Ophiostomatalean Fungi (Ascomycota, Ophiostomatales) Associated with Three Beetles from Pinus sylvestris var. mongolica in Heilongjiang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. Morphological Analyses
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analysis
3. Results
3.1. Sampling Collection and Fungal Isolation
3.2. Phylogenetic Analysis
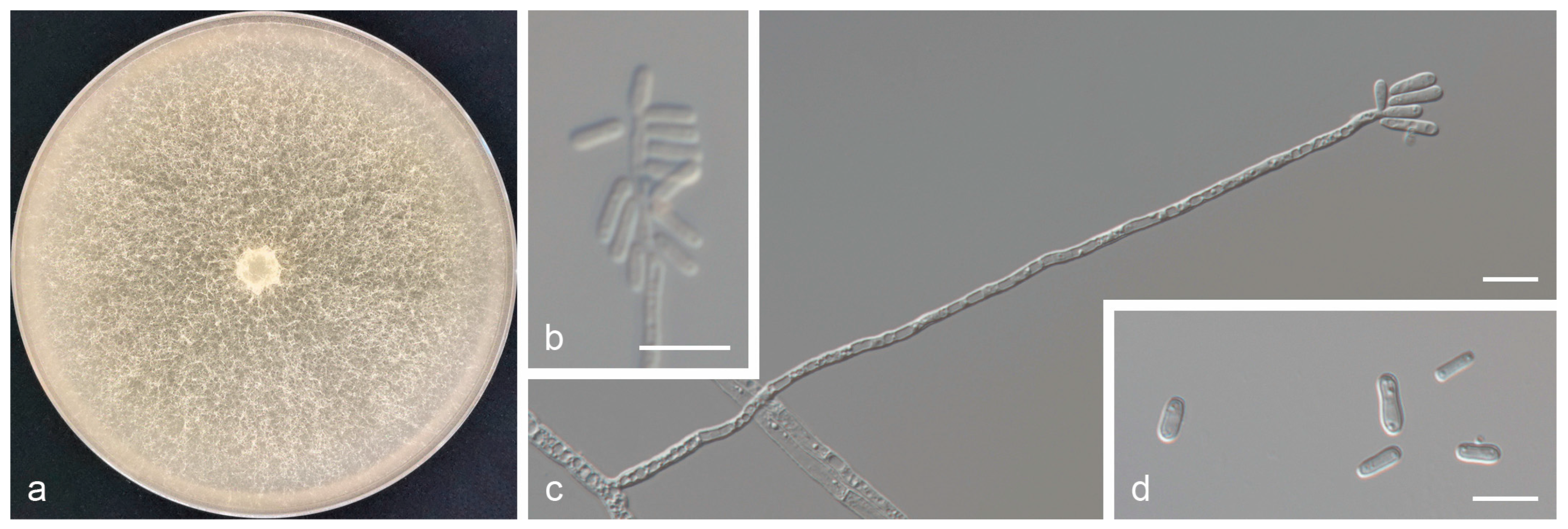
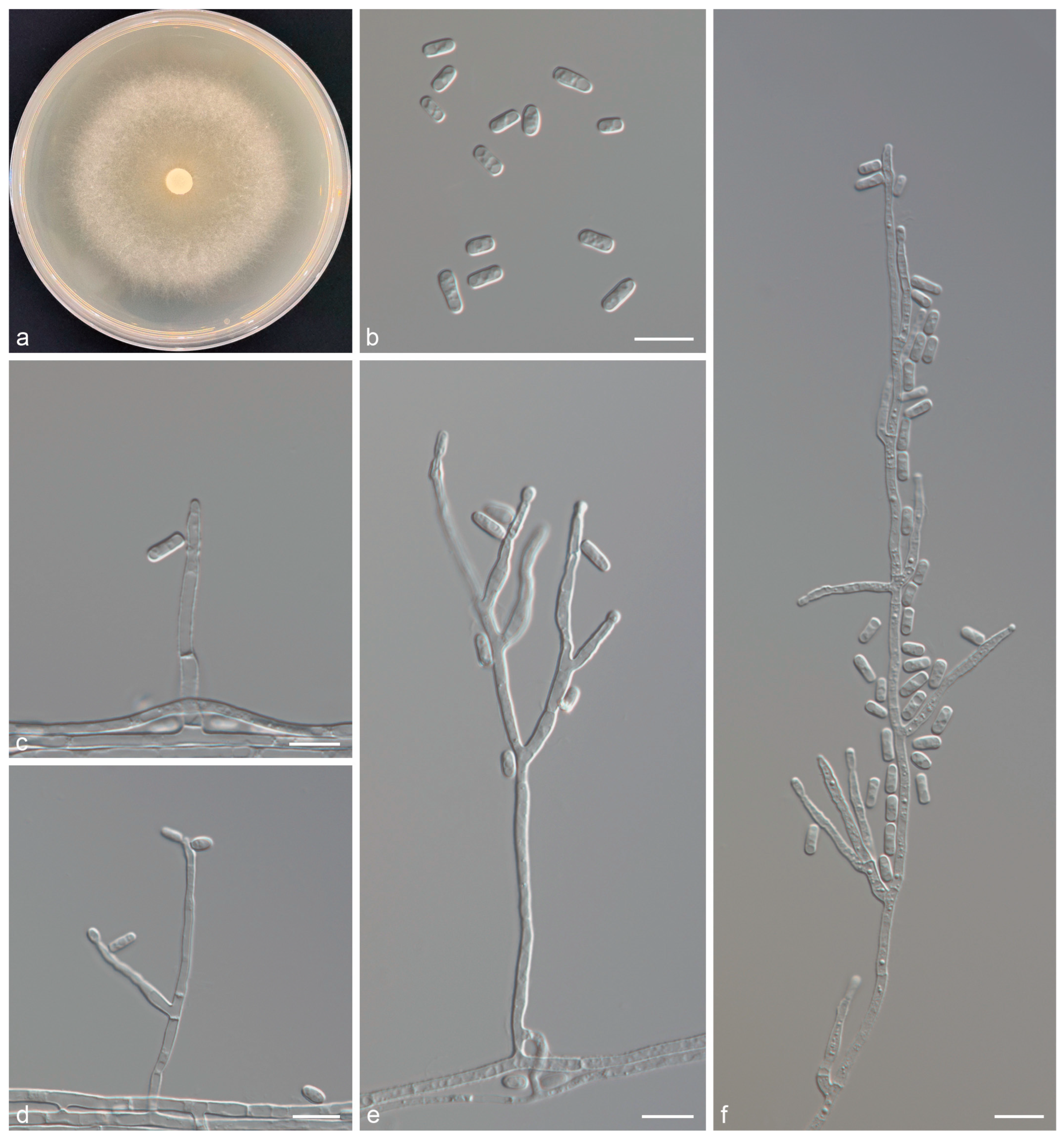
3.3. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Dang, H.; Han, H.; Zhang, X.; Chen, S.; Li, M.; Liu, C. Key strategies underlying the adaptation of Mongolian scots pine (Pinus sylvestris var. mongolica) in sandy land under climate change: A review. Forests 2022, 13, 846. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Sun, S.; Hu, X.; Guan, C.; Gao, W. Differences in growth and wood anatomy in relation to meteorological variables between healthy and declining Mongolian pine trees. Chin. J. Ecol. 2024, in press. [Google Scholar]
- Lu, W.W.; Wu, B.; Bai, J.H.; Song, X.D.; Shi, Z.J.; Dang, H.Z.; Li, H.; Yin, M. Causes and research prospects of the decline of Pinus sylvestris var. mongolica plantation. Chin. Sci. Bull. 2023, 68, 1286–1297. [Google Scholar] [CrossRef]
- Liu, B.; Yan, J.; Wang, D.; Wang, Y.; Zhou, Y.; Chen, Y. Occurrence of major forest pests in China in 2023 and prediction for trend in 2024. Pest Dis. 2024, 43, 41–45. [Google Scholar]
- de Beer, Z.W.; Procter, M.; Wingfield, M.J.; Marincowitz, S.; Duong, T.A. Generic boundaries in the Ophiostomatales reconsidered and revised. Stud. Mycol. 2022, 101, 57–120. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Tie, Y.; Wang, Z.; Kong, L.; Liu, H.; Lu, Q. Three Ophiostomatalean fungi associated with bark beetles from Pinus thunbergii infested by Bursaphelenchus xylophilus in Laoshan Mountain (Shandong, China). Forests 2024, 15, 1990. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Yue, F.; Yan, D.H.; Lu, Q. Identification of ophiostomatalean fungi associated with Tomicus pilifer infesting Pinus koraiensis in Northeastern China. Front. Microbiol. 2022, 13, 919302. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, T.; Liu, Y.; Zeng, F.; Zhang, H.; Decock, C.; Zhang, X.; Lu, Q. Diversity of ophiostomatoid fungi associated with Dendroctonus armandi infesting Pinus armandii in western China. J. Fungi 2022, 8, 214. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, L.; Wang, H.; Decock, C.; Lu, Q. Ophiostomatoid fungi associated with Ips bark beetles in China. Fungal Divers. 2024, 129, 283–364. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, C.; Song, X.; Tie, Y.; Wang, H.; Liu, H.; Lu, Q. Ophiostomatalean fungi associated with Polygraphus bark beetles in the Qinghai-Tibet Plateau, China. MycoKeys 2024, 110, 93–115. [Google Scholar] [CrossRef]
- DiGuistini, S.; Wang, Y.; Liao, N.Y.; Taylor, G.; Tanguay, P.; Feau, N.; Henrissat, B.; Chan, S.K.; Hesse-Orce, U.; Alamouti, S.M.; et al. Genome and transcriptome analyses of the mountain pine beetle fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Natl. Acad. Sci. USA 2011, 108, 2504–2509. [Google Scholar] [CrossRef]
- Lu, Q.; Decock, C.; Zhang, X.Y.; Maraite, H. Ophiostomatoid fungi (Ascomycota) associated with Pinus tabuliformis infested by Dendroctonus valens (Coleoptera) in northern China and an assessment of their pathogenicity on mature trees. Antonie Van. Leeuwenhoek 2009, 96, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Liu, C.; Liu, Z.; Liang, L.; Lu, Q. Morphological and phylogenetic analyses reveal a new species of Ceratocystiopsis (Ophiostomataceae, Ophiostomatales) associated with Ips subelongatus in Inner Mongolia (China) with weak host pathogenicity. Forests 2021, 12, 1795. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Wang, H.; Meng, X.; Liu, X.; Decock, C.; Zhang, X.; Lu, Q. Ophiostomatoid fungi associated with Ips subelongatus, including eight new species from northeastern China. IMA Fungus 2020, 11, 1–29. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.J.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.; Bergdahl, D.R.; Wingfield, M.J.; Halik, S.; Seifert, K.A.; Bright, D.E.; Wingfield, B.D. Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycol. Res. 2004, 108, 411–418. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RaxML Version 8: A tool phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Jankowiak, R.; Solheim, H.; Bilański, P.; Marincowitz, S.; Wingfield, M.J. Seven new species of Graphilbum from conifers in Norway, Poland, and Russia. Mycologia 2020, 112, 1240–1262. [Google Scholar] [CrossRef]
- Romón, P.; De Beer, Z.W.; Zhou, X.D.; Duong, T.A.; Wingfield, B.D.; Wingfield, M.J. Multigene phylogenies of Ophiostomataceae associated with Monterey pine bark beetles in Spain reveal three new fungal species. Mycologia 2014, 106, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Burgess, T.I.; De Beer, Z.W.; Lieutier, F.; Yart, A.; Klepzig, K.; Carnegie, A.; Portales, J.M.; Wingfield, B.D.; Wingfield, M.J. High intercontinental migration rates and population admixture in the sapstain fungus Ophiostoma ips. Mol. Ecol. 2007, 16, 89–99. [Google Scholar] [CrossRef]
- Lu, M.; Zhou, X.D.; De Beer, Z.W.; Wingfield, M.J.; Sun, J.H. Ophiostomatoid fungi associated with the invasive pine-infesting bark beetle, Dendroctonus valens, in China. Fungal Divers. 2009, 38, 133–145. [Google Scholar]
- Chang, R.; Duong, T.A.; Taerum, S.J.; Wingfield, M.J.; Zhou, X.; de Beer, Z.W. Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in Yunnan, China. MycoKeys 2017, 28, 19–64. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Zhang, X.; Si, H.; Zhao, G.; Yuan, X.; Liu, T.; Bose, T.; Dai, M. Ophiostomatoid species associated with pine trees (Pinus spp.) infested by Cryphalus piceae from eastern China, including five new species. MycoKeys 2021, 83, 181–208. [Google Scholar] [CrossRef]
- Pan, Y.; Lu, J.; Chen, P.; Yu, Z.; Zhang, H.; Ye, H.; Zhao, T. Ophiostomatales (Ascomycota) associated with Tomicus species in southwestern China with an emphasis on Ophiostoma canum. J. For. Res. 2020, 31, 2549–2562. [Google Scholar] [CrossRef]
- Zheng, G.; You, M.; Li, X.; Zhou, Q.; Wang, Z.; Wang, H.; Lu, Q. Diversity of fungi associated with Monochamus alternatus larval habitats in Bursaphelenchus xylophilus-infected Pinus massoniana and identification of two new ophiostomatalean species (Ascomycota, Ophiostomatales). MycoKeys 2022, 92, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Ren, N.; You, C. Community structure of endophytic fungi in asymptomatic and symptomatic Pinus sylvestris var. mongolica infected by diplodia tip blight. J. Beijing For. Univ. 2024, 46, 119–131. [Google Scholar]

| Gene Fragment | Primers | PCR Conditions | References |
|---|---|---|---|
| ITS | ITS1-F/ITS4 | 94 °C for 3 min, 35 cycles of 94 °C for 1 min, 55 °C for 45 s, and 72 °C for 1 min, 72 °C for 8 min | [16,17] |
| tub2 | Bt2a/Bt2b | 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, 72 °C for 8 min | [18] |
| tef1-α | EF1F/EF2R | 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, 72 °C for 8 min | [19] |
| Species | Taxon | Isolate No. 1 | Other No. 2 | Insect Vector 3 | GenBank Accession No. | ||
|---|---|---|---|---|---|---|---|
| ITS | tub2 | tef1-α | |||||
| Gra. griseum sp. nov. | 1 | CFCC71126 | CXY3358 | A. griseus | PQ623400 | - | PQ619704 |
| CFCC71130 | CXY3359 | A. griseus | PQ623401 | - | PQ619705 | ||
| CFCC71124 | CXY3360 | A. griseus | PQ623402 | - | PQ619706 | ||
| Gra. nitidum sp. nov. | 2 | CFCC71144 | CXY3361 | P. nitidus | PQ623403 | - | PQ619707 |
| CFCC71143 | CXY3362 | P. nitidus | PQ623404 | PQ619697 | PQ619708 | ||
| Graphilbum sp. | 3 | CFCC71127 | CXY3363 | P. nitidus | PQ623405 | - | PQ619709 |
| CFCC71139 | CXY3364 | I. chinensis | PQ623406 | - | PQ619710 | ||
| Ophiostoma ips | 4 | CFCC71129 | CXY3365 | A. griseus | PQ623407 | PQ619698 | - |
| CFCC71135 | CXY3366 | A. griseus | PQ623408 | PQ619699 | - | ||
| CFCC71128 | CXY3367 | I. chinensis | PQ623409 | PQ619700 | - | ||
| CFCC71136 | CXY3368 | I. chinensis | PQ623410 | PQ619701 | - | ||
| CFCC71137 | CXY3369 | P. nitidus | PQ623411 | PQ619702 | - | ||
| CFCC71138 | CXY3370 | P. nitidus | PQ623412 | PQ619703 | - | ||
| Taxon | Species | Numbers of Isolates 1 | Total | Total Percentage | ||
|---|---|---|---|---|---|---|
| A. griseus | I. chinensis | P. nitidus | ||||
| 1 | Graphilbum griseum | 21 | 21 | 7.17% | ||
| 2 | Gra. nitidum | 6 | 6 | 2.05% | ||
| 3 | Graphilbum sp. | 25 | 26 | 51 | 17.41% | |
| 4 | Ophiostoma ips | 136 | 34 | 45 | 215 | 73.38% |
| Total | 157 | 59 | 77 | 293 | 100.00% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, C.; Tie, Y.; Song, X.; Wang, H.; Lu, Q. Ophiostomatalean Fungi (Ascomycota, Ophiostomatales) Associated with Three Beetles from Pinus sylvestris var. mongolica in Heilongjiang, China. J. Fungi 2025, 11, 27. https://doi.org/10.3390/jof11010027
Wang Z, Liu C, Tie Y, Song X, Wang H, Lu Q. Ophiostomatalean Fungi (Ascomycota, Ophiostomatales) Associated with Three Beetles from Pinus sylvestris var. mongolica in Heilongjiang, China. Journal of Fungi. 2025; 11(1):27. https://doi.org/10.3390/jof11010027
Chicago/Turabian StyleWang, Zheng, Caixia Liu, Yingjie Tie, Xiuyue Song, Huimin Wang, and Quan Lu. 2025. "Ophiostomatalean Fungi (Ascomycota, Ophiostomatales) Associated with Three Beetles from Pinus sylvestris var. mongolica in Heilongjiang, China" Journal of Fungi 11, no. 1: 27. https://doi.org/10.3390/jof11010027
APA StyleWang, Z., Liu, C., Tie, Y., Song, X., Wang, H., & Lu, Q. (2025). Ophiostomatalean Fungi (Ascomycota, Ophiostomatales) Associated with Three Beetles from Pinus sylvestris var. mongolica in Heilongjiang, China. Journal of Fungi, 11(1), 27. https://doi.org/10.3390/jof11010027






