Metabolomic Profiling and Biological Investigation of the Marine Sponge-Derived Fungus Aspergillus sp. SYPUF29 in Response to NO Condition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Samples
2.2. UHPLC-MS/MS Analysis
2.3. Data Processing and Metabolite Identification
2.4. Data Analysis of Non-Targeted Metabolomics
2.5. Determination of Cell Viability
2.6. NO Production Bioassay
2.7. ELISA Assessment of TNF-α, IL-6, and IL-1β in Mouse Serum
2.8. RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
2.9. Western Blot Analysis
3. Results
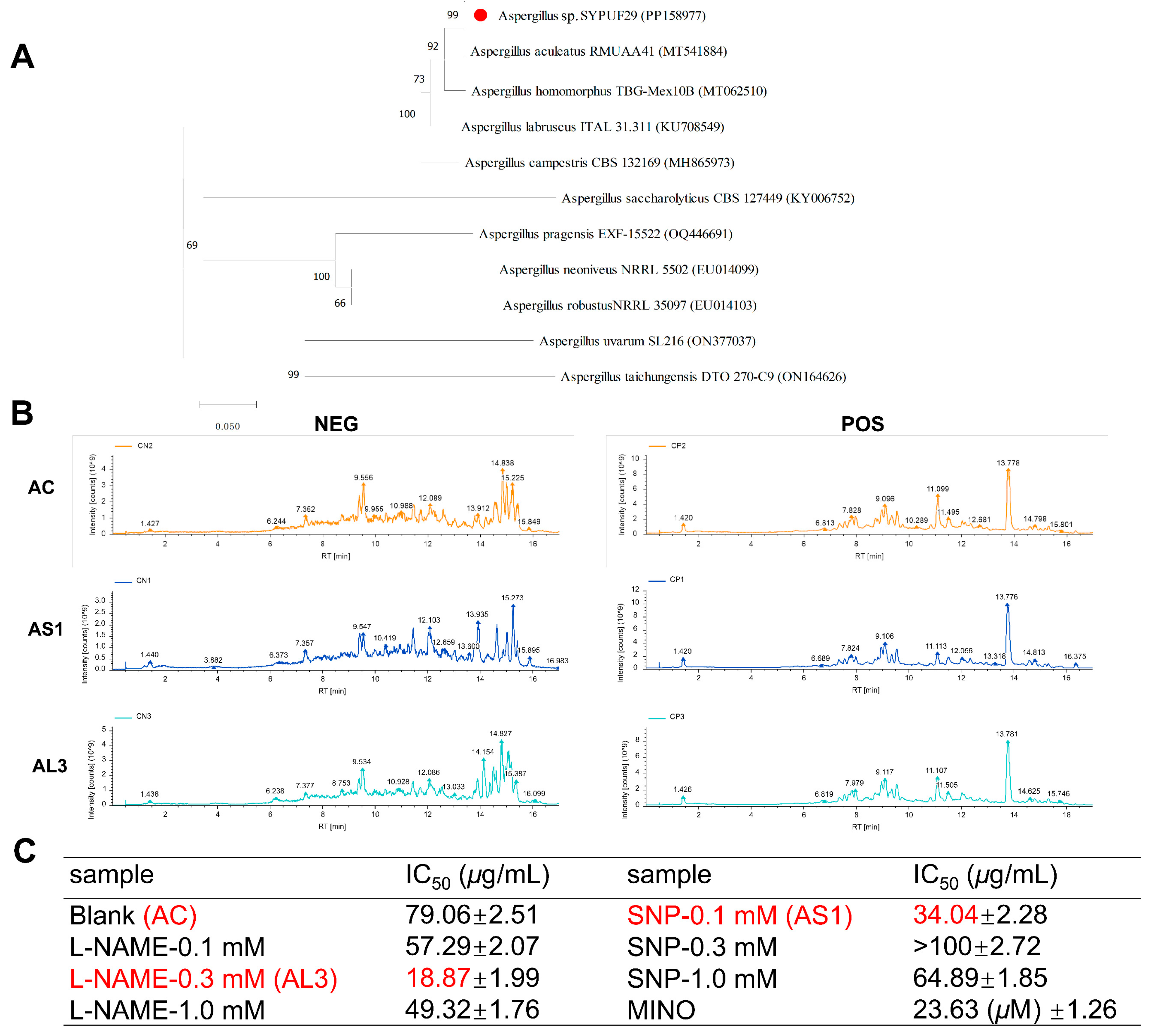
3.1. Phylogenetic Analysis
3.2. Fermentation Conditions for Aspergillus sp. SYPUF29 to Produce the Differential Metabolites under an Additional Source of Nitrogen
3.3. Multivariate Data Analysis for Extract Samples
3.4. Effect of SNP and L-NAME on the Metabolites in the Aspergillus sp. SYPUF29
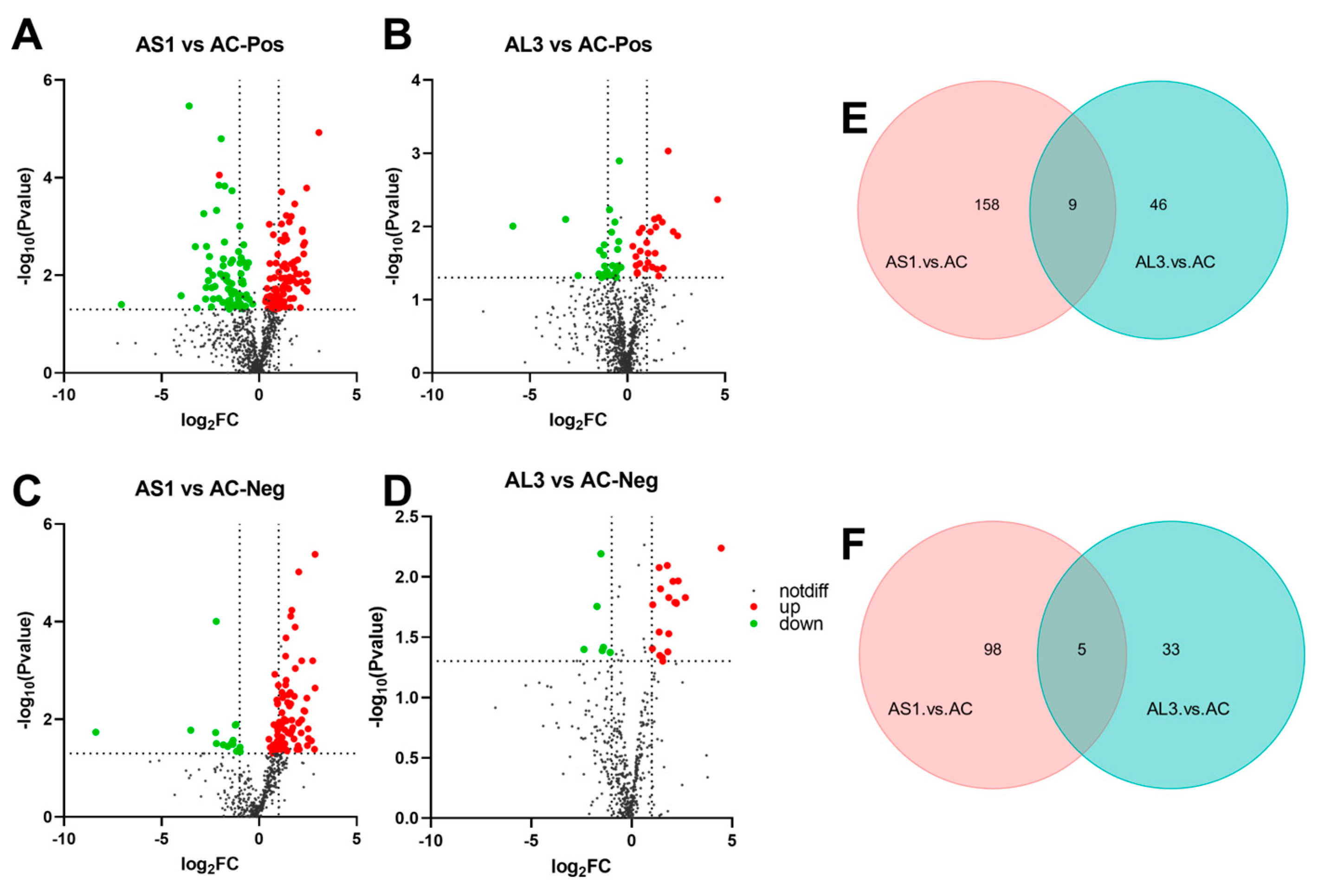
3.5. Difference in Metabolites between Three Extracts
3.6. KEGG-Enriched Pathway Analysis
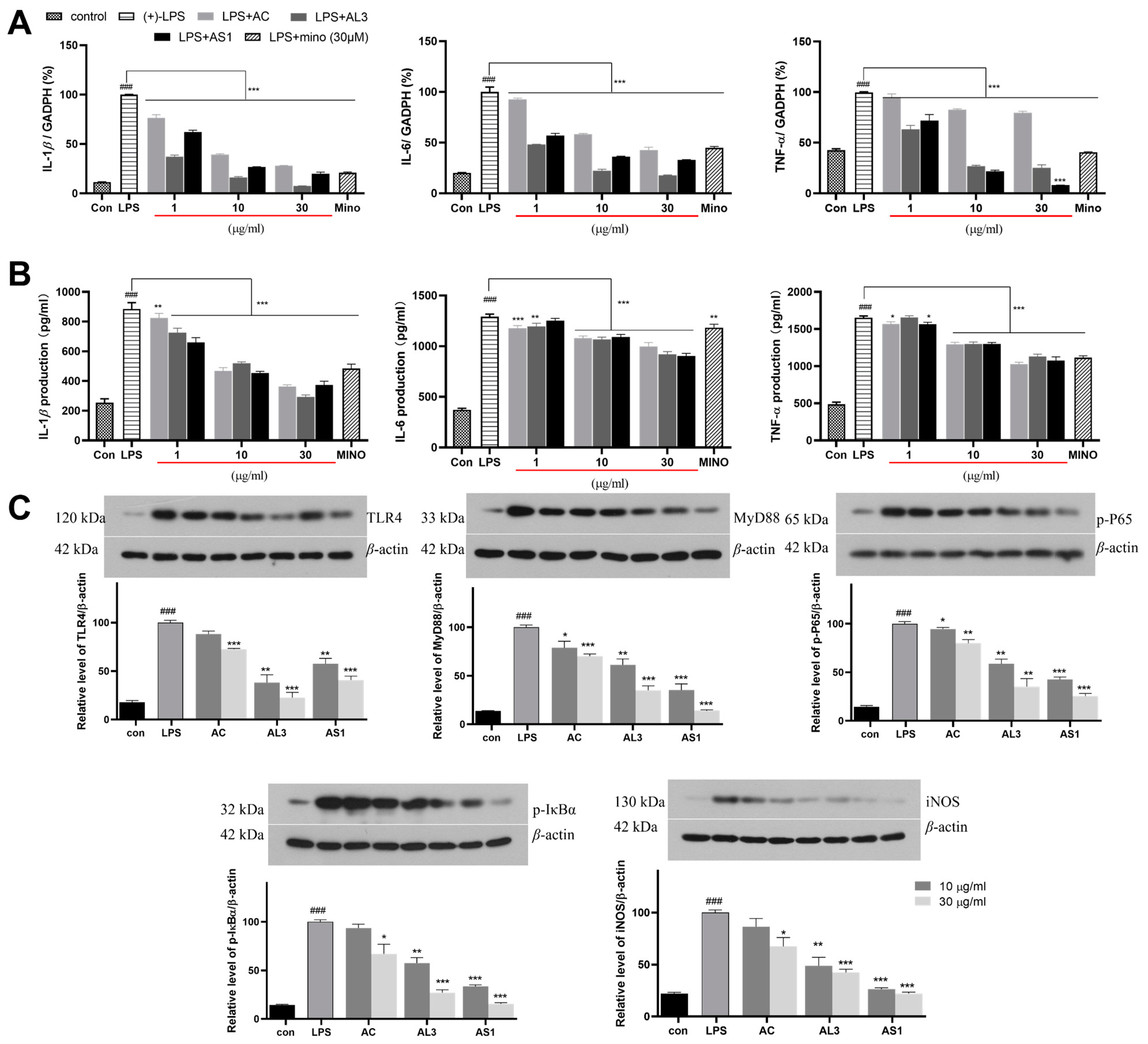
3.7. The Anti-Neuroinflammatory Effects of AS1 and AL3 in LPS-Treated BV2 Cells
3.8. The Chemical Basis of Aspergillus sp. SYPUF29 after SNP Was Administrated
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dansokho, C.; Heneka, M.T. Neuroinflammatory Responses in Alzheimer’s Disease. J. Neural Transm. 2018, 125, 771–779. [Google Scholar] [CrossRef]
- Lelekov-Boissard, T.; Chapuisat, G.; Boissel, J.-P.; Grenier, E.; Dronne, M.-A. Exploration of Beneficial and Deleterious Effects of Inflammation in Stroke: Dynamics of Inflammation Cells. Philos. Trans. A Math. Phys. Eng. Sci. 2009, 367, 4699–4716. [Google Scholar] [CrossRef]
- Rezaeian, A.-H.; Wei, W.; Inuzuka, H. The Regulation of Neuronal Autophagy and Cell Survival by MCL1 in Alzheimer’s Disease. Acta Mater. Med. 2022, 1, 42–55. [Google Scholar] [CrossRef] [PubMed]
- DiSabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The Devil Is in the Details. J. Neurochem. 2016, 139 (Suppl. S2), 136–153. [Google Scholar] [CrossRef]
- Suk, K.; Ock, J. Chemical Genetics of Neuroinflammation: Natural and Synthetic Compounds as Microglial Inhibitors. Inflammopharmacology 2012, 20, 151–158. [Google Scholar] [CrossRef]
- Yeo, I.J.; Yun, J.; Son, D.J.; Han, S.-B.; Hong, J.T. Antifungal Drug Miconazole Ameliorated Memory Deficits in a Mouse Model of LPS-Induced Memory Loss through Targeting iNOS. Cell Death Dis. 2020, 11, 623. [Google Scholar] [CrossRef]
- Doherty, G.H. Nitric Oxide in Neurodegeneration: Potential Benefits of Non-Steroidal Anti-Inflammatories. Neurosci. Bull. 2011, 27, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, H.; Liu, J.; Wei, W.; Rezaeian, A.-H. PROTAC Technology for the Treatment of Alzheimer’s Disease: Advances and Perspectives. Acta Mater. Medica 2022, 1, 24–41. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, W.; Xu, H. Drug Discovery Is an Eternal Challenge for the Biomedical Sciences. Acta Mater. Medica 2022, 1, 1–3. [Google Scholar] [CrossRef]
- Shinde, P.; Banerjee, P.; Mandhare, A. Marine Natural Products as Source of New Drugs: A Patent Review (2015–2018). Expert. Opin. Ther. Pat. 2019, 29, 283–309. [Google Scholar] [CrossRef]
- Huang, L.; Ding, L.; Li, X.; Wang, N.; Yan, Y.; Yang, M.; Cui, W.; Benjamin Naman, C.; Cheng, K.; Zhang, W.; et al. A New Lateral Root Growth Inhibitor from the Sponge-Derived Fungus Aspergillus sp. LS45. Bioorg. Med. Chem. Lett. 2019, 29, 1593–1596. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, Y.; Yang, Y.; Liu, J.; Lin, B.; Hou, Y.; Chen, G.; Li, N. 1H NMR-Guided Isolation of Hasubanan Alkaloids from the Alkaloidal Extract of Stephania longa. Bioorg. Chem. 2023, 139, 106717. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, Y.; Yang, Y.; Liu, J.; Chen, G.; Lin, B.; Hou, Y.; Li, N. Natural Potential Neuroinflammatory Inhibitors from Stephania epigaea H.S. Lo. Bioorg. Chem. 2021, 107, 104597. [Google Scholar] [CrossRef]
- Xiao, J.; Hao, T.; Chen, G.; Song, J.; Lin, B.; Li, W.; Xu, J.; Liu, J.; Hou, Y.; Li, N. Natural Neuroprotective Alkaloids from Stephania japonica (Thunb.) Miers. Bioorg. Chem. 2019, 91, 103175. [Google Scholar] [CrossRef]
- Xiao, J.; Song, J.Y.; Lin, B.; Li, W.; Yang, Y.Q.; Liu, J.Y.; Hou, Y.; Chen, G.; Li, N. Amide-Iminoate Isomerism in Antineuroinflammatory Isoquinoline Alkaloids from Stephania Cepharantha. J. Nat. Prod. 2020, 83, 864–872. [Google Scholar] [CrossRef]
- Wang, W.J.; Li, X.P.; Shen, W.H.; Huang, Q.Y.; Cong, R.P.; Zheng, L.P.; Wang, J.W. Nitric Oxide Mediates Red Light-Induced Perylenequinone Production in Shiraia Mycelium Culture. Bioresour. Bioprocess. 2024, 11, 2. [Google Scholar] [CrossRef]
- Daroodi, Z.; Taheri, P. Function of the Endophytic Fungus Acrophialophora jodhpurensis, Methionine, and Nitric Oxide in Wheat Resistance Induction against Fusarium graminearum via Interplay of Reactive Oxygen Species and Iron. Physiol. Mol. Plant Pathol. 2023, 128, 102132. [Google Scholar] [CrossRef]
- Modolo, L.V.; Cunha, F.Q.; Braga, M.R.; Salgado, I. Nitric Oxide Synthase-Mediated Phytoalexin Accumulation in Soybean Cotyledons in Response to the Diaporthe phaseolorumf sp. meridionalis Elicitor. Plant Physiol. 2002, 130, 1288–1297. [Google Scholar] [CrossRef]
- Gong, X.; Fu, Y.; Jiang, D.; Li, G.; Yi, X.; Peng, Y. L-Arginine Is Essential for Conidiation in the Filamentous Fungus Coniothyrium minitans. Fungal Genet. Biol. 2007, 44, 1368–1379. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Hou, Y.; Li, N.; Xie, G.; Wang, J.; Yuan, Q.; Jia, C.; Liu, X.; Li, G.; Tang, Y.; Wang, B. Pterostilbene Exerts Anti-Neuroinflammatory Effect on Lipopolysaccharide-Activated Microglia via Inhibition of MAPK Signalling Pathways. J. Funct. Foods 2015, 19, 676–687. [Google Scholar] [CrossRef]
- Guo, C.; Wang, P.; Lin, X.; Salendra, L.; Kong, F.; Liao, S.; Yang, B.; Zhou, X.; Wang, J.; Liu, Y. Phloroglucinol Heterodimers and Bis-Indolyl Alkaloids from the Sponge-Derived Fungus Aspergillus sp. SCSIO 41018. Org. Chem. Front. 2019, 6, 3053–3059. [Google Scholar] [CrossRef]
- Shchennikova, A.V.; Beletsky, A.V.; Filyushin, M.A.; Slugina, M.A.; Gruzdev, E.V.; Mardanov, A.V.; Kochieva, E.Z.; Ravin, N.V. Nepenthes × Ventrata Transcriptome Profiling Reveals a Similarity between the Evolutionary Origins of Carnivorous Traps and Floral Organs. Front. Plant Sci. 2021, 12, 643137. [Google Scholar] [CrossRef]
- Ke, C.; Li, A.; Hou, Y.; Sun, M.; Yang, K.; Cheng, J.; Wang, J.; Ge, T.; Zhang, F.; Li, Q.; et al. Metabolic Phenotyping for Monitoring Ovarian Cancer Patients. Sci. Rep. 2016, 6, 23334. [Google Scholar] [CrossRef]
- Liu, L.; Xu, Y.; Dai, H.; Tan, S.; Mao, X.; Chen, Z. Dynorphin Activation of Kappa Opioid Receptor Promotes Microglial Polarization toward M2 Phenotype via TLR4/NF-κB Pathway. Cell Biosci. 2020, 10, 42. [Google Scholar] [CrossRef]
- Lee, H.-H.; Shin, J.-S.; Chung, K.-S.; Kim, J.-M.; Jung, S.-H.; Yoo, H.-S.; Hassan, A.H.E.; Lee, J.K.; Inn, K.-S.; Lee, S.; et al. 3′,4′-Dihydroxyflavone Mitigates Inflammatory Responses by Inhibiting LPS and TLR4/MD2 Interaction. Phytomedicine 2023, 109, 154553. [Google Scholar] [CrossRef]
- Xu, J.; Liu, J.; Mi, Y.; Zhao, T.; Mu, D.; Meng, Q.; Wang, F.; Li, N.; Hou, Y. Triad3A-Dependent TLR4 Ubiquitination and Degradation Contributes to the Anti-Inflammatory Effects of Pterostilbene on Vascular Dementia. J. Agric. Food Chem. 2022, 70, 5896–5910. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, S.; Yang, F.; Dong, S. Marine Indole Alkaloids-Isolation, Structure and Bioactivities. Mar. Drugs 2021, 19, 658. [Google Scholar] [CrossRef]
- Munekata, P.E.S.; Pateiro, M.; Conte-Junior, C.A.; Domínguez, R.; Nawaz, A.; Walayat, N.; Movilla Fierro, E.; Lorenzo, J.M. Marine Alkaloids: Compounds with In Vivo Activity and Chemical Synthesis. Mar. Drugs 2021, 19, 374. [Google Scholar] [CrossRef]
- Kochanowska-Karamyan, A.J.; Hamann, M.T. Marine Indole Alkaloids: Potential New Drug Leads for the Control of Depression and Anxiety. Chem. Rev. 2010, 110, 4489–4497. [Google Scholar] [CrossRef]
- Behl, T.; Rana, T.; Sehgal, A.; Makeen, H.A.; Albratty, M.; Alhazmi, H.A.; Meraya, A.M.; Bhatia, S.; Sachdeva, M. Phytochemicals Targeting Nitric Oxide Signaling in Neurodegenerative Diseases. Nitric Oxide 2023, 130, 1–11. [Google Scholar] [CrossRef]
- Liy, P.M.; Puzi, N.N.A.; Jose, S.; Vidyadaran, S. Nitric Oxide Modulation in Neuroinflammation and the Role of Mesenchymal Stem Cells. Exp. Biol. Med. 2021, 246, 2399–2406. [Google Scholar] [CrossRef]
- Carlström, M. Nitric Oxide Signalling in Kidney Regulation and Cardiometabolic Health. Nat. Rev. Nephrol. 2021, 17, 575–590. [Google Scholar] [CrossRef]
- Gas, E.; Flores-Pérez, Ú.; Sauret-Güeto, S.; Rodríguez-Concepción, M. Hunting for Plant Nitric Oxide Synthase Provides New Evidence of a Central Role for Plastids in Nitric Oxide Metabolism. Plant Cell 2009, 21, 18–23. [Google Scholar] [CrossRef]
- Astuti, R.I.; Nasuno, R.; Takagi, H. Nitric Oxide Signalling in Yeast. In Advances in Microbial Physiology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 72, pp. 29–63. ISBN 978-0-12-814413-8. [Google Scholar]
- Conceição, P.M.; Chaves, A.F.A.; Navarro, M.V.; Castilho, D.G.; Calado, J.C.P.; Haniu, A.E.C.J.; Xander, P.; Batista, W.L. Cross-Talk between the Ras GTPase and the Hog1 Survival Pathways in Response to Nitrosative Stress in Paracoccidioides brasiliensis. Nitric Oxide 2019, 86, 1–11. [Google Scholar] [CrossRef]
- Filippovich, S.Y.; Onufriev, M.V.; Bachurina, G.P.; Kritsky, M.S. The Role of Nitrogen Oxide in Photomorphogenesis in Neurospora crassa. Appl. Biochem. Microbiol. 2019, 55, 427–433. [Google Scholar] [CrossRef]
- Tirpude, N.V.; Sharma, A.; Bhardwaj, N. Agnuside Mitigates OVA-LPS Induced Perturbed Lung Homeostasis via Modulating Inflammatory, Autophagy, Apoptosis-Fibrosis Response and Myeloid Lineages in Mice Model of Allergic Asthma. Int. Immunopharmacol. 2022, 106, 108579. [Google Scholar] [CrossRef]
- Juergens, L.J.; Racké, K.; Tuleta, I.; Stoeber, M.; Juergens, U.R. Anti-Inflammatory Effects of 1,8-Cineole (Eucalyptol) Improve Glucocorticoid Effects in Vitro: A Novel Approach of Steroid-Sparing Add-on Therapy for COPD and Asthma? Synergy 2017, 5, 1–8. [Google Scholar] [CrossRef]
- Shukla, P.K.; Meena, A.S.; Dalal, K.; Canelas, C.; Samak, G.; Pierre, J.F.; Rao, R. Chronic Stress and Corticosterone Exacerbate Alcohol-Induced Tissue Injury in the Gut-Liver-Brain Axis. Sci. Rep. 2021, 11, 826. [Google Scholar] [CrossRef]
- Ahn, H.J.; You, H.J.; Park, M.S.; Li, Z.; Choe, D.; Johnston, T.V.; Ku, S.; Ji, G.E. Microbial Biocatalysis of Quercetin-3-Glucoside and Isorhamnetin-3-Glucoside in Salicornia herbacea and Their Contribution to Improved Anti-Inflammatory Activity. RSC Adv. 2020, 10, 5339–5350. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Y.; Xiang, H.; Guo, M.; Li, S.; Liu, M.; Yao, J. The Tryptophan Metabolite Indole-3-Carboxaldehyde Alleviates Mice with DSS-Induced Ulcerative Colitis by Balancing Amino Acid Metabolism, Inhibiting Intestinal Inflammation, and Improving Intestinal Barrier Function. Molecules 2023, 28, 3704. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Wang, W.; Hu, H.; Zhang, X. Identification of New Arylamine N-Acetyltransferases and Enhancing 2-Acetamidophenol Production in Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2020, 19, 105. [Google Scholar] [CrossRef]
- Yu, D.; Qi, S.; Guan, X.; Yu, W.; Yu, X.; Cai, M.; Li, Q.; Wang, W.; Zhang, W.; Qin, J.-J. Inhibition of STAT3 Signaling Pathway by Terphenyllin Suppresses Growth and Metastasis of Gastric Cancer. Front. Pharmacol. 2022, 13, 870367. [Google Scholar] [CrossRef]
- Yurchenko, E.A.; Menchinskaya, E.S.; Pislyagin, E.A.; Chingizova, E.A.; Girich, E.V.; Yurchenko, A.N.; Aminin, D.L.; Mikhailov, V.V. Cytoprotective Activity of P-Terphenyl Polyketides and Flavuside B from Marine-Derived Fungi against Oxidative Stress in Neuro-2a Cells. Molecules 2021, 26, 3618. [Google Scholar] [CrossRef]
- Takagi, M.; Motohashi, K.; Shin-ya, K. Isolation of 2 New Metabolites, JBIR-74 and JBIR-75, from the Sponge-Derived Aspergillus sp. fS14. J. Antibiot. 2010, 63, 393–395. [Google Scholar] [CrossRef]
- Lopez, J.A.V.; Nogawa, T.; Futamura, Y.; Aono, H.; Hashizume, D.; Osada, H. N-Acetyl-α-Hydroxy-β-Oxotryptamine, a Racemic Natural Product Isolated from Streptomyces sp. 80H647. J. Antibiot. 2021, 74, 477–479. [Google Scholar] [CrossRef]
- Ren, L.; Wang, Y.-Z.; Zhang, W.; Zhou, R.; Zhao, M.; Tang, Z.-S.; Sun, J.; Zhang, D.-B. Triculata A, a Novel Compound from Tricyrtis Maculata (D. Don) J. F. Macbr. with Biological Properties. Nat. Prod. Res. 2021, 35, 3729–3737. [Google Scholar] [CrossRef]
- Pedras, M.S.; Khan, A.Q. Biotransformation of the Phytoalexin Camalexin by the Phytopathogen Rhizoctonia solani. Phytochemistry 2000, 53, 59–69. [Google Scholar] [CrossRef]
- Pei, S.L.; Chen, L.; Xu, J.L.; Shao, C.L. Secondary Metabolites and Their Biological Activities of Two Actinomycetes Streptomyces Coelicoflavus and Nocardiopsis Dassonvillei Associated with Ascidians Styela Clava and Botryllus Schlosseri. Chin. J. Mar. Drugs 2017, 36, 55–60. [Google Scholar] [CrossRef]
- Jin, H.G.; Liu, K.Y.; Qu, W.H.; Li, T.J.; Liao, L.; Yu, J.M. Isolation and Structure Identification of Chemical Constituents from the Fruits of Akebiae quinata. Nat. Prod. Res. Dev. 2019, 31, 2077–2081. [Google Scholar] [CrossRef]
- Marchelli, R.; Vining, L.C. Terphenyllin, a Novel p-Terphenyl Metabolite from Aspergillus candidus. J. Antibiot. 1975, 28, 328–331. [Google Scholar] [CrossRef]
- Wang, F.; Fang, Y.; Zhang, M.; Lin, A.; Zhu, T.; Gu, Q.; Zhu, W. Six New Ergosterols from the Marine-Derived Fungus Rhizopus sp. Steroids 2008, 73, 19–26. [Google Scholar] [CrossRef]


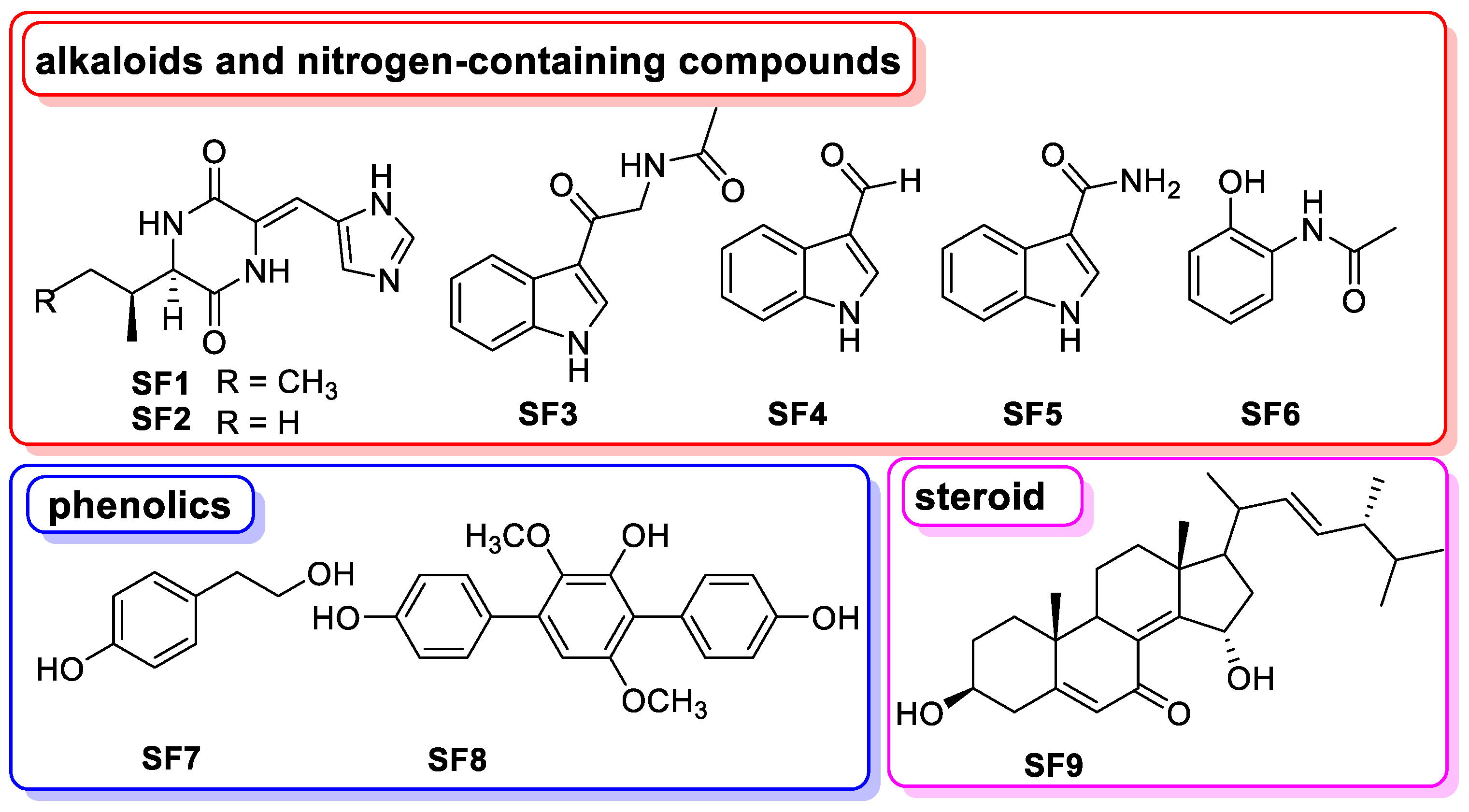
| Types | AS1-AC-Pos | AL3-AC-Pos | AS1-AC-Neg | AL3-AC-Neg |
|---|---|---|---|---|
| Alkaloids and Nitrogen-containing compounds | 14 | 9 | 3 | 2 |
| Flavonoids | 1 | 1 | 0 | 2 |
| Amino acids and their derivatives | 3 | 1 | 0 | 3 |
| Nucleotides and their derivatives | 0 | 1 | 1 | 0 |
| Phenolic acids | 4 | 2 | 1 | 1 |
| Esters | 0 | 0 | 1 | 0 |
| Terpenes | 0 | 2 | 0 | 0 |
| Steroids | 3 | 2 | 2 | 0 |
| others | 1 | 0 | 0 | 0 |
| Phospholipids | 0 | 0 | 2 | 3 |
| Lipids | 0 | 2 | 2 | 4 |
| Coumarins | 1 | 0 | 0 | 0 |
| Organic acids | 1 | 0 | 0 | 1 |
| Total | 28 | 20 | 12 | 16 |
| Compounds | IC50 (μM) | Compounds | IC50 (μM) |
|---|---|---|---|
| SF1 | 13.19 | SF6 | >40 |
| SF2 | 1.73 | SF7 | >40 |
| SF3 | 38.99 | SF8 | 2.78 |
| SF4 | >40 | SF9 | >40 |
| SF5 | >40 | Mino | 23.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Lin, X.; Yang, Y.; Yu, Y.; Li, Y.; Xu, M.; Liu, Y. Metabolomic Profiling and Biological Investigation of the Marine Sponge-Derived Fungus Aspergillus sp. SYPUF29 in Response to NO Condition. J. Fungi 2024, 10, 636. https://doi.org/10.3390/jof10090636
Xiao J, Lin X, Yang Y, Yu Y, Li Y, Xu M, Liu Y. Metabolomic Profiling and Biological Investigation of the Marine Sponge-Derived Fungus Aspergillus sp. SYPUF29 in Response to NO Condition. Journal of Fungi. 2024; 10(9):636. https://doi.org/10.3390/jof10090636
Chicago/Turabian StyleXiao, Jiao, Xiuping Lin, Yanqiu Yang, Yingshu Yu, Yinyin Li, Mengjie Xu, and Yonghong Liu. 2024. "Metabolomic Profiling and Biological Investigation of the Marine Sponge-Derived Fungus Aspergillus sp. SYPUF29 in Response to NO Condition" Journal of Fungi 10, no. 9: 636. https://doi.org/10.3390/jof10090636
APA StyleXiao, J., Lin, X., Yang, Y., Yu, Y., Li, Y., Xu, M., & Liu, Y. (2024). Metabolomic Profiling and Biological Investigation of the Marine Sponge-Derived Fungus Aspergillus sp. SYPUF29 in Response to NO Condition. Journal of Fungi, 10(9), 636. https://doi.org/10.3390/jof10090636








