New Promoters for Metabolic Engineering of Ashbya gossypii
Abstract
1. Introduction
2. Materials and Methods
2.1. Ashbya gossypii Strains and Growth Conditions
2.2. Assembly and Genomic Integration of the Cassettes for Renilla and Firefly Luciferase Expression
2.3. Dual Luciferase Reporter (DLR) Assay for Promoter Analysis
2.4. Construction and Integration of MSN2 Expression Cassettes
2.5. Quantitative Real-Time PCR
2.6. RNAseq
2.7. Sporulation Analysis
3. Results and Discussion
3.1. Adaptation of a Luciferase Reporter Assay for Promoter Analysis in A. gossypii
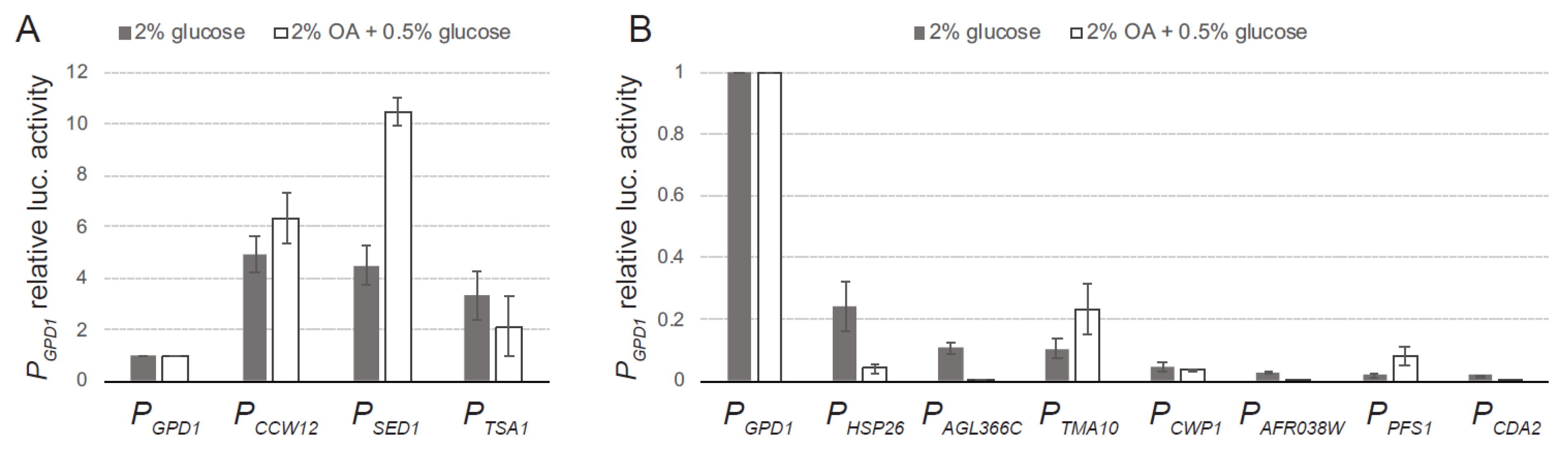
3.2. Dual luciferase Assay of Selected Promoter Sequences
3.3. In Vivo Analysis of New Promoter Sequences
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aguiar, T.Q.; Silva, R.; Domingues, L. New biotechnological applications for Ashbya gossypii: Challenges and perspectives. Bioengineered 2016, 8, 309–315. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Ledesma-Amaro, R.; Lozano-Martinez, P.; Díaz-Fernández, D.; Martinez-Buey, R.; Jiménez, A. Bioproduction of riboflavin: A bright yellow history. J. Ind. Microbiol. Biotechnol. 2017, 44, 659–665. [Google Scholar] [CrossRef]
- Serrano-Amatriain, C.; Ledesma-Amaro, R.; López-Nicolás, R.; Ros, G.; Jiménez, A.; Revuelta, J.L. Folic Acid Production by Engineered Ashbya gossypii. Metab. Eng. 2016, 38, 473–482. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Buey, R.M.; Revuelta, J.L. Increased production of inosine and guanosine by means of metabolic engineering of the purine pathway in Ashbya gossypii. Microb. Cell Factories 2015, 14, 1–8. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Lozano-Martínez, P.; Jiménez, A.; Revuelta, J.L. EngineeringAshbya gossypiifor efficient biolipid production. Bioengineered 2015, 6, 119–123. [Google Scholar] [CrossRef]
- Aguiar, T.Q.; Silva, R.; Domingues, L. Ashbya gossypii beyond industrial riboflavin production: A historical perspective and emerging biotechnological applications. Biotechnol. Adv. 2015, 33, 1774–1786. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; Aguiar, T.Q.; Coelho, E.; Jiménez, A.; Revuelta, J.L.; Domingues, L. Metabolic engineering of Ashbya gossypii for deciphering the de novo biosynthesis of γ-lactones. Microb. Cell Factories 2019, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Fernández, D.; Aguiar, T.Q.; Martín, V.I.; Romaní, A.; Silva, R.; Domingues, L.; Revuelta, J.L.; Jiménez, A. Microbial lipids from industrial wastes using xylose-utilizing Ashbya gossypii strains. Bioresour. Technol. 2019, 293, 122054. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Martínez, P.; Martinez-Buey, R.; Ledesma-Amaro, R.; Jiménez, A.; Revuelta, J.L. Engineering Ashbya gossypii strains for de novo lipid production using industrial by-products. Microb. Biotechnol. 2016, 10, 425–433. [Google Scholar] [CrossRef]
- Schwechheimer, S.K.; Park, E.Y.; Revuelta, J.L.; Becker, J.; Wittmann, C. Biotechnology of riboflavin. Appl. Microbiol. Biotechnol. 2016, 100, 2107–2119. [Google Scholar] [CrossRef]
- Jiménez, A.; Muñoz-Fernández, G.; Ledesma-Amaro, R.; Martinez-Buey, R.; Revuelta, J.L. One-vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii. Microb. Biotechnol. 2019, 12, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Hoff, B.; Revuelta, J.L. Multiplex genome editing in Ashbya gossypii using CRISPR-Cpf1. New Biotechnol. 2020, 57, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Jiménez, A.; Revuelta, J.L. Pathway Grafting for Polyunsaturated Fatty Acids Production in Ashbya gossypii through Golden Gate Rapid Assembly. ACS Synth. Biol. 2018, 7, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Ro, D.-K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nat. Cell Biol. 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Sharpe, P.L.; Hong, S.-P.; Yadav, N.S.; Xie, D.; Short, D.R.; Damude, H.G.; Rupert, R.A.; Seip, J.E.; Wang, J.; et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 2013, 31, 734–740. [Google Scholar] [CrossRef]
- Gassler, T.; Sauer, M.; Gasser, B.; Egermeier, M.; Troyer, C.; Causon, T.; Hann, S.; Mattanovich, D.; Steiger, M.G. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 2020, 38, 210–216. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Serrano-Amatriain, C.; Jiménez, A.; Revuelta, J.L. Metabolic engineering of riboflavin production in Ashbya gossypii through pathway optimization. Microb. Cell Factories 2015, 14, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Santos, M.A.; Revuelta, J.L. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol. 2008, 8, 67. [Google Scholar] [CrossRef]
- Dünkler, A.; Wendland, J. Use of MET3 promoters for regulated gene expression in Ashbya gossypii. Curr. Genet. 2007, 52, 1–10. [Google Scholar] [CrossRef]
- Kaufmann, A. A plasmid collection for PCR-based gene targeting in the filamentous ascomycete Ashbya gossypii. Fungal Genet. Biol. 2009, 46, 595–603. [Google Scholar] [CrossRef]
- Jiménez, A.; Santos, M.A.; Pompejus, M.; Revuelta, J.L. Metabolic Engineering of the Purine Pathway for Riboflavin Production in Ashbya gossypii. Appl. Environ. Microbiol. 2005, 71, 5743–5751. [Google Scholar] [CrossRef]
- Aguiar, T.Q.; Dinis, C.; Domingues, L. Cre-loxP-based system for removal and reuse of selection markers in Ashbya gossypii targeted engineering. Fungal Genet. Biol. 2014, 68, 1–8. [Google Scholar] [CrossRef]
- Mateos, L.; Jiménez, A.; Revuelta, J.L.; Santos, M.A. Purine Biosynthesis, Riboflavin Production, and Trophic-Phase Span Are Controlled by a Myb-Related Transcription Factor in the Fungus Ashbya gossypii. Appl. Environ. Microbiol. 2006, 72, 5052–5060. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Santos, M.A.; Jiménez, A.; Revuelta, J.L. Strain Design of Ashbya gossypii for Single-Cell Oil Production. Appl. Environ. Microbiol. 2014, 80, 1237–1244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stahmann, K.-P.; Revuelta, J.L.; Seulberger, H. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 2000, 53, 509–516. [Google Scholar] [CrossRef]
- Díaz-Fernández, D.; Lozano-Martínez, P.; Buey, R.M.; Revuelta, J.L.; Jiménez, A. Utilization of xylose by engineered strains of Ashbya gossypii for the production of microbial oils. Biotechnol. Biofuels 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Wendland, J. Sporulation in Ashbya gossypii. J. Fungi 2020, 6, 157. [Google Scholar] [CrossRef]
- Anderson, C.A.; Roberts, S.; Zhang, H.; Kelly, C.M.; Kendall, A.; Lee, C.; Gerstenberger, J.; Koenig, A.B.; Kabeche, R.; Gladfelter, A.S. Ploidy variation in multinucleate cells changes under stress. Mol. Biol. Cell 2015, 26, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Gurvitz, A.; Rottensteiner, H. The biochemistry of oleate induction: Transcriptional upregulation and peroxisome proliferation. Biochim. Biophys. Acta (BBA)-Bioenerg. 2006, 1763, 1392–1402. [Google Scholar] [CrossRef]
- Fleißner, A.; Dersch, P.; Fleissner, A. Expression and export: Recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 2010, 87, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Polli, F.; Meijrink, B.; Bovenberg, R.A.; Driessen, A.J. New promoters for strain engineering of Penicillium chrysogenum. Fungal Genet. Biol. 2016, 89, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Fitz, E.; Wanka, F.; Seiboth, B. The Promoter Toolbox for Recombinant Gene Expression in Trichoderma reesei. Front. Bioeng. Biotechnol. 2018, 6, 135. [Google Scholar] [CrossRef]
- Umemura, M.; Kuriiwa, K.; Dao, L.V.; Okuda, T.; Terai, G. Promoter tools for further development of Aspergillus oryzae as a platform for fungal secondary metabolite production. Fungal Biol. Biotechnol. 2020, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
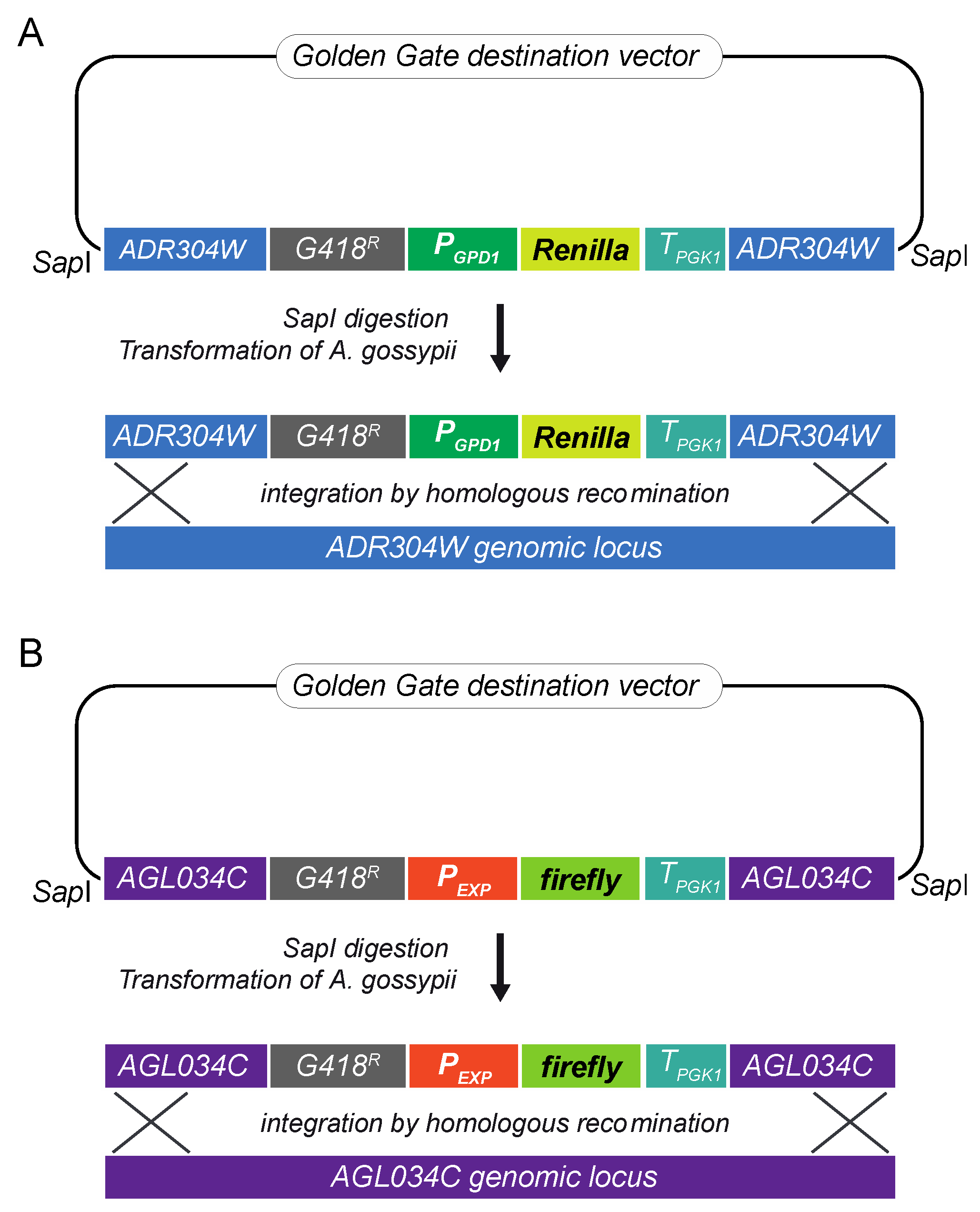


| Gene | Standard Name | S. cerevisiae Homolog | RNAseq FKPM * | GO Description | Promoter Length (bp) |
|---|---|---|---|---|---|
| AGR049W | CCW12 | YLR110C | 102847.2 | Cell wall mannoprotein | 432 |
| AFR505C | TMA10 | YLR327C | 71752.9 | Protein of unknown function that associates with ribosomes | 258 |
| ACR272C | CWP1 | YKL096W | 28833.9 | Cell wall mannoprotein that localizes to birth scars of daughter cells | 950 |
| AER312W | TSA1 | YML028W | 25764.9 | Thioredoxin peroxidase. Panther family PTHR10681 | 255 |
| AER031C | GDP (TDH3) | YGR192C | 15480.9 | Glyceraldehyde-3-phosphate dehydrogenase. Panther family PTHR10836 | 373 |
| AGL366C | No homolog | 14317.3 | 672 | ||
| AGR138W | SED1 | YDR077W | 13184.5 | Major stress-induced structural GPI-cell wall glycoprotein. Panther family PTHR35523 | 711 |
| ADL036C | CDA2 | YLR308W | 12871.6 | Chitin deacetylase. Panther family PTHR10587 | 325 |
| AGR408W | HSP26 | YBR072W | 5158.0 | Small heat shock protein (sHSP) with chaperone activity | 1000 |
| AFR038W | YHR138C | 2681.4 | Protein of unknown function | 202 | |
| AFR132C | PFS1 | YHR185C | 72.8 | Sporulation protein required for prospore membrane formation | 176 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Fernández, G.; Montero-Bullón, J.-F.; Revuelta, J.L.; Jiménez, A. New Promoters for Metabolic Engineering of Ashbya gossypii. J. Fungi 2021, 7, 906. https://doi.org/10.3390/jof7110906
Muñoz-Fernández G, Montero-Bullón J-F, Revuelta JL, Jiménez A. New Promoters for Metabolic Engineering of Ashbya gossypii. Journal of Fungi. 2021; 7(11):906. https://doi.org/10.3390/jof7110906
Chicago/Turabian StyleMuñoz-Fernández, Gloria, Javier-Fernando Montero-Bullón, José Luis Revuelta, and Alberto Jiménez. 2021. "New Promoters for Metabolic Engineering of Ashbya gossypii" Journal of Fungi 7, no. 11: 906. https://doi.org/10.3390/jof7110906
APA StyleMuñoz-Fernández, G., Montero-Bullón, J.-F., Revuelta, J. L., & Jiménez, A. (2021). New Promoters for Metabolic Engineering of Ashbya gossypii. Journal of Fungi, 7(11), 906. https://doi.org/10.3390/jof7110906






