Highly Clonal Structure and Abundance of One Haplotype Characterise the Diplodia sapinea Populations in Europe and Western Asia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Disease Severity
2.2. Fungal Isolations, DNA Extraction and Isolate Identification
2.3. ITS Sequencing
2.4. Species-Specific Conventional PCR Primer Design
2.5. Haplotype Determination
2.6. Genetic Diversity and Differentiation of Populations
2.7. Mating Type Determination and Random Mating
2.8. Isolation by Distance
2.9. Population Clustering
3. Results
3.1. Identification of Diplodia sapinea with Conventional PCR Primers
3.2. Multilocus Haplotypes
3.3. Population Differentiation
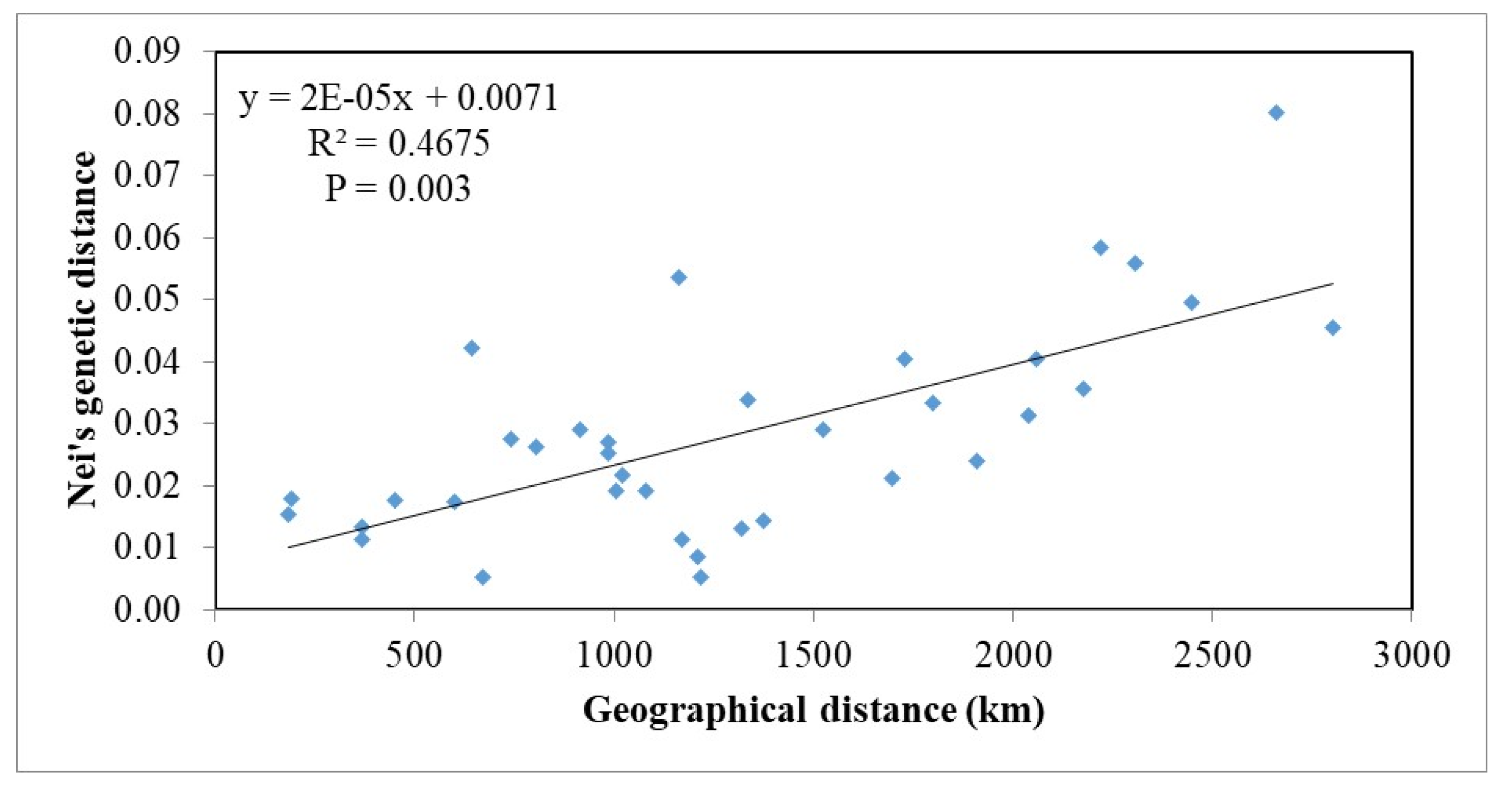
3.4. Isolation by Distance and Clustering Analysis
3.5. Genetic Diversity and Population Statistics
3.6. Mating Type Distribution and Haploid Linkage Disequilibrium
3.7. Haplotypic Diversity at Small Spatial Scale
3.8. Allele Polymorphism in Different Loci in North America and Europe
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, A.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.; Groenewald, J.; Crous, P. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef] [Green Version]
- Slippers, B.; Boissin, E.; Phillips, A.; Groenewald, J.; Lombard, L.; Wingfield, M.; Postma, A.; Burgess, T.; Crous, P. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud. Mycol. 2013, 76, 31–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brookhouser, L.W.; Peterson, G.W. Infection of Austrian, Scots, and ponderosa pines by Diplodia pinea. Phytopathology 1971, 61, 409–414. [Google Scholar] [CrossRef]
- Peterson, G.W. Infection, epidemiology, and control of Diplodia blight of Austrian ponderosa and Scots pines. Phytopathology 1977, 67, 511–514. [Google Scholar] [CrossRef]
- Swart, W.J.; Knox-Davis, P.S.; Wingfield, M.J. Sphaeropsis sapinea, with special reference to its occurrence on Pinus spp. in South Africa. South Africa For. J. 1985, 35, 1–8. [Google Scholar]
- Blodgett, J.T.; Stanosz, G.R. Sphaeropsis sapinea Morphotypes Differ in Aggressiveness, but Both Infect Nonwounded Red or Jack Pines. Plant Dis. 1997, 81, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Swart, W.J.; Wingfield, M.J. Biology and Control of Sphaeropsis sapinea on Pinus species in South Mrica. Detail 1991, 30, 40. [Google Scholar]
- Bihon, W.; Slippers, B.; Burgess, T.; Wingfield, M.J.; Wingfield, B. Sources of Diplodia pinea endophytic infections in Pinus patula and P. radiata seedlings in South Africa. For. Pathol. 2011, 41, 370–375. [Google Scholar] [CrossRef] [Green Version]
- Drenkhan, T.; Voolma, K.; Adamson, K.; Sibul, I. The large pine weevil Hylobius abietis (L.) as a potential vector of the pathogenic fungus Diplodia sapinea (Fr.) Fuckel. Agric. For. Èntomol. 2016, 19, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Bußkamp, J.; Langer, G.J.; Langer, E.J. Sphaeropsis sapinea and fungal endophyte diversity in twigs of Scots pine (Pinus sylvestris) in Germany. Mycol. Prog. 2020, 19, 985–999. [Google Scholar] [CrossRef]
- Oliva, J.; Ridley, M.; Redondo, M.A.; Caballol, M. Competitive exclusion amongst endophytes determines shoot blight severity on pine. Funct. Ecol. 2020, 35, 239–254. [Google Scholar] [CrossRef]
- Stanosz, G.R.; Blodgett, J.T.; Smith, D.R.; Kruger, E.L. Water stress and Sphaeropsis sapinea as a latent pathogen of red pine seedlings. New Phytol. 2001, 149, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Flowers, J.; Hartman, J.; Vaillancourt, L. Detection of Latent Sphaeropsis sapinea Infections in Austrian Pine Tissues Using Nested-Polymerase Chain Reaction. Phytopathology 2003, 93, 1471–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luchi, N.; Capretti, P.; Surico, G.; Orlando, C.; Pazzagli, M.; Pinzani, P. A Real-Time Quantitative PCR Assay for the Detection of Sphaeropsis sapinea from Inoculated Pinus nigra Shoots. J. Phytopathol. 2005, 153, 37–42. [Google Scholar] [CrossRef]
- Luchi, N.; Pratesi, N.; Simi, L.; Pazzagli, M.; Capretti, P.; Scala, A.; Slippers, B.; Pinzani, P. High-Resolution Melting Analysis: A new molecular approach for the early detection of Diplodia pinea in Austrian pine. Fungal Biol. 2011, 115, 715–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.R.; Stanosz, G.R. A Species-Specific PCR Assay for Detection of Diplodia pinea and D. scrobiculata in Dead Red and Jack Pines with Collar Rot Symptoms. Plant Dis. 2006, 90, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Desprez-Loustau, M.-L.; Marçais, B.; Nageleisen, L.-M.; Piou, D.; Vannini, A. Interactive effects of drought and pathogens in forest trees. Ann. For. Sci. 2006, 63, 597–612. [Google Scholar] [CrossRef] [Green Version]
- Fabre, B.; Marçais, B.; Desprez-Loustau, M.; Piou, D. Can the emergence of pine Diplodia shoot blight in France be explained by changes in pathogen pressure linked to climate change? Glob. Chang. Biol. 2011, 17, 3218–3227. [Google Scholar] [CrossRef] [Green Version]
- Langer, G.; Bressem, U.; Habermann, M. Diplodia-Triebsterben der Kiefer und endophytischer Nachweis des Erregers Sphaeropsis sapinea. AFZ-Der Wald 2011, 11, 28–31. [Google Scholar]
- De Urbina, E.O.; Mesanza, N.; Aragonés, A.; Raposo, R.; Elvira-Recuenco, M.; Boqué, R.; Patten, C.; Aitken, J.; Iturritxa, E. Emerging Needle Blight Diseases in Atlantic Pinus Ecosystems of Spain. Forests 2017, 8, 18. [Google Scholar] [CrossRef] [Green Version]
- Fries, E.M. Systema Mycologicum, 2, 620; Gryphiswaldia: Lund, Sweden; Berlin, Germany, 1823. [Google Scholar]
- Oliva, J.; Boberg, J.; Stenlid, J. First report of Sphaeropsis sapinea on Scots pine (Pinus sylvestris) and Austrian pine (P. nigra) in Sweden. New Dis. Rep. 2013, 27, 23. [Google Scholar] [CrossRef] [Green Version]
- Adamson, K.; Klavina, D.; Drenkhan, R.; Gaitnieks, T.; Hanso, M. Diplodia sapinea is colonizing the native Scots pine (Pinus sylvestris) in the northern Baltics. Eur. J. Plant Pathol. 2015, 143, 343–350. [Google Scholar] [CrossRef]
- Brodde, L.; Adamson, K.; Camarero, J.J.; Castaño, C.; Drenkhan, R.; Lehtijärvi, A.; Luchi, N.; Migliorini, D.; Sánchez-Miranda, A.; Stenlid, J.; et al. Diplodia Tip Blight on Its Way to the North: Drivers of Disease Emergence in Northern Europe. Front. Plant Sci. 2019, 9, 1818. [Google Scholar] [CrossRef] [Green Version]
- Hanso, M.; Drenkhan, R. Diplodia pinea is a new pathogen on Austrian pine (Pinus nigra) in Estonia. Plant Pathol. 2009, 58, 797. [Google Scholar] [CrossRef]
- Müller, M.M.; Hantula, J.; Wingfield, M.; Drenkhan, R. Diplodia sapinea found on Scots pine in Finland. For. Pathol. 2019, 49, e12483. [Google Scholar] [CrossRef] [Green Version]
- Burgess, T.; Wingfield, M.J. Quarantine is important in restricting the spread of exotic seed-borne tree pathogens in the Southern hemisphere. Intern. For. Rev. 2002, 4, 56–65. [Google Scholar]
- Wingfield, M.J.; Slippers, B.; Roux, J.; Wingfield, B.D. Worldwide movement of exotic forest fungi, especially in the tropics and the southern hemisphere: This article examines the impact of fungal pathogens introduced in plantation forestry. Bioscience 2001, 51, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Adamson, K.; Drenkhan, R.; Hanso, M. Invasive brown spot needle blight caused by Lecanosticta acicola in Estonia. Scand. J. For. Res. 2015, 30, 587–593. [Google Scholar] [CrossRef]
- Drenkhan, R.; Riit, T.; Adamson, K.; Hanso, M. The earliest samples of Hymenoscyphus albidus vs. H. fraxineus in Estonian mycological herbaria. Mycol. Prog. 2016, 15, 835–844. [Google Scholar] [CrossRef]
- Hanso, M.; Drenkhan, R. First observations of Mycosphaerella pini Estonia. Plant Pathol. 2008, 57, 1177. [Google Scholar] [CrossRef]
- Jürisoo, L.; Adamson, K.; Padari, A.; Drenkhan, R. Health of elms and Dutch elm disease in Estonia. Eur. J. Plant Pathol. 2019, 154, 823–841. [Google Scholar] [CrossRef]
- Lutter, R.; Drenkhan, R.; Tullus, A.; Jürimaa, K.; Tullus, T.; Tullus, H. First record of Entoleuca mammata in hybrid aspen plantations in hemiboreal Estonia and stand–environmental factors affecting its prevalence. Eur. J. For. Res. 2019, 138, 263–274. [Google Scholar] [CrossRef]
- Mullett, M.S.; Adamson, K.; Bragança, H.; Bulgakov, T.; Georgieva, M.; Henriques, J.; Jürisoo, L.; Laas, M.; Drenkhan, R. New country and regional records of the pine needle blight pathogens Lecanosticta acicola, Dothistroma septosporum and Dothistroma pini. For. Pathol. 2018, 48, e12440. [Google Scholar] [CrossRef]
- Bihon, W.; Burgess, T.; Slippers, B.; Wingfield, M.J.; Wingfield, B. Distribution of Diplodia pinea and its genotypic diversity within asymptomatic Pinus patula trees. Australas. Plant Pathol. 2011, 40, 540–548. [Google Scholar] [CrossRef]
- Bihon, W.; Burgess, T.; Slippers, B.; Wingfield, M.J.; Wingfield, B.D. High levels of genetic diversity and cryptic recombination is widespread in introduced Diplodia pinea populations. Australas. Plant Pathol. 2012, 41, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Bihon, W.; Wingfield, M.J.; Slippers, B.; Duong, T.; Wingfield, B. MAT gene idiomorphs suggest a heterothallic sexual cycle in a predominantly asexual and important pine pathogen. Fungal Genet. Biol. 2014, 62, 55–61. [Google Scholar] [CrossRef] [Green Version]
- McDonald, B.A.; Linde, C. Pathogen populationgenetics, Evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [Green Version]
- De Wet, J.; Wingfield, M.J.; Coutinho, T.; Wingfield, B. Characterization of Sphaeropsis sapinea Isolates from South Africa, Mexico, and Indonesia. Plant Dis. 2000, 84, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.R.; Stanosz, G.R. Confirmation of two distinct populations of Sphaeropsis sapinea in the north central United States using RAPDs. Phytopathology 1995, 85, 699–704. [Google Scholar] [CrossRef]
- Hausner, G.; Reid, J.; Hopkin, A.; Davis, C. Variation in culture and rDNA among isolates of Sphaeropsis sapinea from Ontario and Manitoba. Can. J. Plant Pathol. 1999, 21, 256–264. [Google Scholar] [CrossRef]
- Burgess, T.; Wingfield, B.D.; Wingfield, M.J. Comparison of genotypic diversity in native and introduced populations of Sphaeropsis sapinea isolated from Pinus radiata. Mycol. Res. 2001, 105, 1331–1339. [Google Scholar] [CrossRef] [Green Version]
- Manzanos, T.; Aragonés, A.; Iturritxa, E. Genotypic diversity and distribution of Sphaeropsis sapinea within Pinus radiata trees from northern Spain. For. Pathol. 2019, 49, e12550. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; De Wet, J.; Coutinho, T. Genotypic Diversity of Sphaeropsis sapinea from South Africa and Northern Sumatra. Plant Dis. 2000, 84, 139–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bihon, W.; Slippers, B.; Burgess, T.; Wingfield, M.J.; Wingfield, B. Diverse sources of infection and cryptic recombination revealed in South African Diplodia pinea populations. Fungal Biol. 2012, 116, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Burgess, T.; Wingfield, M.J.; Wingfield, B. Simple Sequence Repeat Markers Distinguish among Morphotypes of Sphaeropsis sapinea. Appl. Environ. Microbiol. 2001, 67, 354–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, T.I.; Wingfield, M.J.; Wingfield, B.D. Global distribution of Diplodia pinea genotypes revealed using simple sequence repeat (SSR) markers. Australas. Plant Pathol. 2004, 33, 513–519. [Google Scholar] [CrossRef]
- Zlatkovic, M.; Wingfield, M.J.; Jami, F.; Slippers, B. Genetic uniformity characterizes the invasive spread of Neofusicoccum parvum and Diplodia sapinea in the Western Balkans. For. Pathol. 2019, 49, e12491. [Google Scholar] [CrossRef]
- Caudullo, G.; Welk, E.; San-Miguel-Ayanz, J. Chorological data for the main European woody species. Data Brief 2020, 12, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Mullett, M.; Barnes, I. Dothistroma Isolation and Molecular Identification Methods; 2012; 22p. Available online: https://www.forestresearch.gov.uk/documents/305/DIAROD_052012_Isolation_and_indentification_97fNCCI.pdf (accessed on 30 July 2021).
- Keriö, S.; Terhonen, E.; LeBoldus, J.M. Safe DNA-extraction Protocol Suitable for Studying Tree-fungus Interactions. Bio-Protocol 2020, 10, e3634. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J. PCR Protocols: A Guide to Methods and Applications: Part Three—Genetics and Evolution; Chapter 38, Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Drenkhan, R.; Adamson, K.; Jürimaa, K.; Hanso, M. Dothistroma septosporumon firs (Abiesspp.) in the northern Baltics. For. Pathol. 2014, 44, 250–254. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT; Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Peakall, R.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Zhan, J.; Pettway, R.; McDonald, B. The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol. 2003, 38, 286–297. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Jakobsson, M.; Rosenberg, N. ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 2008, 24, 2498–2504. [Google Scholar] [CrossRef] [Green Version]
- Kivelä, M.; Arnaud-Haond, S.; Saramäki, J. Eden etworks: A user-friendly software to build and analyse networks in biogeography, ecology and population genetics. Mol. Ecol. Res. 2015, 15, 117–122. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Wen, W.; Falush, D. Documentation for STRUCTURE Software: Version 2.3; University of Chicago, Department of Human Genetics: Chicago, IL, USA, 2010. [Google Scholar]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamson, K.; Mullett, M.; Solheim, H.; Barnes, I.; Müller, M.M.; Hantula, J.; Vuorinen, M.; Kacergius, A.; Markovskaja, S.; Musolin, D.L.; et al. Looking for relationships between the populations of Dothistroma septosporum in northern Europe and Asia. Fungal Genet. Biol. 2018, 110, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Drenkhan, R.; Hantula, J.; Vuorinen, M.; Jankovský, L.; Müller, M.M. Genetic diversity of Dothistroma septosporum in Estonia, Finland and Czech Republic. Eur. J. Plant Pathol. 2013, 136, 71–85. [Google Scholar] [CrossRef]
- Drenkhan, R.; Hanso, M. Recent invasion of foliage fungi of pines (Pinus spp.) to the Northern Baltics. For. Stud. 2009, 51, 49–64. [Google Scholar] [CrossRef]
- Sutton, B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute. 1980. Available online: https://www.cabdirect.org/?target=%2fcabdirect%2fabstract%2f19801366283 (accessed on 16 June 2021).
- McDonald, B.; McDermott, J.M. Population Genetics of Plant Pathogenic Fungi. Bioscience 1993, 43, 311–319. [Google Scholar] [CrossRef]
- Lundquist, J.E. A History of Five Forest Diseases in South Africa. South Afr. For. J. 1987, 140, 51–59. [Google Scholar] [CrossRef]
- Terhonen, E.-L.; Babalola, J.; Kasanen, R.; Jalkanen, R.; Blumenstein, K. Sphaeropsis sapinea found as symptomless endophyte in Finland. Silva Fenn. 2021, 55. [Google Scholar] [CrossRef]





| Primer Pair | Locus | Fluorescent Label | Annealing Temp (°C) | Reference |
|---|---|---|---|---|
| TB1 and TB2-2 | SS1 | ATTO550 | 58 | [46] |
| TB5 and TB6 | SS2 | ATTO532 | 58 | [46] |
| TB19 and TB20-2 | SS5 | FAM | 62 | [46] |
| TB23 and TB24 | SS7 | ATTO532 | 62 | [46] |
| TB35-2 and TB36 | SS8 | ATTO565 | 62 | [46] |
| TB37 and TB38 | SS9 | ATTO550 | 62 | [46] |
| TB41 and TB42 | SS10 | FAM | 62 | [46] |
| TB43 and TB44 | SS11 | ATTO565 | 58 | [46] |
| WB1-a and WB1-b | SS12 | ATTO565 | 66 | [35] |
| WB2-a and WB2-b | SS13 | FAM | 57 | [35] |
| WB4-a and WB4-b | SS14 | ATTO550 | 60 | [35] |
| WB7-a and WB7-b | SS15 | ATTO550 | 55 | [35] |
| WB8-a and WB8-b | SS16 | ATTO532 | 55 | [35] |
| MLH No. | BEL | EST | FIN | GEO | GER | ITA | LAT | MAC | NOR | POL | RUS | SER | SLO | SWI | UKR | Haplotype Frequency | Percent of Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | (1) | - | - | - | - | - | - | - | - | - | - | - | - | - | (1) | 0.0 |
| 2 | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 2 | 0.6 |
| 3 | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 4 | - | (1) | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 (1) | 0.3 |
| 5 | - | (1) | - | - | - | - | - | - | - | - | - | - | - | - | - | (1) | 0.0 |
| 6 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 7 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 8 | - | 1 (7) | - | - | 24 | - | - | - | - | - | - | - | - | - | - | 25 (7) | 7.3 |
| 9 | - | 1 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 2 | 0.6 |
| 10 | - | (1) | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 (1) | 0.3 |
| 11 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 12 | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 13 | - | (1) | - | - | - | - | - | - | - | - | - | - | - | - | - | (1) | 0.3 |
| 14 | - | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | 1 | 0.3 |
| 15 | - | (3) | 2 | - | - | - | 1 | - | - | - | - | 2 | - | - | - | 5 (3) | 1.5 |
| 16 | - | (1) | - | - | - | - | - | - | - | - | - | - | - | - | 2 | 2 (1) | 0.6 |
| 17 | - | - | - | 2 | - | - | - | - | - | - | - | - | - | - | - | 2 | 0.6 |
| 18 | - | - | - | 7 | - | - | - | - | - | - | - | - | - | - | - | 7 | 2.0 |
| 19 | - | - | - | 10 | - | - | - | - | - | - | - | 6 | 3 (2) | - | - | 19 (2) | 5.6 |
| 20 | - | - | - | 16 | - | - | - | - | - | - | - | - | - | - | - | 16 | 4.7 |
| 21 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 1 | 0.3 |
| 22 | - | - | 1 | - | - | - | 3 | - | - | 1 | - | - | - | - | 3 | 8 | 2.3 |
| 23 | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 24 | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | 1 | 0.3 |
| 25 | - | - | - | 4 | - | - | - | - | - | - | - | - | - | - | - | 4 | 1.2 |
| 26 | - | - | - | - | - | - | - | - | - | - | - | - | 1 (1) | - | - | 1 (1) | 0.3 |
| 27 | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 28 | - | - | - | - | - | - | - | - | - | - | - | - | 1 (2) | - | - | 1 (2) | 0.3 |
| 29 | 3 | 5 (14) | 3 | 3 | 41 | - | 7 | 4 | 5 | 27 | - | 14 | 36 (16) | 1 | 6 | 155 (30) | 45.3 |
| 30 | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 31 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 32 | - | - | - | - | - | - | - | - | - | - | - | 5 | 2 | - | - | 7 | 2.0 |
| 33 | 2 | 1 | 2 | 1 | 2 | 1 | - | - | - | 1 | - | - | 8 (12) | - | 7 | 25 (12) | 7.3 |
| 34 | - | - | - | - | - | 1 | - | 2 | - | - | - | - | - | - | - | 3 | 0.9 |
| 35 | - | - | - | - | - | 7 | - | - | - | - | - | - | - | - | - | 7 | 2.0 |
| 36 | - | - | - | - | 2 | - | - | - | - | - | - | - | - | - | - | 2 | 0.6 |
| 37 | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 38 | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 39 | - | - | - | - | - | - | - | - | - | - | - | - | (1) | - | - | (1) | 0.0 |
| 40 | - | 1 | - | - | 1 | 1 | - | - | - | - | - | - | 1 (2) | 1 | - | 5 (2) | 1.5 |
| 41 | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 42 | - | (1) | - | - | - | - | - | - | - | - | - | - | - | - | - | (1) | 0.0 |
| 43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 1 | 0.3 |
| 44 | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 45 | - | 1 (5) | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 (5) | 0.3 |
| 46 | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 47 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | 1 | 0.3 |
| 48 | - | 2 (5) | - | - | - | - | - | - | - | - | - | - | - | - | - | 2 (5) | 0.6 |
| 49 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 1 | 0.3 |
| 50 | 4 | (4) | 2 | - | 2 | - | 1 | - | - | 3 | 1 | 1 | 2 (2) | - | 1 | 17 (6) | 5.0 |
| 51 | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 0.3 |
| 52 | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | 1 | 0.3 |
| N | 10 | 14 (45) | 11 | 46 | 80 | 13 | 14 | 7 | 5 | 32 | 2 | 29 | 54 (38) | 2 | 23 | 342 (83) | 100 |
| Subpopulation Code | N of Sampling Sites | N of Sampled Trees | N of Sampled Insects | N of Isolates | N of Haplotypes cc | N of Private Haplotypes cc | Clonal Fraction |
|---|---|---|---|---|---|---|---|
| BEL | 2 | 10 | - | 10 | 4 | 1 | 0.60 |
| EST | 11 | 14 | - | 14 | 9 | 2 | 0.69 |
| FIN | 3 | 11 | - | 11 | 6 | 1 | 0.45 |
| GEO | 2 | 46 | - | 46 | 10 | 3 | 0.78 |
| GER | 5 | 80 | - | 80 | 13 | 7 | 0.84 |
| ITA | 1 | 13 | - | 13 | 7 | 3 | 0.46 |
| LAT | 8 | 14 | - | 14 | 6 | 2 | 0.57 |
| MAC | 1 | 7 | - | 7 | 3 | 1 | 0.57 |
| NOR | 1 | - | 5 | 5 | 1 | 0 | 0.80 |
| POL | 2 | 32 | - | 32 | 4 | 0 | 0.88 |
| RUS | 1 | 2 | - | 2 | 2 | 1 | 0.00 |
| SER | 1 | 29 | - | 29 | 6 | 1 | 0.79 |
| SLO | 10 | 54 | - | 54 | 8 | 2 | 0.85 |
| SWI | 1 | 2 | - | 2 | 2 | 0 | 0.00 |
| UKR | 3 | 23 | - | 23 | 9 | 2 | 0.61 |
| Subpopulation Code | BEL | EST | FIN | GEO | GER | ITA | LAT | MAC | POL | RUS | SER | SLO | SWI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EST | 0.093 | ||||||||||||
| FIN | 0.411 | 0.435 | |||||||||||
| GEO | 0.150 | 0.004 | 0.127 | ||||||||||
| GER | 0.138 | 0.422 | 0.253 | 0.001 | |||||||||
| ITA | 0.140 | 0.470 | 0.327 | 0.013 | 0.041 | ||||||||
| LAT | 0.241 | 0.454 | 0.461 | 0.062 | 0.249 | 0.337 | |||||||
| MAC | 0.144 | 0.429 | 0.318 | 0.122 | 0.167 | 0.439 | 0.504 | ||||||
| POL | 0.234 | 0.413 | 0.380 | 0.140 | 0.339 | 0.384 | 0.481 | 0.457 | |||||
| RUS | 0.332 | 0.055 | 0.287 | 0.046 | 0.047 | 0.078 | 0.177 | 0.096 | 0.333 | ||||
| SER | 0.388 | 0.167 | 0.423 | 0.390 | 0.054 | 0.393 | 0.445 | 0.531 | 0.483 | 0.322 | |||
| SLO | 0.430 | 0.439 | 0.375 | 0.163 | 0.399 | 0.489 | 0.438 | 0.349 | 0.368 | 0.117 | 0.411 | ||
| SWI | 0.268 | 0.399 | 0.355 | 0.257 | 0.399 | 0.332 | 0.429 | 0.598 | 0.539 | 0.334 | 0.284 | 0.438 | |
| UKR | 0.378 | 0.145 | 0.397 | 0.152 | 0.042 | 0.097 | 0.394 | 0.409 | 0.407 | 0.381 | 0.392 | 0.428 | 0.430 |
| Subpopulation Code | EST | FIN | GEO | GER | ITA | LAT | SER | SLO | UKR |
|---|---|---|---|---|---|---|---|---|---|
| EST | - | 183 | 2307 | 1217 | 1908 | 190 | 1524 | 1209 | 987 |
| FIN | 0.015 | - | 2446 | 1318 | 2057 | 368 | 1697 | 1374 | 1169 |
| GEO | 0.056 | 0.049 | - | 2803 | 2659 | 2222 | 2039 | 2177 | 1801 |
| GER | 0.005 | 0.013 | 0.045 | - | 914 | 1079 | 985 | 672 | 1004 |
| ITA | 0.024 | 0.040 | 0.080 | 0.029 | - | 1727 | 643 | 742 | 1164 |
| LAT | 0.018 | 0.013 | 0.058 | 0.019 | 0.040 | - | 1334 | 1021 | 803 |
| SER | 0.029 | 0.021 | 0.031 | 0.025 | 0.042 | 0.034 | - | 368 | 601 |
| SLO | 0.008 | 0.014 | 0.036 | 0.005 | 0.027 | 0.022 | 0.011 | - | 451 |
| UKR | 0.027 | 0.011 | 0.033 | 0.019 | 0.054 | 0.026 | 0.017 | 0.018 | - |
| Subpopulation Code | N of Isolates | N of Haplotypes cc | Total No of Alleles | Private Alleles | Allelic Richness Ar (SE) 1 cc | Private Allelic Richness Par (SE) 1 cc | Mean Number of Different Alleles Na (SE) 1 cc | Mean Unbiased Diversity uh (SE) 1 cc | Mean Haploid Genetic Diversity h (SE) 1 cc | Disease Severity 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| BEL | 10 | 4 | 17 | 1 | NC | NC | NC | NC | NC | 2 |
| EST | 14 | 9 | 17 | 1 | 1.224 (0.124) | 0.043 (0.043) | 1.231 (0.163) | 0.072 (0.051) | 0.060 (0.042) | 1 |
| FIN | 11 | 6 | 19 | 1 | 1.333 (0.154) | 0.064 (0.064) | 1.385 (0.180) | 0.138 (0.063) | 0.115 (0.052) | 1 |
| GEO | 46 | 10 | 22 | 3 | 1.478 (0.257) | 0.152 (0.103) | 1.462 (0.243) | 0.149 (0.079) | 0.124 (0.066) | 2 |
| GER | 80 | 13 | 22 | 2 | 1.363 (0.159) | 0.077 (0.048) | 1.385 (0.180) | 0.138 (0.063) | 0.115 (0.052) | 2 |
| ITA | 13 | 7 | 22 | 4 | 1.560 (0.192) | 0.321 (0.147) | 1.462 (0.183) | 0.179 (0.069) | 0.150 (0.057) | 2 |
| LAT | 14 | 6 | 20 | 0 | 1.410 (0.159) | 0.047 (0.029) | 1.462 (0.183) | 0.169 (0.065) | 0.140 (0.054) | 1 |
| MAC | 7 | 3 | 16 | 0 | NC | NC | NC | NC | NC | 2 |
| NOR | 5 | 1 | 14 | 0 | NC | NC | NC | NC | NC | 1 |
| POL | 32 | 4 | 17 | 0 | NC | NC | NC | NC | NC | 2 |
| RUS | 2 | 2 | 13 | 0 | NC | NC | NC | NC | NC | 2 |
| SER | 29 | 6 | 19 | 0 | 1.372 (0.171) | 0.022 (0.022) | 1.385 (0.180) | 0.174 (0.078) | 0.143 (0.064) | 2 |
| SLO | 54 | 8 | 19 | 0 | 1.472 (0.187) | 0.032 (0.032) | 1.538 (0.215) | 0.185 (0.071) | 0.154 (0.059) | 2 |
| SWI | 2 | 2 | 15 | 0 | NC | NC | NC | NC | NC | 2 |
| UKR | 23 | 9 | 20 | 1 | 1.378 (0.203) | 0.043 (0.043) | 1.308 (0.175) | 0.118 (0.065) | 0.098 (0.054) | 1 |
| Subpopulation Code | MAT1-1-1 Non-cc | MAT1-2-1 Non-cc | p-Value of Exact Binomial Test Non-cc | Index of Association IA Non-cc | p-Value of IA Non-cc | MAT1-1-1 cc | MAT1-2-1 cc | p-Value of Exact Binomial Test cc | Index of Association IA cc | p-Value of IA cc |
|---|---|---|---|---|---|---|---|---|---|---|
| BEL | 5 | 5 | 0.623 | 1.369 | 0.052 | 2 | 2 | 0.688 | 1.583 | 0.339 |
| EST | 7 | 5 | 0.387 | 1.384 | 0.923 | 4 | 4 | 0.637 | 1.443 | 1.000 |
| FIN | 6 | 5 | 0.500 | 1.640 | 0.462 | 3 | 3 | 0.656 | 1.628 | 0.824 |
| GEO | 9 | 36 | 0.000 | 1.707 | 0.001 | 4 | 6 | 0.377 | 1.855 | 0.169 |
| GER | 45 | 32 | 0.086 | 2.081 | 0.738 | 10 | 4 | 0.090 | 1.576 | 0.975 |
| ITA | 2 | 11 | 0.011 | 2.795 | 0.002 | 2 | 5 | 0.228 | 2.377 | 0.346 |
| LAT | 5 | 8 | 0.291 | 1.397 | 0.260 | 2 | 4 | 0.344 | 1.626 | 0.721 |
| MAC | 0 | 7 | 0.007 | 4.168 | 0.011 | 0 | 3 | 0.125 | 6.210 | 0.045 |
| NOR | 5 | 0 | 0.031 | NC | NC | 1 | 0 | 0.500 | NC | NC |
| POL | 20 | 12 | 0.108 | 1.604 | 0.005 | 3 | 1 | 0.313 | 1.546 | 0.254 |
| RUS | 2 | 0 | 0.250 | NC | NC | 2 | 0 | 0.250 | NA | NA |
| SER | 14 | 15 | 0.500 | 1.774 | 0.001 | 4 | 2 | 0.344 | 2.342 | 0.156 |
| SLO | 19 | 35 | 0.020 | 1.259 | 0.050 | 3 | 6 | 0.254 | 1.167 | 0.890 |
| SWI | 1 | 1 | 0.750 | NC | NC | 1 | 1 | 0.750 | NC | NC |
| UKR | 13 | 9 | 0.262 | 1.715 | 0.185 | 7 | 2 | 0.090 | 2.144 | 0.738 |
| Country | Location | Sampling Date | Host Species | Substrate | No. of Trees | No. of Isolates | Clonal Fraction | MLH Nos. |
|---|---|---|---|---|---|---|---|---|
| Estonia | Järvselja nursery | 17.05.2012; 28.06.2013; 16.02.2018 | Pinus nigra | Cone, shoot | 1 | 16 | 0.38 | 4; 5; 8; 10; 16; 29; 29; 29; 29; 33; 42; 48; 50; 50; 50; 50 |
| Estonia | Muhu | 25.10.2012 | P. nigra | Cone | 1 stand | 14 | 0.71 | 8; 15; 29; 29; 29; 29; 29; 29; 45; 45; 45; 45; 45; 45 |
| Estonia | Vormsi | 06.09.2012 | P. sylvestris | Cone, needle | 6 | 10 | 0.60 | 2; 8; 8; 29; 48; 48; 48; 48; 48; 48 |
| Slovakia | Arboretum Mlyňany | 20.08.2019 | P. sylvestris | Cone | 1 | 2 | 0.50 | 32; 32 |
| Slovakia | Borová Hora | 03.09.2019 | P. sylvestris | Cone | 1 | 2 | 0.50 | 29; 29 |
| Slovakia | Borová Hora | 03.09.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Galanta | 15.08.2019 | P. nigra | Cone | 1 | 2 | 0.50 | 29; 29 |
| Slovakia | Galanta | 15.08.2019 | P. nigra | Cone | 1 | 2 | 0.50 | 33; 33 |
| Slovakia | Galanta | 15.08.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 33; 33; 33 |
| Slovakia | Galanta | 15.08.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 50; 50; 50 |
| Slovakia | Galanta | 15.08.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Galanta | 15.08.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 33; 33; 33 |
| Slovakia | Galanta | 15.08.2019 | P. sylvestris | Cone | 1 | 4 | 0.50 | 39; 40; 40; 40 |
| Slovakia | Hlohovec | 13.08.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 28; 28; 28 |
| Slovakia | Kežmarok | 04.10.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Kežmarok | 04.10.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Kežmarok | 03.11.2019 | P. sylvestris | Cone | 1 | 2 | 0.50 | 29; 29 |
| Slovakia | Kežmarok | 03.11.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Nová Lehota | 01.10.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 33; 33; 33 |
| Slovakia | Palárikovo | 13.08.2019 | P. nigra | Cone | 1 | 2 | 0.50 | 26; 26 |
| Slovakia | Palárikovo | 13.08.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 19; 19; 19 |
| Slovakia | Palárikovo | 13.08.2019 | P. sylvestris | Cone | 1 | 3 | 0.67 | 33; 33; 33 |
| Slovakia | Stupava | 21.03.2019 | P. nigra | Cone | 1 | 2 | 0.50 | 29; 29 |
| Slovakia | Stupava | 21.03.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Slovakia | Tematín | 01.10.2019 | P. nigra | Cone | 1 | 2 | 0.50 | 33; 33 |
| Slovakia | Vlčany | 13.08.2019 | P. nigra | Cone | 1 | 3 | 0.67 | 29; 29; 29 |
| Publication Region | N. America [42] * | N. America [47] * | Europe | Europe [48] | Europe [43] | Present Study Europe |
|---|---|---|---|---|---|---|
| N of isolates | 16 | 51 | 27 | 85 | 86 | 425 |
| Locus | ||||||
| SS1 | 5 | 1 | 1 | 1 | 1 | 1 |
| SS2 | 2 | 2 | 1 | 1 | - | 1 |
| SS3 | 2 | 1 | 1 | - | - | - |
| SS4 | 2 | 1 | 1 | - | - | - |
| SS5 | 7 | 4 | 2 | - | 1 | 5 |
| SS6 | 1 | 1 | 1 | - | - | - |
| SS7 | 5 | 2 | 1 | 1 | - | 3 |
| SS8 | 8 | 1 | 1 | 1 | - | 2 |
| SS9 | 5 | 4 | 1 | - | 3 | 4 |
| SS10 | 2 | 2 | 1 | 1 | 2 | 2 |
| SS11 | 7 | 7 | 2 | 1 | 1 | 1 |
| SS12 | - | 4 | 1 | - | 2 | 9 |
| SS13 | - | - | - | 3 | 1 | 2 |
| SS14 | - | - | - | 4 | 3 | 5 |
| SS15 | - | - | - | 2 | 2 | 2 |
| SS16 | - | - | - | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamson, K.; Laas, M.; Blumenstein, K.; Busskamp, J.; Langer, G.J.; Klavina, D.; Kaur, A.; Maaten, T.; Mullett, M.S.; Müller, M.M.; et al. Highly Clonal Structure and Abundance of One Haplotype Characterise the Diplodia sapinea Populations in Europe and Western Asia. J. Fungi 2021, 7, 634. https://doi.org/10.3390/jof7080634
Adamson K, Laas M, Blumenstein K, Busskamp J, Langer GJ, Klavina D, Kaur A, Maaten T, Mullett MS, Müller MM, et al. Highly Clonal Structure and Abundance of One Haplotype Characterise the Diplodia sapinea Populations in Europe and Western Asia. Journal of Fungi. 2021; 7(8):634. https://doi.org/10.3390/jof7080634
Chicago/Turabian StyleAdamson, Kalev, Marili Laas, Kathrin Blumenstein, Johanna Busskamp, Gitta J. Langer, Darta Klavina, Anu Kaur, Tiit Maaten, Martin S. Mullett, Michael M. Müller, and et al. 2021. "Highly Clonal Structure and Abundance of One Haplotype Characterise the Diplodia sapinea Populations in Europe and Western Asia" Journal of Fungi 7, no. 8: 634. https://doi.org/10.3390/jof7080634
APA StyleAdamson, K., Laas, M., Blumenstein, K., Busskamp, J., Langer, G. J., Klavina, D., Kaur, A., Maaten, T., Mullett, M. S., Müller, M. M., Ondrušková, E., Padari, A., Pilt, E., Riit, T., Solheim, H., Soonvald, L., Tedersoo, L., Terhonen, E., & Drenkhan, R. (2021). Highly Clonal Structure and Abundance of One Haplotype Characterise the Diplodia sapinea Populations in Europe and Western Asia. Journal of Fungi, 7(8), 634. https://doi.org/10.3390/jof7080634






