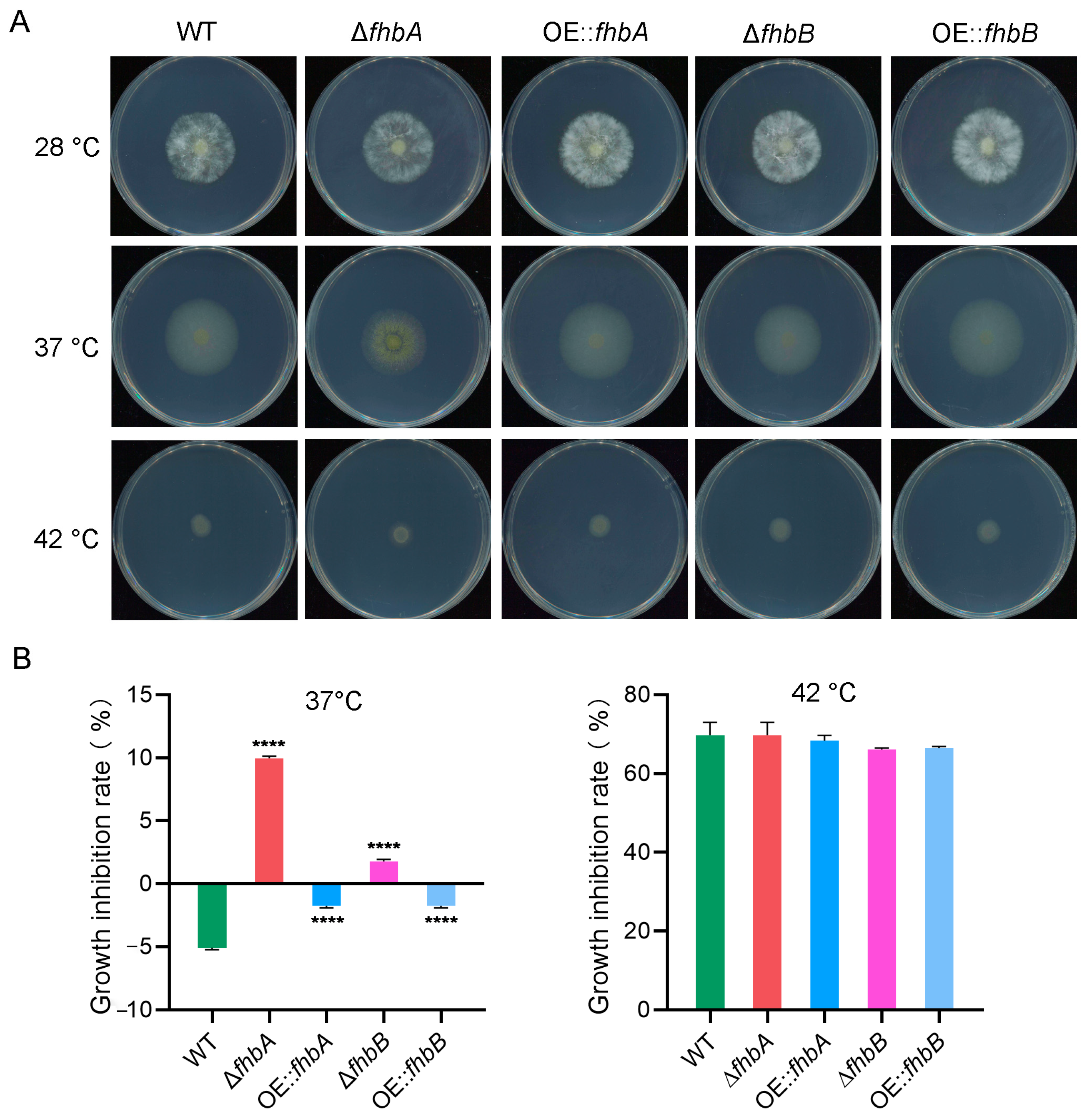
3.10. Both fhbA and fhbB Response to High Temperatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans; International Agency for Research on Cancer; World Health Organization. Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene (No.82); World Health Organization: Geneva, Switzerland, 2002. [Google Scholar]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Aflatoxins: A global concern for food safety, uuman health and their management. Front. Microbiol. 2016, 7, 2170. [Google Scholar] [CrossRef] [PubMed]
- Mahato, D.K.; Lee, K.E.; Kamle, M.; Devi, S.; Dewangan, K.N.; Kumar, P.; Kang, S.G. Aflatoxins in food and feed: An overview on prevalence, detection and control strategies. Front. Microbiol. 2019, 10, 2266. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.O.; Weitzberg, E. Nitric oxide signaling in health and disease. Cell 2022, 185, 2853–2878. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, L.; Wang, W.; Gao, H. Nitric oxide, nitric oxide formers and their physiological impacts in bacteria. Int. J. Mol. Sci. 2022, 23, 10778. [Google Scholar] [CrossRef] [PubMed]
- Brouquisse, R. Multifaceted roles of nitric oxide in plants. J. Exp. Bot. 2019, 70, 4319–4322. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tang, R.H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jing, L.; Yang, Z.; Chen, L.; Guo, F.; et al. Nitric oxide represses the Arabidopsis floral transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, D.; Marcos, J.F.; Marcos, A.T.; Strauss, J. Nitric oxide in fungi: Is there NO light at the end of the tunnel? Curr. Genet. 2016, 62, 513–518. [Google Scholar] [CrossRef]
- Astuti, R.I.; Nasuno, R.; Takagi, H. Nitric oxide signalling in yeast. In Nitric Oxide and Other Small Signalling Molecules; Advances in Microbial Physiology; Academic Press: Cambridge, MA, USA, 2018; pp. 29–63. [Google Scholar]
- Zhao, Y.; Lim, J.; Xu, J.; Yu, J.H.; Zheng, W. Nitric oxide as a developmental and metabolic signal in filamentous fungi. Mol. Microbiol. 2020, 113, 872–882. [Google Scholar] [CrossRef]
- Pengkit, A.; Jeon, S.S.; Son, S.J.; Shin, J.H.; Baik, K.Y.; Choi, E.H.; Park, G. Identification and functional analysis of endogenous nitric oxide in a filamentous fungus. Sci. Rep. 2016, 6, 30037. [Google Scholar] [CrossRef]
- Baidya, S.; Cary, J.W.; Grayburn, W.S.; Calvo, A.M. Role of nitric oxide and flavohemoglobin homolog genes in Aspergillus nidulans sexual development and mycotoxin production. Appl. Environ. Microbiol. 2011, 77, 5524–5528. [Google Scholar] [CrossRef] [PubMed]
- Turrion-Gomez, J.L.; Benito, E.P. Flux of nitric oxide between the necrotrophic pathogen Botrytis cinerea and the host plant. Mol. Plant Pathol. 2011, 12, 606–616. [Google Scholar] [CrossRef]
- Samalova, M.; Johnson, J.; Illes, M.; Kelly, S.; Fricker, M.; Gurr, S. Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytol. 2013, 197, 207–222. [Google Scholar] [CrossRef]
- Astuti, R.I.; Watanabe, D.; Takagi, H. Nitric oxide signaling and its role in oxidative stress response in Schizosaccharomyces pombe. Nitric Oxide 2016, 52, 29–40. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, S.; Wu, J.; Sun, X.; Ma, A. Heat stress in macrofungi: Effects and response mechanisms. Appl. Microbiol. Biotechnol. 2021, 105, 7567–7576. [Google Scholar] [CrossRef]
- Ridnour, L.A.; Thomas, D.D.; Mancardi, D.; Espey, M.G.; Miranda, K.M.; Paolocci, N.; Feelisch, M.; Fukuto, J.; Wink, D.A. The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species. Putting perspective on stressful biological situations. Biol. Chem. 2004, 385, 1–10. [Google Scholar] [CrossRef]
- Ilari, A.; Boffi, A. Structural studies on flavohemoglobins. Methods Enzymol. 2008, 436, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Forrester, M.T.; Foster, M.W. Protection from nitrosative stress: A central role for microbial flavohemoglobin. Free Radic. Biol. Med. 2012, 52, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Dou, Y.; Li, T.; Olson, J.S.; Zhu, H.; Riggs, A.F. Nitric-oxide dioxygenase activity and function of flavohemoglobins. sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 2000, 275, 31581–31587. [Google Scholar] [CrossRef]
- Liu, L.; Zeng, M.; Hausladen, A.; Heitman, J.; Stamler, J.S. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 2000, 97, 4672–4676. [Google Scholar] [CrossRef]
- Nakano, M.M. Essential role of flavohemoglobin in long-term anaerobic survival of Bacillus subtilis. J. Bacteriol. 2006, 188, 6415–6418. [Google Scholar] [CrossRef] [PubMed]
- Cassanova, N.; O’Brien, K.M.; Stahl, B.T.; McClure, T.; Poyton, R.O. Yeast flavohemoglobin, a nitric oxide oxidoreductase, is located in both the cytosol and the mitochondrial matrix: Effects of respiration, anoxia, and the mitochondrial genome on its intracellular level and distribution. J. Biol. Chem. 2005, 280, 7645–7653. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.J.; Raitt, D.; Burke, P.V.; Clewell, A.S.; Kwast, K.E.; Poyton, R.O. Function and expression of flavohemoglobin in Saccharomyces cerevisiae. Evidence for a role in the oxidative stress response. J. Biol. Chem. 1996, 271, 25131–25138. [Google Scholar] [CrossRef]
- Ullmann, B.D.; Myers, H.; Chiranand, W.; Lazzell, A.L.; Zhao, Q.; Vega, L.A.; Lopez-Ribot, J.L.; Gardner, P.R.; Gustin, M.C. Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot. Cell 2004, 3, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Fushinobu, S.; Kim, S.W.; Nakanishi, Y.; Wakagi, T.; Shoun, H. Aspergillus oryzae flavohemoglobins promote oxidative damage by hydrogen peroxide. Biochem. Biophys. Res. Commun. 2010, 394, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Schinko, T.; Berger, H.; Lee, W.; Gallmetzer, A.; Pirker, K.; Pachlinger, R.; Buchner, I.; Reichenauer, T.; Güldener, U.; Strauss, J. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol. Microbiol. 2010, 78, 720–738. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Li, R.W.; Arroyo-Manzanares, N.; De Saeger, S.; Diana Di Mavungu, J. Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 2017, 104, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Zhao, Q.; Pei, H.; Zhou, X.; Zhao, K.; Yu, M.; Han, G.; Fan, J.; Tao, F. Systematic characterization of bZIP transcription factors required for development and aflatoxin generation by high-throughput gene knockout in Aspergillus flavus. J. Fungi 2022, 8, 356. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Shao, Q.; Li, C.; Zhao, K.; Jiang, L.; Fan, J.; Jiang, H.; Tao, F. An efficient Agrobacterium-mediated transformation method for aflatoxin generation fungus Aspergillus flavus. J. Microbiol. 2018, 56, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Zhao, K.; Zhao, Q.; Xiang, F.; Han, G. A novel site-specific integration system for genetic modification of Aspergillus flavus. G3-Genes Genomes Genet. 2020, 10, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhou, X.; Chen, D.; Jiao, Y.; Han, G.; Tao, F. HacA, a key transcription factor for the unfolded protein response, is required for fungal development, aflatoxin biosynthesis and pathogenicity of Aspergillus flavus. Int. J. Food Microbiol. 2024, 417, 110693. [Google Scholar] [CrossRef] [PubMed]
- Zoupa, E.; Pitsikas, N. The nitric oxide (NO) donor sodium nitroprusside (SNP) and its potential for the schizophrenia therapy: Lights and shadows. Molecules 2021, 26, 3196. [Google Scholar] [CrossRef] [PubMed]
- Xiang, F.; Zhao, Q.; Zhao, K.; Pei, H.; Tao, F. The efficacy of composite essential oils against aflatoxigenic fungus Aspergillus flavus in Maize. Toxins 2020, 12, 562. [Google Scholar] [CrossRef] [PubMed]
- Bonamore, A.; Boffi, A. Flavohemoglobin: Structure and reactivity. IUBMB Life 2008, 60, 19–28. [Google Scholar] [CrossRef] [PubMed]
- de Jesús-Berríos, M.; Liu, L.; Nussbaum, J.C.; Cox, G.M.; Stamler, J.S.; Heitman, J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 2003, 13, 1963–1968. [Google Scholar] [CrossRef] [PubMed]
- Turrion-Gomez, J.L.; Eslava, A.P.; Benito, E.P. The flavohemoglobin BCFHG1 is the main NO detoxification system and confers protection against nitrosative conditions but is not a virulence factor in the fungal necrotroph Botrytis cinerea. Fungal Genet. Biol. 2010, 47, 484–496. [Google Scholar] [CrossRef]
- Zhou, S.; Fushinobu, S.; Nakanishi, Y.; Kim, S.W.; Wakagi, T.; Shoun, H. Cloning and characterization of two flavohemoglobins from Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2009, 381, 7–11. [Google Scholar] [CrossRef]
- Zhou, S.; Fushinobu, S.; Kim, S.W.; Nakanishi, Y.; Maruyama, J.; Kitamoto, K.; Wakagi, T.; Shoun, H. Functional analysis and subcellular location of two flavohemoglobins from Aspergillus oryzae. Fungal Genet. Biol. 2011, 48, 200–207. [Google Scholar] [CrossRef] [PubMed]
- te Biesebeke, R.; Levasseur, A.; Boussier, A.; Record, E.; van den Hondel, C.A.; Punt, P.J. Phylogeny of fungal hemoglobins and expression analysis of the Aspergillus oryzae flavohemoglobin gene fhbA during hyphal growth. Fungal Biol. 2010, 114, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hao, Z.; Chai, R.; Qiu, H.; Wang, J.; Wang, Y.; Sun, G. The flavohemoglobin gene MoFHB1 is involved in the endurance against nitrosative stress in Magnaporthe oryzae. FEMS Microbiol. Lett. 2022, 369, fnab162. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Luo, Y.; Sun, T.; Qiu, H.; Geng, Q.; Li, Y.; Liu, M.; Keller, N.P.; Song, F.; Tian, J. Nitric oxide-mediated regulation of Aspergillus flavus asexual development by targeting TCA cycle and mitochondrial function. J. Hazard. Mater. 2024, 471, 134385. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Kawahara, N.; Takagi, H. The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 2013, 430, 137–143. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Chen, D.; Yu, M.; Jiao, Y.; Tao, F. Role of Flavohemoglobins in the Development and Aflatoxin Biosynthesis of Aspergillus flavus. J. Fungi 2024, 10, 437. https://doi.org/10.3390/jof10060437
Zhou X, Chen D, Yu M, Jiao Y, Tao F. Role of Flavohemoglobins in the Development and Aflatoxin Biosynthesis of Aspergillus flavus. Journal of Fungi. 2024; 10(6):437. https://doi.org/10.3390/jof10060437
Chicago/Turabian StyleZhou, Xiaoling, Dongyue Chen, Min Yu, Yuan Jiao, and Fang Tao. 2024. "Role of Flavohemoglobins in the Development and Aflatoxin Biosynthesis of Aspergillus flavus" Journal of Fungi 10, no. 6: 437. https://doi.org/10.3390/jof10060437




