Expression and Purification along with Evaluation of Serological Response and Diagnostic Potential of Recombinant Sap2 Protein from C. parapsilosis for Use in Systemic Candidiasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Strains
2.2. Mice Information
2.3. Human Patients and Control Subjects
2.4. Cloning and Expression of Recombinant Sap2 Protein from C. parapsilosis
2.5. Refolding of Recombinant Sap2 Protein from C. parapsilosis
2.6. CD Spectroscopy
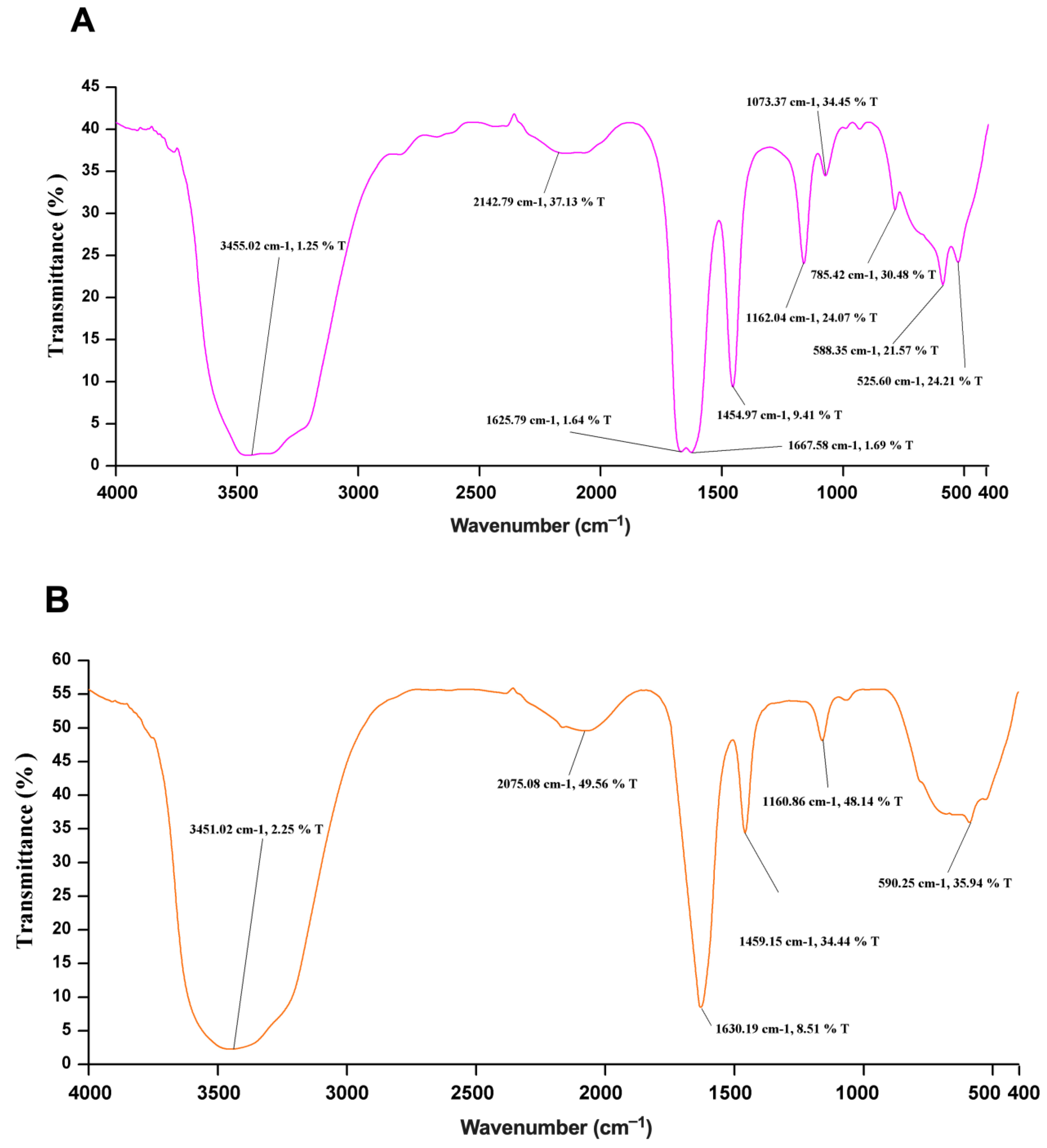
2.7. FTIR Spectroscopy
2.8. Animal Immunization
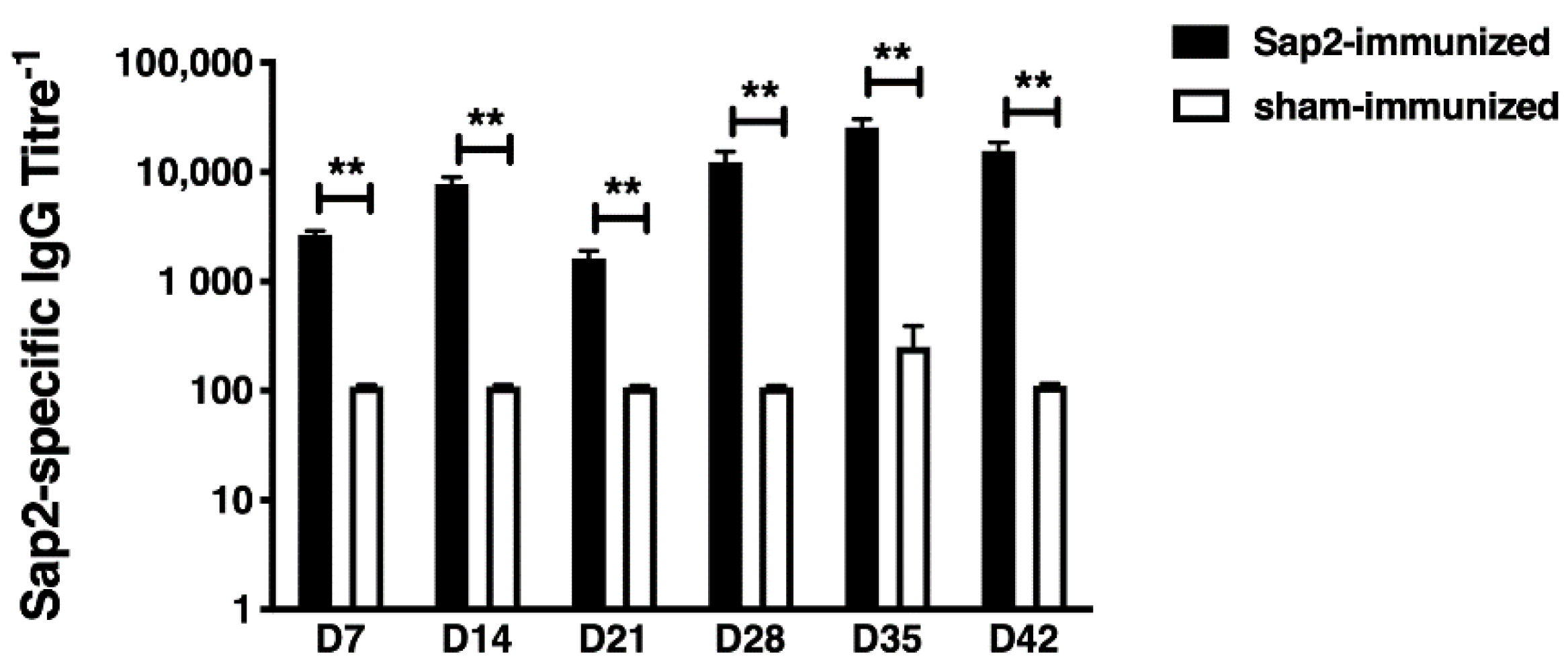
2.9. Evaluation of Sap2-Specific Antibody Response in Mice
2.10. Detection of Sap2-Antigens Using Human Serum Samples by Western Blot
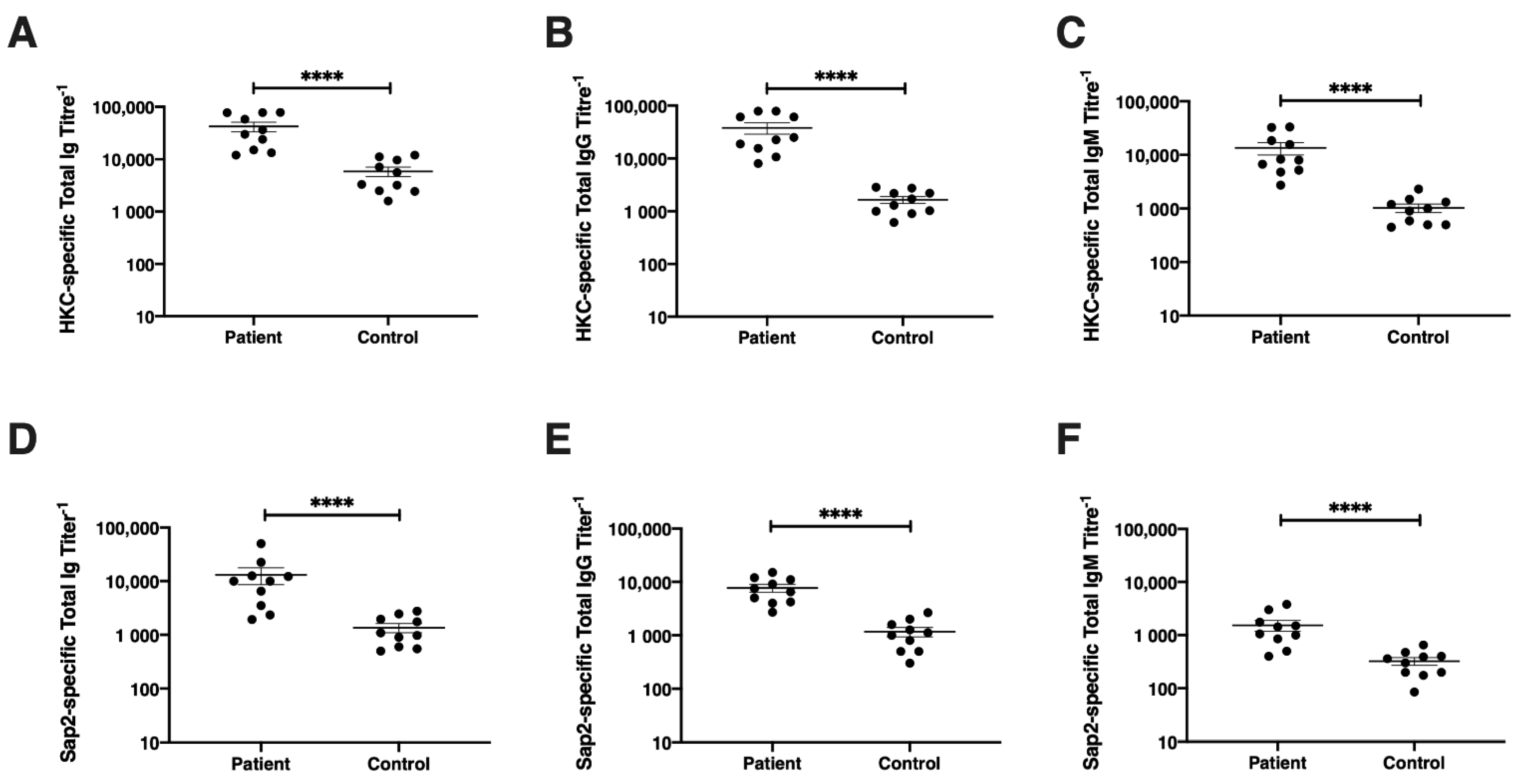
2.11. Detection of Sap2-Specific and Candida-Specific Antibodies by ELISA in Human Serum
2.12. Statistical Analysis
3. Results
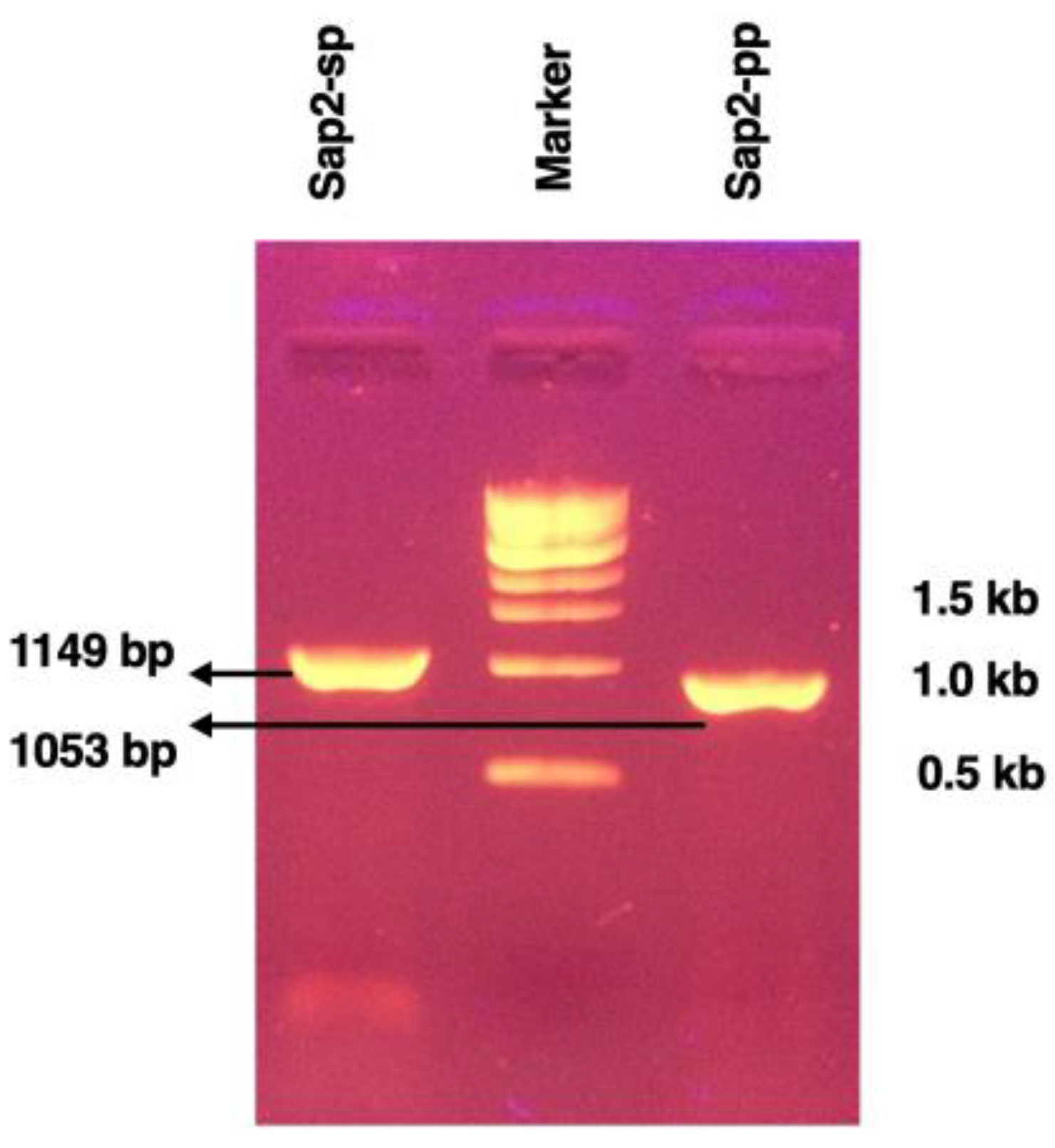
3.1. Cloning of Recombinant Sap2 Protein
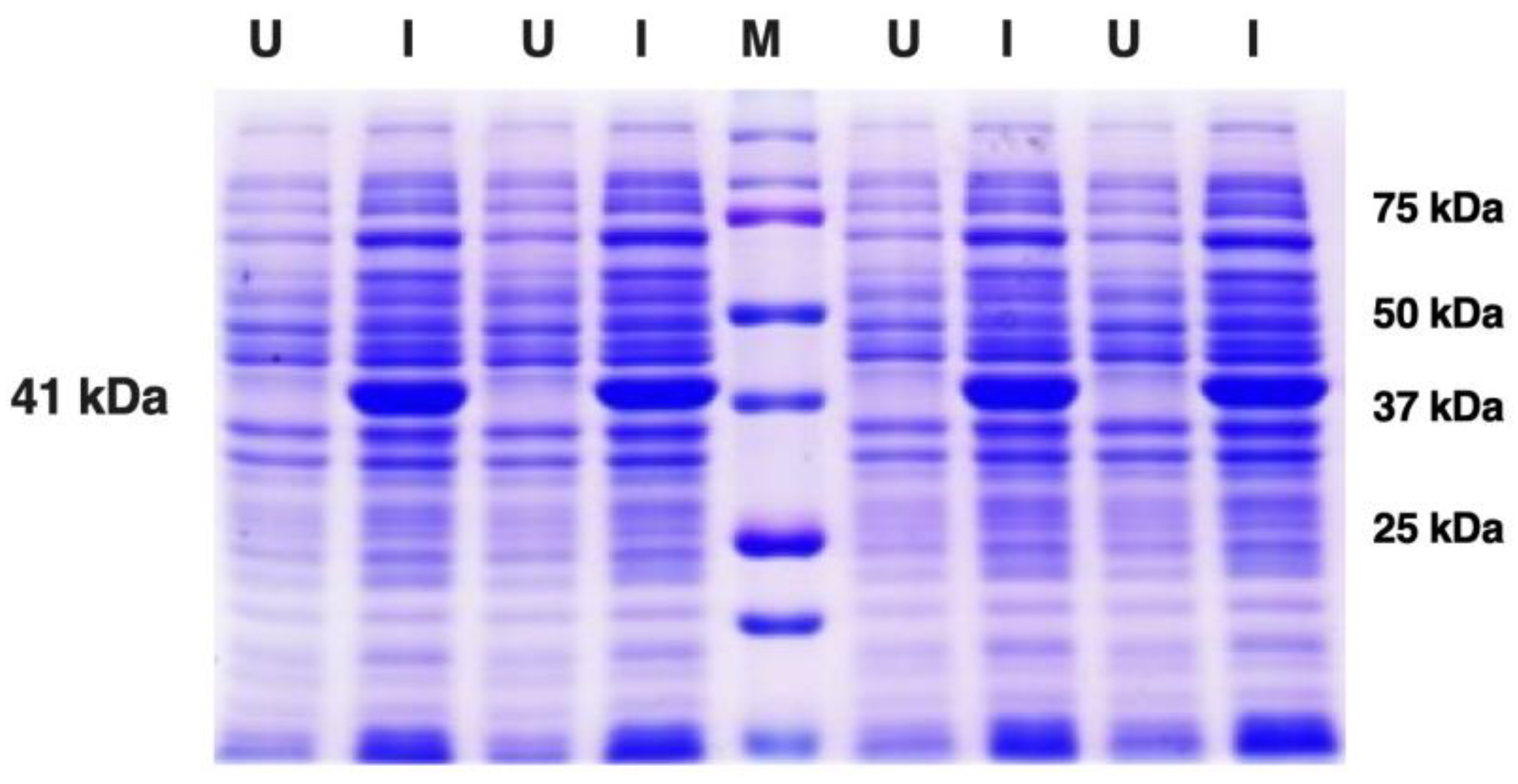
3.2. Expression and Purification of Recombinant Sap2 Protein
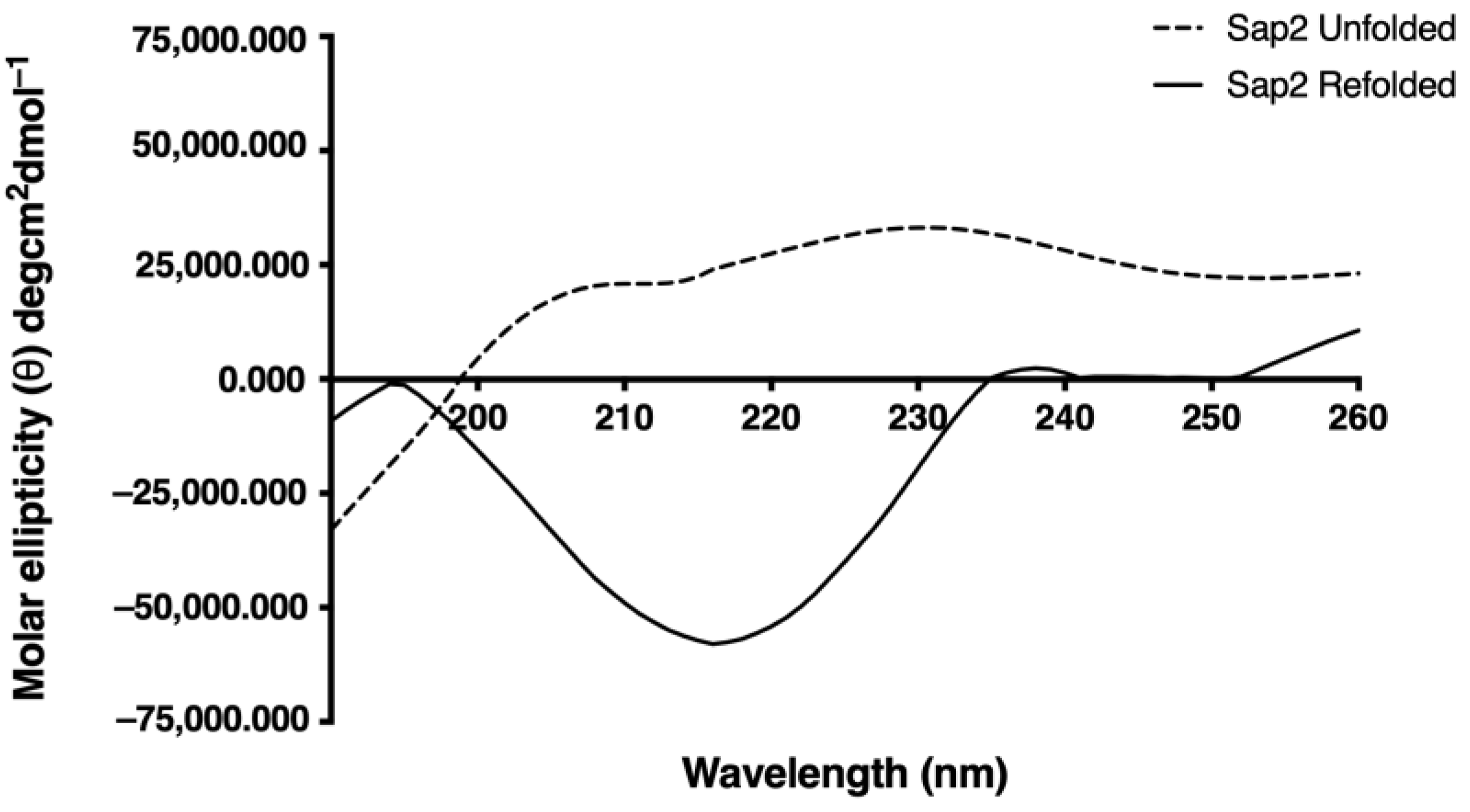
3.3. In Vitro Refolding of Recombinant Sap2 Protein
3.4. Structural Analysis of Recombinant Sap2 Protein
3.5. Evaluation of rSap2 Serological Response in Mice
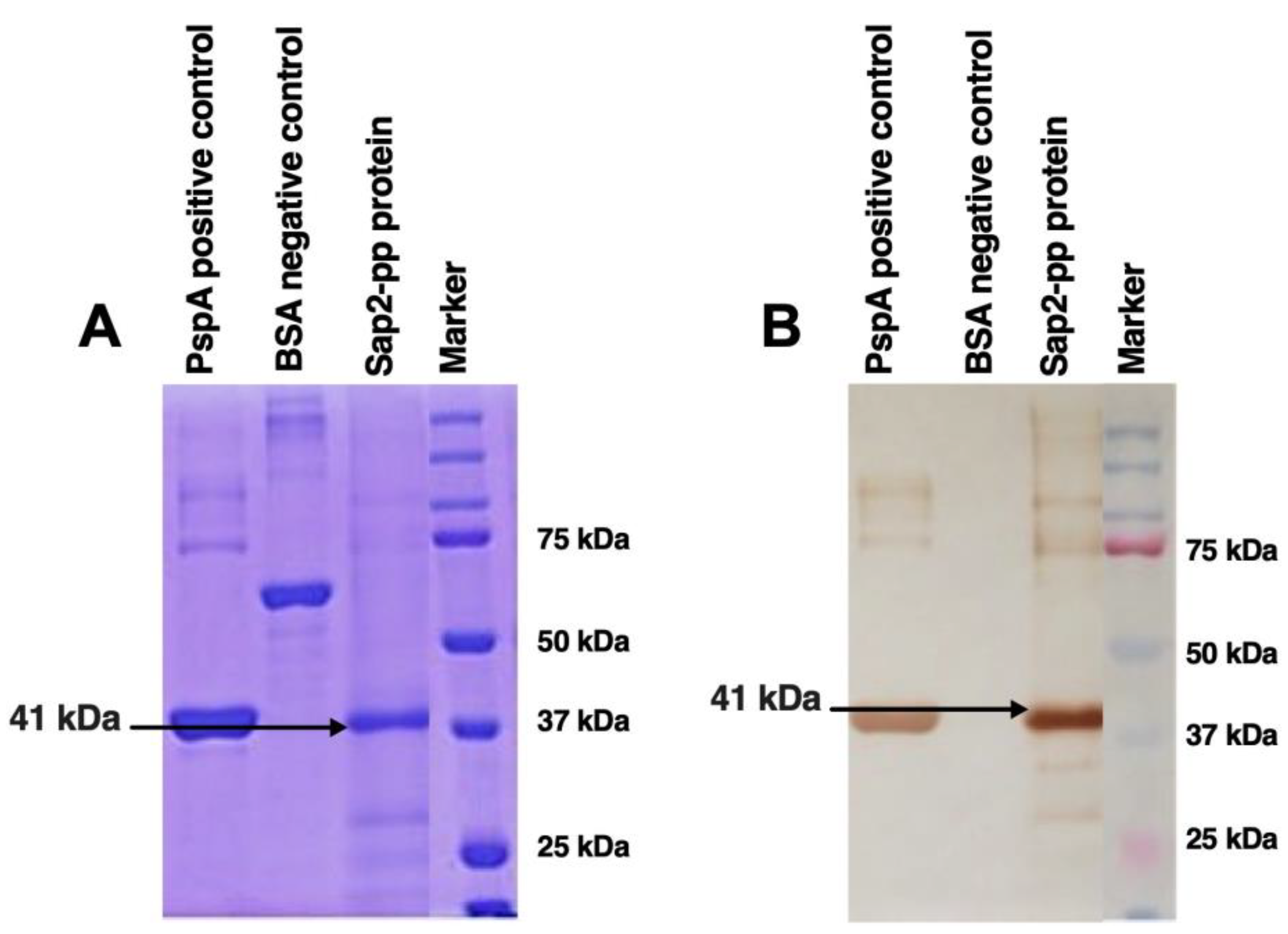
3.6. Diagnostic Potential of rSap2-Protein Using Immunoblotting
3.7. Diagnostic Potential of rSap2-Protein Using ELISA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 1–20. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.H.; Chakrabarti, A.; Li, R.Y.; Patel, A.K.; Watcharananan, S.P.; Liu, Z.; Chindamporn, A.; Tan, A.L.; Sun, P.L.; Wu, U.I.; et al. Incidence and species distribution of candidaemia in Asia: A laboratory-based surveillance study. Clin. Microbiol. Infect. 2015, 21, 946–953. [Google Scholar] [CrossRef] [Green Version]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calandra, T.; Roberts, J.A.; Antonelli, M.; Bassetti, M.; Vincent, J.L. Diagnosis and management of invasive candidiasis in the ICU: An updated approach to an old enemy. Crit. Care 2016, 20, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; de Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal biofilms and polymicrobial diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef]
- Seyoum, E.; Bitew, A.; Mihret, A. Distribution of Candida albicans and non-albicans Candida species isolated in different clinical samples and their in vitro antifungal suscetibity profile in Ethiopia. BMC Infect. Dis. 2020, 20, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberoi, J.K.; Wattal, C.; Goel, N.; Raveendran, R.; Datta, S.; Prasad, K. Non-albicans candida species in blood stream infections in a tertiary care hospital at New Delhi, India. Indian J. Med. Res. 2012, 136, 997–1003. [Google Scholar]
- Papon, N.; Courdavault, V.; Clastre, M.; Bennett, R.J.; Nac, N.-C. Emerging and Emerged Pathogenic Candida Species: Beyond the Candida albicans Paradigm. PLoS Pathog. 2013, 9, e1003550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaller, M.A.; Messer, S.A.; Jones, R.N.; Castanheira, M. Antifungal susceptibilities of Candida, Cryptococcus neoformans and Aspergillus fumigatus from the Asia and Western Pacific region: Data from the SENTRY antifungal surveillance program (2010–2012). J. Antibiot. 2015, 68, 556–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenn, J.A.P. Update of medically important yeasts and a practical approach to their identification. Lab. Med. 2007, 38, 178–183. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2015, 62, e1–e50. [Google Scholar] [CrossRef]
- Ademe, M.; Girma, F. Candida auris: From multidrug resistance to pan-resistant strains. Infect. Drug Resist. 2020, 13, 1287–1294. [Google Scholar] [CrossRef]
- Garcia-bustos, V.; Cabanero-navalon, M.D.; Ruiz-saur, A.; Ruiz-gait, A.C. What Do We Know about Candida auris? State of the Art, Knowledge Gaps, and Future Directions. Mcroorganisms 2021, 9, 2177. [Google Scholar] [CrossRef]
- Drug-Resistant Candida Species; CDC’s Antibiotic Resistance Threats Report; CDC: Atlanta, GA, USA, 2019.
- Chiurlo, M.; Mastrangelo, A.; Ripa, M.; Scarpellini, P. Invasive fungal infections in patients with COVID-19: A review on pathogenesis, epidemiology, clinical features, treatment, and outcomes. New Microbiol. 2021, 44, 71–83. [Google Scholar]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef] [Green Version]
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: A multi-institutional study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Khan, Z. Invasive candidiasis: A review of nonculture-based laboratory diagnostic methods. Indian J. Med. Microbiol. 2012, 30, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Ellepola, A.N.B.; Morrison, C.J. Laboratory diagnosis of invasive candidiasis. J. Microbiol. 2005, 43, 65–84. [Google Scholar] [PubMed]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoompoung, P.; Chayakulkeeree, M. Recent Progress in the Diagnosis of Pathogenic Candida Species in Blood Culture. Mycopathologia 2016, 181, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Yera, H.; Sendid, B.; Francois, N.; Camus, D.; Poulain, D. Contribution of serological tests and blood culture to the early diagnosis of systemic candidiasis. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Dunyach, C.; Bertout, S.; Phelipeau, C.; Drakulovski, P.; Reynes, J.; Mallié, M. Detection and identification of Candida spp. in human serum by LightCycler® real-time polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2008, 60, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Hui, M.; Cheung, S.W.; Chin, M.L.; Chu, K.C.; Chan, R.C.Y.; Cheng, A.F.B. Development and application of a rapid diagnostic method for invasive Candidiasis by the detection of D-/L-arabinitol using gas chromatography/mass spectrometry. Diagn. Microbiol. Infect. Dis. 2004, 49, 117–123. [Google Scholar] [CrossRef]
- Philip, A.; Odabasi, Z.; Matiuzzi, G.; Paetznick, V.L.; Tan, S.W.; Warmington, J.; Rex, J.H.; Ostrosky-Zeichner, L. Syscan3, a kit for detection of anti-Candida antibodies for diagnosis of invasive candidiasis. J. Clin. Microbiol. 2005, 43, 4834–4835. [Google Scholar] [CrossRef] [Green Version]
- Mikulska, M.; Calandra, T.; Sanguinetti, M.; Poulain, D.; Viscoli, C. The use of mannan antigen and anti-mannan antibodies in the diagnosis of invasive candidiasis: Recommendations from the Third European Conference on Infections in Leukemia. Crit. Care 2010, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Huppler, A.R.; Fisher, B.T.; Lehrnbecher, T.; Walsh, T.J.; Steinbach, W.J. Role of Molecular Biomarkers in the Diagnosis of Invasive Fungal Diseases in Children. J. Pediatr. Infect. Dis. Soc. 2017, 6, S32–S44. [Google Scholar] [CrossRef] [Green Version]
- Clancy, C.J.; Nguyen, M.H. Diagnosing invasive candidiasis. J. Clin. Microbiol. 2018, 56, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pemán, J.; Zaragoza, R. Current diagnostic approaches to invasive candidiasis in critical care settings. Mycoses 2010, 53, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Sendid, B.; Poirot, J.L.; Tabouret, M.; Bonnin, A.; Caillot, D.; Camus, D.; Poulain, D. Combined detection of mannanaemia and anti-mannan antibodies as a strategy for the diagnosis of systemic infection caused by pathogenic Candida species. J. Med. Microbiol. 2002, 51, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutschlechner, W.; Karall, D.; Hartmann, C.; Streiter, B.; Baumgartner-Sigl, S.; Orth-Höller, D.; Lass-Flörl, C. Mammary candidiasis: Molecular-based detection of candida species in human milk samples. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1309–1313. [Google Scholar] [CrossRef]
- Nabili, M.; Ashrafi, M.; Janbabaie, G.; Hedayati, M.T.; Ali-Moghaddam, K.; Shokohi, T. Quantification and optimization of Candida albicans DNA in blood samples using Real- Time PCR. Rep. Biochem. Mol. Biol. 2013, 2, 42–47. [Google Scholar] [PubMed]
- Mylonakis, E.; Clancy, C.J.; Ostrosky-Zeichner, L.; Garey, K.W.; Alangaden, G.J.; Vazquez, J.A.; Groeger, J.S.; Judson, M.A.; Vinagre, Y.M.; Heard, S.O.; et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: A clinical trial. Clin. Infect. Dis. 2015, 60, 892–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honarvar, B.; Lankarani, K.B.; Taghavi, M.; Vahedi, G.; Mortaz, E. Biomarker-guided antifungal stewardship policies for patients with invasive candidiasis. Curr. Med. Mycol. 2018, 4, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Hsieh, W.H.; Liu, T.S.; Lee, S.H.; Wang, C.H.; Chou, H.C.; Yeo, Y.H.; Tseng, C.P.; Lee, C.C. Multiplex PCR System for Rapid Detection of Pathogens in Patients with Presumed Sepsis—A Systemic Review and Meta-Analysis. PLoS ONE 2013, 8, e62323. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Wolk, D.M.; Lowery, T.J. T2MR and T2Candida: Novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol. 2016, 11, 103–117. [Google Scholar] [CrossRef]
- Graus, M.S.; Neumann, A.K.; Timlin, J.A. Hyperspectral fluorescence microscopy detects autofluorescent factors that can be exploited as a diagnostic method for Candida species differentiation. J. Biomed. Opt. 2017, 22, 016002. [Google Scholar] [CrossRef] [Green Version]
- Laín, A.; Elguezabal, N.; Brena, S.; García-Ruiz, J.C.; Del Palacio, A.; Moragues, M.D.; Pontón, J. Diagnosis of invasive candidiasis by enzyme-linked immunosorbent assay using the N-terminal fragment of Candida albicans hyphal wall protein 1. BMC Microbiol. 2007, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mikulska, M.; Giacobbe, D.R.; Furfaro, E.; Mesini, A.; Marchese, A.; Del Bono, V.; Viscoli, C. Lower sensitivity of serum (1,3)-β-D-glucan for the diagnosis of candidaemia due to Candida parapsilosis. Clin. Microbiol. Infect. 2016, 22, 646.e5–646.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lass-Flörl, C.; Samardzic, E.; Knoll, M. Serology anno 2021—Fungal infections: From invasive to chronic. Clin. Microbiol. Infect. 2021, 27, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Fusek, M.; Smith, E.A.; Dunn, B.M.; Foundling, S.I.; Monod, M.; Dunn, B.M. Extracellular Aspartic Proteinases from Candida albicans, Candida tropicalis, and Candida parapsilosis Yeasts Differ Substantially in Their Specificities. Biochemistry 1994, 33, 9791–9799. [Google Scholar] [CrossRef]
- Na, B.K.; Song, C.Y. Use of monoclonal antibody in diagnosis of candidiasis caused by Candida albicans: Detection of circulating aspartyl proteinase antigen. Clin. Diagn. Lab. Immunol. 1999, 6, 924–929. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, X.; Zhi Gang, J.U.; Liu, J.; Dong, S.; Wang, L. Detection of Candida albicans Sap2 in cancer patient serum samples by an indirect competitive enzyme-linked immunosorbent assay for the diagnosis of candidiasis. Indian J. Pathol. Microbiol. 2013, 56, 243–247. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, V.K.; Vasam, M.; Patil, P.S.; Dhaked, R.K.; Ansari, A.S.; Lohiya, N.K.; Parashar, D.; Tapryal, S. An in vitro refolding method to produce oligomers of anti-CHIKV, E2-IgM Fc fusion subunit vaccine candidates expressed in E. coli. J. Immunol. Methods 2020, 487, 112869. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Rohatgi, S. Vaccination with secreted aspartyl proteinase 2 protein from candida parapsilosis can enhance survival of mice during c. tropicalis-mediated systemic candidiasis. Infect. Immun. 2020, 88, e00312-20. [Google Scholar] [CrossRef]
- Corrêa, D.; Ramos, C. The use of circular dichroism spectroscopy to study protein folding, form and function. Afr. J Biochem. Res. 2009, 3, 164–173. [Google Scholar]
- Dostál, J.; Pecina, A.; Hrušková-Heidingsfeldová, O.; Marečková, L.; Pichová, I.; Řezáčová, P.; Lepšík, M.; Brynda, J. Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2494–2504. [Google Scholar] [CrossRef]
- Pitarch, A.; Nombela, C.; Gil, C. Diagnosis of Invasive Candidiasis: From Gold Standard Methods to Promising Leading-edge Technologies. Curr. Top. Med. Chem. 2018, 18, 1375–1392. [Google Scholar] [CrossRef]
- Trovato, L.; Astuto, M.; Castiglione, G.; Scalia, G.; Oliveri, S. Diagnostic surveillance by Candida albicans germ tube antibody in intensive care unit patients. J. Microbiol. Immunol. Infect. 2020, 53, 778–784. [Google Scholar] [CrossRef]
- Kondori, N.; Edebo, L.; Mattsby-Baltzer, I. Circulating β (1-3) Glucan and Immunoglobulin G Subclass Antibodies to Candida albicans Cell Wall Antigens in Patients with Systemic Candidiasis. Clin. Diagn. Lab. Immunol. 2004, 11, 344–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar, W.; Hecker, H. Diagnosis of systemic Candida infections in patients of the intensive care unit. Mycoses 2002, 45, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Sendid, B.; Caillot, D.; Baccouch-Humbert, B.; Klingspor, L.; Grandjean, M.; Bonnin, A.; Poulain, D. Contribution of the Platelia Candida-specific antibody and antigen tests to early diagnosis of systemic Candida tropicalis infection in neutropenic adults. J. Clin. Microbiol. 2003, 41, 4551–4558. [Google Scholar] [CrossRef] [Green Version]
- Clancy, C.J.; Nguyen, M.L.; Cheng, S.; Huang, H.; Fan, G.; Jaber, R.A.; Wingard, J.R.; Cline, C.; Nguyen, M.H. Immunoglobulin G responses to a panel of Candida albicans antigens as accurate and early markers for the presence of systemic candidiasis. J. Clin. Microbiol. 2008, 46, 1647–1654. [Google Scholar] [CrossRef] [Green Version]
- Sendid, B.; Dotan, N.; Nseir, S.; Savaux, C.; Vandewalle, P.; Standaert, A.; Zerimech, F.; Guery, B.P.; Dukler, A.; Colombe, J.F.; et al. Antibodies against glucan, chitin, and Saccharomyces cerevisiae mannan as new biomarkers of Candida albicans infection that complement tests based on C. albicans mannan. Clin. Vaccine Immunol. 2008, 15, 1868–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clancy, C.J.; Nguyen, M.H. Undiagnosed invasive candidiasis: Incorporating non-culture diagnostics into rational prophylactic and preemptive antifungal strategies. Expert Rev. Anti. Infect. Ther. 2014, 12, 731–734. [Google Scholar] [CrossRef]
- Morrison, C.J.; Hurst, S.F.; Reiss, E. Competitive binding inhibition enzyme-linked immunosorbent assay that uses the secreted aspartyl proteinase of Candida albicans as an antigenic marker for diagnosis of disseminated candidiasis. Clin. Diagn. Lab. Immunol. 2003, 10, 835–848. [Google Scholar] [CrossRef] [Green Version]
- Díez, A.; Carrano, G.; Bregón-Villahoz, M.; Cuétara, M.S.; García-Ruiz, J.C.; Fernandez-de-Larrinoa, I.; Moragues, M.D. Biomarkers for the diagnosis of invasive candidiasis in immunocompetent and immunocompromised patients. Diagn. Microbiol. Infect. Dis. 2021, 101, 115509. [Google Scholar] [CrossRef]
- Ardizzoni, A.; Posteraro, B.; Baschieri, M.C.; Bugli, F.; Sáez-Rosòn, A.; Manca, L.; Cacaci, M.; Paroni Sterbini, F.; De Waure, C.; Sevilla, M.J.; et al. An antibody reactivity-based assay for diagnosis of invasive candidiasis using protein array. Int. J. Immunopathol. Pharmacol. 2014, 27, 403–412. [Google Scholar] [CrossRef]
- Laín, A.; Elguezabal, N.; Amutio, E.; Fernández De Larrinoa, I.; Moragues, M.D.; Pontón, J. Use of recombinant antigens for the diagnosis of invasive candidiasis. Clin. Dev. Immunol. 2008, 2008, 72195. [Google Scholar] [CrossRef] [Green Version]
- Laín, A.; Moragues, M.D.; García Ruiz, J.C.; Mendoza, J.; Camacho, A.; Del Palacio, A.; Pontón, J. Evaluation of a novel enzyme-linked immunosorbent assay to detect immunoglobulin G antibody to enolase for serodiagnosis of invasive candidiasis. Clin. Vaccine Immunol. 2007, 14, 318–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quanping, S.; Yanyan, H.; Yicun, W.; Zhigang, J.; Yuling, G.; Li, W. The use of hybrid phage displaying antigen epitope and recombinant protein in the diagnosis of systemic Candida albicans infection in rabbits and cancer patients. Diagn. Microbiol. Infect. Dis. 2010, 68, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Pitarch, A.; Jiménez, A.; Nombela, C.; Gill, C. Serological proteome analysis to identify systemic candidiasis patients in the intensive care unit: Analytical, diagnostic and prognostic validation of anti-Candida enolase antibodies on quantitative clinical platforms. Proteom. Clin. Appl. 2008, 2, 596–618. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-Q.; Ma, C.-F.; Shi, L.-N.; Lu, J.-F.; Wang, Y.; Huang, M.; Kong, Q.-Q. Diagnostic value of immunoglobulin G antibodies against Candida enolase and fructose-bisphosphate aldolase for candidemia. BMC Infect. Dis. 2013, 13, 253. [Google Scholar] [CrossRef] [Green Version]
- He, Z.X.; Chen, J.; Li, W.; Cheng, Y.; Zhang, H.P.; Zhang, L.N.; Hou, T.W. Serological response and diagnostic value of recombinant candida cell wall protein enolase, phosphoglycerate kinase, and β-glucosidase. Front. Microbiol. 2015, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kretschmar, M.; Hube, B.; Bertsch, T.; Sanglard, D.; Merker, R.; Schröder, M.; Hof, H.; Nichterlein, T. Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect. Immun. 1999, 67, 6637–6642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, M.; Borelli, C.; Korting, H.C.; Hube, B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 2005, 48, 365–377. [Google Scholar] [CrossRef]
- Schaller, M.; Korting, H.C.; Schäfer, W.; Bastert, J.; Chen, W.C.; Hube, B. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 1999, 34, 169–180. [Google Scholar] [CrossRef]
- Schaller, M.; Bein, M.; Korting, H.C.; Baur, S.; Hamm, G.; Monod, M.; Beinhauer, S.; Hube, B. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect. Immun. 2003, 71, 3227–3234. [Google Scholar] [CrossRef] [Green Version]
- Wan Tso, G.H.; Reales-Calderon, J.A.; Pavelka, N. The Elusive Anti-Candida Vaccine: Lessons from the past and opportunities for the future. Front. Immunol. 2018, 9, 897. [Google Scholar] [CrossRef] [Green Version]
- De Bernardis, F.; Arancia, S.; Morelli, L.; Hube, B.; Sanglard, D.; Schäfer, W.; Cassone, A. Evidence that Members of the Secretory Aspartyl Proteinase Gene Family, in Particular SAP2, are Virulence Factors for Candida Vaginitis. J. Infect. Dis. 1999, 179, 201–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, M.; Korting, H.C.; Borelli, C.; Hamm, G.; Hube, B. Candida albicans-secreted aspartic proteinases modify the epithelial cytokine response in an in vitro model of vaginal candidiasis. Infect. Immun. 2005, 73, 2758–2765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Su, Q.P.; Wang, G.Y.; Wen, D.Z.; Zhang, Y.H.; Bao, H.Z.; Wang, L. Production of hybrid phage displaying secreted aspartyl proteinase epitope of Candida albicans and its application for the diagnosis of disseminated candidiasis. Mycoses 2007, 50, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Hube, B.; Sanglard, D.; Odds, F.C.; Hess, D.; Brown, A.J.P.; Gow, N.A.R.; Monod, M.; Scha, W. Disruption of Each of the Secreted Aspartyl Proteinase Genes Attenuates Virulence. Infect. Immun. 1997, 65, 3529–3538. [Google Scholar] [CrossRef] [Green Version]
- Ghadjari, A.; Matthews, R.C.; Burnie, J.P. Epitope Mapping Candida Albicans Proteinase (SAP 2). FEMS Immunol. Med. Microbiol. 1997, 19, 115–123. [Google Scholar] [CrossRef]
- Wang, Y.; Su, Q.; Dong, S.; Shi, H.; Gao, X.; Wang, L. Hybrid phage displaying SLAQVKYTSASSI induces protection against Candida albicans challenge in BALB/c mice. Hum. Vaccines Immunother. 2014, 10, 1057–1063. [Google Scholar] [CrossRef] [Green Version]
- Deorukhkar, S.C.; Saini, S. Laboratory approach for diagnosis of candidiasis through ages. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 206–218. [Google Scholar]


| S.N | CR No. | Age/Sex | Hospital Ward | Site of Candida Isolation | Isolate | Lab ID. |
|---|---|---|---|---|---|---|
| 1 | 20180-2398021 | 72Y/F | General surgery/EMG Ward | Peripheral catheter blood | Candida krusei | 3515 |
| 2 | 20170-6584839 | 64Y/F | General surgery/EMG Ward | Peripheral catheter blood | Candida tropicalis | 40685 |
| 3 | 20170-6584840 | 60Y/M | General surgery/EMG Ward | Peripheral catheter blood | Candida albicans | 40561 |
| 4 | 20180-1997525 | 53Y/M | Nephrology/Ward 22 | Peripheral catheter blood | Candida albicans | 40701 |
| 5 | 20180-1654784 | 79Y/M | Advanced Urology ward | Peripheral catheter blood | Candida albicans | 40891 |
| 6 | 20180-1787504 | 17Y/M | Cardiology/Ward A | Peripheral catheter blood | Candida albicans | 1105 |
| 7 | 20180-2525899 | 2Y/M | APC EMG Ward 2B | Peripheral catheter blood | Candida albicans | 106 |
| 8 | 20160-3375341 | 41Y/M | Hepatology/MMW | Peripheral catheter blood | Candida glabrata | 1731 |
| 9 | 20180-2693279 | 43Y/M | Gastroenterology/EMG Ward 5 | Peripheral catheter blood | Candida albicans | 5118 |
| 10 | 20180-284280 | 41Y/F | General surgery/EMG Ward | Peripheral catheter blood | Candida albicans | 5957 |
| Sap2-Regions | Sap2 Amplicons | Primer Sequence with Restriction Enzyme | Base Pairs | Amino Acids b | Position |
|---|---|---|---|---|---|
| Sap2-sp | Sap2 sp FP | CCCCGGATCCGCTAAAAGAGATGATAACCCTG | 1149 bp | 383 aa | (30–412 aa) |
| Sap2 sp RP | CCCCCAAGCTTTTGAATATAACGATTGTAAAAAAAGG | ||||
| Sap2-pp a | Sap2 pp FP | CCCCGGATCCTCTTCTCCATCATCACCATTGTAC | 1053 bp | 351 aa | (62–412 aa) |
| Sap2 pp RP | CCCCCAAGCTTTTGAATATAACGATTGTAAAAAAAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, M.; Chandley, P.; Kaur, H.; Ghosh, A.K.; Rudramurthy, S.M.; Rohatgi, S. Expression and Purification along with Evaluation of Serological Response and Diagnostic Potential of Recombinant Sap2 Protein from C. parapsilosis for Use in Systemic Candidiasis. J. Fungi 2021, 7, 999. https://doi.org/10.3390/jof7120999
Shukla M, Chandley P, Kaur H, Ghosh AK, Rudramurthy SM, Rohatgi S. Expression and Purification along with Evaluation of Serological Response and Diagnostic Potential of Recombinant Sap2 Protein from C. parapsilosis for Use in Systemic Candidiasis. Journal of Fungi. 2021; 7(12):999. https://doi.org/10.3390/jof7120999
Chicago/Turabian StyleShukla, Manisha, Pankaj Chandley, Harsimran Kaur, Anup K. Ghosh, Shivaprakash M. Rudramurthy, and Soma Rohatgi. 2021. "Expression and Purification along with Evaluation of Serological Response and Diagnostic Potential of Recombinant Sap2 Protein from C. parapsilosis for Use in Systemic Candidiasis" Journal of Fungi 7, no. 12: 999. https://doi.org/10.3390/jof7120999









