Diversity and Composition of Fungicolous Fungi Residing in Macrofungi from the Qinling Mountains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. PCR Amplification and Sequencing of Internal Transcribed Spacer Regions
2.4. Preparation of Metabarcoding Libraries and Sequencing
2.5. Bioinformatic Processing and Analysis of Sequencing Data
2.6. Compilation of Metadata
2.7. Statistical Analyses
3. Results
3.1. Identification of Collected Macrofungi
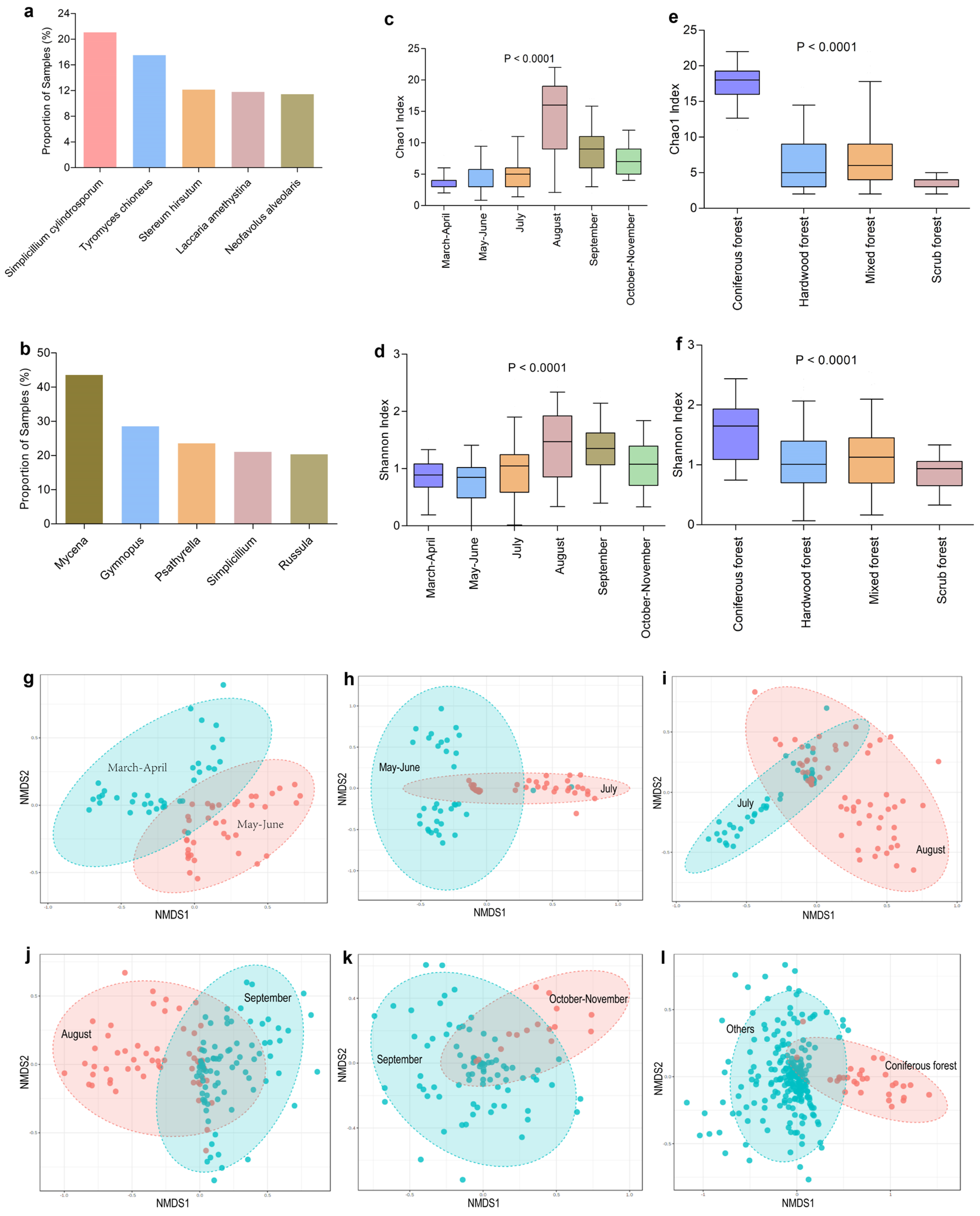
3.2. Distribution of Fungicolous Fungi in Macrofungal Samples
3.3. Diversity and Composition of Fungicolous Fungal Communities Varied with Months of Collection and the Habitats of Host Fungi
3.4. Trophic Modes of Host Macrofungi and Fungicolous Fungi
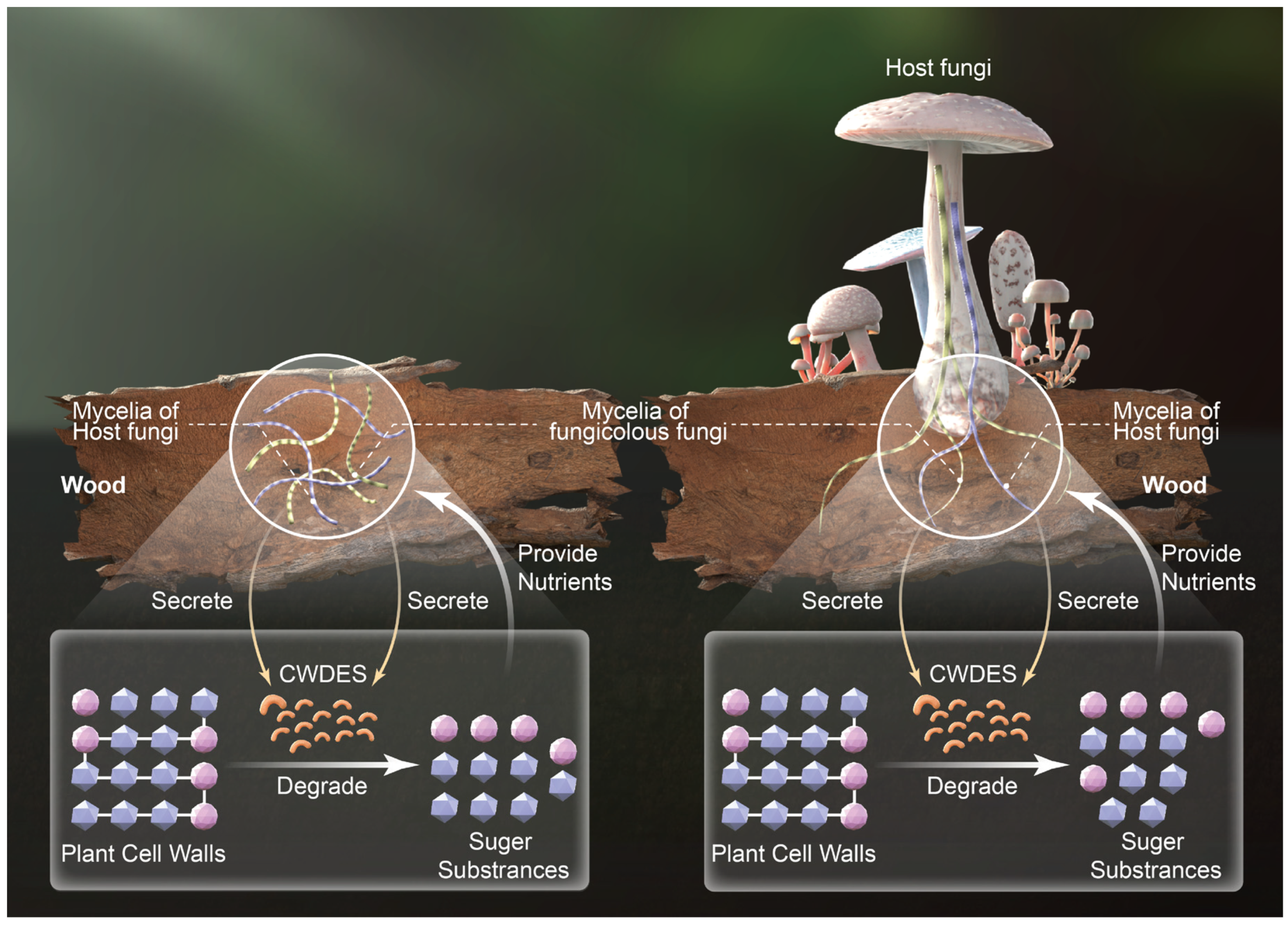
3.5. Carbohydrate-Active Enzyme Diversity of Host Macrofungi and Fungicolous Fungi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koskinen, J.; Roslin, T.; Nyman, T.; Abrego, N.; Michell, C.; Vesterinen, E.J. Finding flies in the mushroom soup: Host specificity of fungus-associated communities revisited with a novel molecular method. Mol. Ecol. 2019, 28, 190–202. [Google Scholar] [CrossRef]
- Pent, M.; Põldmaa, K.; Bahram, M. Bacterial communities in boreal forest mushrooms are shaped both by soil Parameters and host identity. Front. Microbiol. 2017, 8, 836. [Google Scholar] [CrossRef]
- Ma, Y.; Gao, W.; Zhang, F.; Zhu, X.; Kong, W.; Niu, S.; Gao, K.; Yang, H. Community composition and trophic mode diversity of fungi associated with fruiting body of medicinal Sanghuangporus vaninii. BMC Microbiol. 2022, 22, 251. [Google Scholar] [CrossRef] [PubMed]
- Rainey, P. Effect of Pseudomonas putida on hyphal growth of Agaricus bisporus. Mycol. Res. 1991, 95, 699–704. [Google Scholar] [CrossRef]
- Oh, S.-Y.; Kim, M.; Eimes, J.A.; Lim, Y.W. Effect of fruiting body bacteria on the growth of Tricholoma matsutake and its related molds. PLoS ONE 2018, 13, e0190948. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Zheng, L.P.; Wang, J.W. Bacteria associated with Shiraia fruiting bodies influence fungal production of Hypocrellin A. Front. Microbiol. 2019, 10, 2023. [Google Scholar] [CrossRef]
- Barnett, H.L. The nature of mycoparasitism by fungi. Annu. Rev. Microbiol. 1963, 17, 1–14. [Google Scholar] [CrossRef]
- Rudakov, O.L. Physiological groups in mycophilic fungi. Mycologia 1978, 70, 9–150. [Google Scholar] [CrossRef]
- Maurice, S.; Arnault, G.; Nordén, J.; Botnen, S.S.; Miettinen, O.; Kauserud, H. Fungal sporocarps house diverse and host-specific communities of fungicolous fungi. ISME J. 2021, 15, 1445–1457. [Google Scholar] [CrossRef]
- Sun, J.-Z.; Liu, X.-Z.; McKenzie, E.H.C.; Jeewon, R.; Liu, J.-K.; Zhang, X.-L.; Zhao, Q.; Hyde, K.D. Fungicolous fungi: Terminology, diversity, distribution, evolution, and species checklist. Fungal Divers. 2019, 95, 337–430. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiol. Spectr. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Marano, A.V.; Pires-Zottarelli, C.L.A.; Barrera, M.D.; Steciow, M.M.; Gleason, F.H. Diversity, role in decomposition, and succession of zoosporic fungi and straminipiles on submerged decaying leaves in a woodland stream. Hydrobiologia 2011, 659, 93–109. [Google Scholar] [CrossRef]
- Doty, S.L. Endophytic Yeasts: Biology and Applications; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Pospiech, H.; Laukkanen, H.; Hohtola, A.; Pirttilä, A.; Myllylä, R. Two endophytic fungi in different tissues of scots pine buds (Pinus sylvestris L.). Microb. Ecol. 2003, 45, 53–62. [Google Scholar] [CrossRef]
- Spribille, T.; Tuovinen, V.; Resl, P.; Vanderpool, D.; Wolinski, H.; Aime, M.C.; Schneider, K.; Stabentheiner, E.; Toome-Heller, M.; Thor, G.; et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 2016, 353, 488–492. [Google Scholar] [CrossRef]
- Yurkov, A.; Krüger, D.; Begerow, D.; Arnold, N.; Tarkka, M.T. Basidiomycetous Yeasts from Boletales Fruiting Bodies and Their Interactions with the Mycoparasite Sepedonium chrysospermum and the Host Fungus Paxillus. Microb. Ecol. 2012, 63, 295–303. [Google Scholar] [CrossRef]
- Lazarus, K.L.; Benny, G.L.; Ho, H.-M.; Smith, M.E. Phylogenetic systematics of Syncephalis (Zoopagales, Zoopagomycotina), a genus of ubiquitous mycoparasites. Mycologia 2017, 109, 49–333. [Google Scholar] [CrossRef]
- Poldmaa, K.; Samuels, G.J. A phyllophoricolous species of Hypomyces with KOH-negative perithecia. Mycologia 1999, 91, 177–199. [Google Scholar] [CrossRef]
- Poldmaa, K. Three species of Hypomyces growing on basidiomata of Stereaceae. Mycologia 2003, 95, 921–933. [Google Scholar] [CrossRef]
- Reynolds, N.K.; Benny, G.L.; Ho, H.-M.; Hou, Y.-H.; Crous, P.W.; Smith, M.E. Phylogenetic and morphological analyses of the mycoparasitic genus Piptocephalis. Mycologia 2019, 111, 54–68. [Google Scholar] [CrossRef]
- Poinar, G.O.; Buckley, R. Evidence of mycoparasitism and hypermycoparasitism in Early Cretaceous amber. Mycol. Res. 2007, 111, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Pacioni, G.; Leonardi, M.; Aimola, P.; Ragnelli, A.M.; Rubini, A.; Paolocci, F. Isolation and characterization of some mycelia inhabiting Tuber ascomata. Mycol. Res. 2007, 111, 1450–1460. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. 1990, 18, 315–322. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Met. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- He, X.-L.; Huo, W.-Y.; Zhang, L.-G.; Liu, Y.; Qi, P.; Dai, L.; Qiao, T.; Lu, P.; Li, J.-Z. Psilocybe ningshanensis (Hymenogastraceae, Agaricales), a new species from China. Phytotaxa 2022, 545, 175–185. [Google Scholar] [CrossRef]
- He, X.L.; Huo, W.Y.; Zhang, L.G.; Dai, L.; Liu, Y.; Li, J.Z. Two New Species of Helvella (Pezizales, Ascomycota) in the Qinling Mountains, China. J. Fungal Res. 2023. [Google Scholar]
- Huo, W.; Zhang, L.; Liu, Y.; He, X.; Qi, P.; Dai, L.; Qiao, T.; Lu, P.; Li, J. Microstoma Ningshanica, a new species of Microstoma based on molecular, light and scanning electron microscopy analyses from Shaanxi Province, China. All Life 2022, 15, 901–907. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009, 75, 7537. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Met. 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Deshpande, V.; Wang, Q.; Greenfield, P.; Charleston, M.; Porras-Alfaro, A.; Kuske, C.R.; Cole, J.R.; Midgley, D.J.; Tran-Dinh, N. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 2016, 108, 1–5. [Google Scholar] [CrossRef]
- Shuai, W.; Chen, G.; Zhang, H. Carbohydrate-active enzyme (CAZy) database and its new prospect. Chin. J. Bioprocess Eng. 2014, 12, 102–108. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: A review. J. Basic. Microbiol. 2009, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef]



| Months | Widely Distributed Species | Widely Distributed Genera | ||
|---|---|---|---|---|
| Name | Proportion of Samples (%) | Name | Proportion of Samples (%) | |
| March-April | Tubaria praestans | 56.41 | Psathyrella | 61.54 |
| May-June | Laccaria amethystina | 50.00 | Entoloma | 58.33 |
| July | Simplicillium cylindrosporum | 38.30 | Russula | 53.19 |
| August | Agaricus moelleroides | 39.34 | Mycena | 63.93 |
| September | Stereum hirsutum | 30.49 | Mycena | 43.90 |
| October-November | Hypholoma fasciculare | 40.00 | Gymnopus | 73.33 |
| Habitat | Widely Distributed Species | Widely Distributed Genera | ||
|---|---|---|---|---|
| Name | Proportion of Samples (%) | Name | Proportion of Samples (%) | |
| Scrub forest | Tubaria praestans | 70.00 | Tubaria | 70.00 |
| Mixed forest | Stereum hirsutum | 22.90 | Mycena | 32.06 |
| Hardwood forest | Tyromyces chioneus | 35.78 | Mycena | 52.30 |
| Coniferous forest | Agaricus moelleroides | 76.67 | Agaricus | 76.67 |
| Categories | Saprotroph | Pathotroph | Symbiotroph | Others | |
|---|---|---|---|---|---|
| Month | March–April | 0.53 ± 0.11 *** | 0.05 ± 0.05 * | 0.03 ± 0.03 *** | 0.39 ± 0.08 |
| May–June | 0.47 ± 0.1 *** | 0.06 ± 0.05 * | 0.06 ± 0.07 *** | 0.41 ± 0.08 | |
| July | 0.28 ± 0.11 *** | 0.07 ± 0.06 * | 0.25 ± 0.14 *** | 0.4 ± 0.09 | |
| August | 0.34 ± 0.1 *** | 0.09 ± 0.08 * | 0.15 ± 0.09 *** | 0.42 ± 0.08 | |
| September | 0.35 ± 0.13 *** | 0.08 ± 0.07 * | 0.15 ± 0.09 *** | 0.42 ± 0.09 | |
| October–November | 0.39 ± 0.05 *** | 0.07 ± 0.04 * | 0.13 ± 0.06 *** | 0.41 ± 0.06 | |
| Habitat | Coniferous forest | 0.39 ± 0.05 *** | 0.07 ± 0.03 | 0.11 ± 0.03 *** | 0.44 ± 0.06 * |
| Hardwood forest | 0.44 ± 0.15 *** | 0.07 ± 0.07 | 0.1 ± 0.1 *** | 0.39 ± 0.09 * | |
| Mingled forest | 0.32 ± 0.11 *** | 0.08 ± 0.07 | 0.18 ± 0.12 *** | 0.42 ± 0.08 * | |
| Scrub forest | 0.46 ± 0.13 *** | 0.1 ± 0.09 | 0.03 ± 0.06 *** | 0.41 ± 0.06 * | |
| Substrate | Wood | 0.42 ± 0.15 *** | 0.08 ± 0.08 | 0.09 ± 0.08 *** | 0.41 ± 0.1 |
| Ground | 0.35 ± 0.12 *** | 0.07 ± 0.05 | 0.17 ± 0.12 *** | 0.41 ± 0.08 | |
| Altitude | Intermediate | 0.38 ± 0.15 | 0.08 ± 0.07 | 0.14 ± 0.13 | 0.41 ± 0.09 |
| High | 0.38 ± 0.11 | 0.07 ± 0.05 | 0.13 ± 0.08 | 0.41 ± 0.08 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, W.; Cui, L.; Yan, P.; He, X.; Zhang, L.; Liu, Y.; Dai, L.; Qi, P.; Hu, S.; Qiao, T.; et al. Diversity and Composition of Fungicolous Fungi Residing in Macrofungi from the Qinling Mountains. J. Fungi 2024, 10, 601. https://doi.org/10.3390/jof10090601
Huo W, Cui L, Yan P, He X, Zhang L, Liu Y, Dai L, Qi P, Hu S, Qiao T, et al. Diversity and Composition of Fungicolous Fungi Residing in Macrofungi from the Qinling Mountains. Journal of Fungi. 2024; 10(9):601. https://doi.org/10.3390/jof10090601
Chicago/Turabian StyleHuo, Wenyan, Langjun Cui, Pengdong Yan, Xuelian He, Liguang Zhang, Yu Liu, Lu Dai, Peng Qi, Suying Hu, Ting Qiao, and et al. 2024. "Diversity and Composition of Fungicolous Fungi Residing in Macrofungi from the Qinling Mountains" Journal of Fungi 10, no. 9: 601. https://doi.org/10.3390/jof10090601
APA StyleHuo, W., Cui, L., Yan, P., He, X., Zhang, L., Liu, Y., Dai, L., Qi, P., Hu, S., Qiao, T., & Li, J. (2024). Diversity and Composition of Fungicolous Fungi Residing in Macrofungi from the Qinling Mountains. Journal of Fungi, 10(9), 601. https://doi.org/10.3390/jof10090601






