Immunomodulating Activity of Pleurotus eryngii Mushrooms Following Their In Vitro Fermentation by Human Fecal Microbiota
Abstract
:1. Introduction
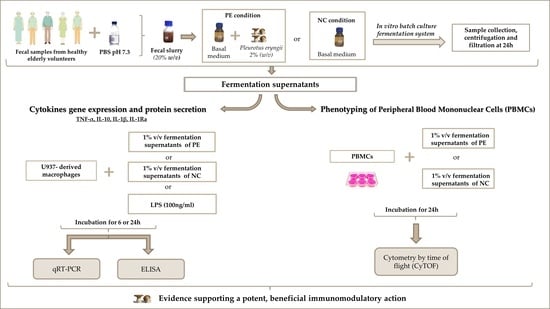
2. Materials and Methods
2.1. Fungal Strains, Mushroom Cultivation, and Determination of Glucan Content
2.2. Fecal Sample Collection and In Vitro Static Batch Culture Fermentations
2.3. Cell Culture and Treatment
2.4. Quantification of Cytokine Gene Expression in FS-Treated Macrophages
2.5. Quantification of Cytokine Secretion by FS-Treated Macrophages
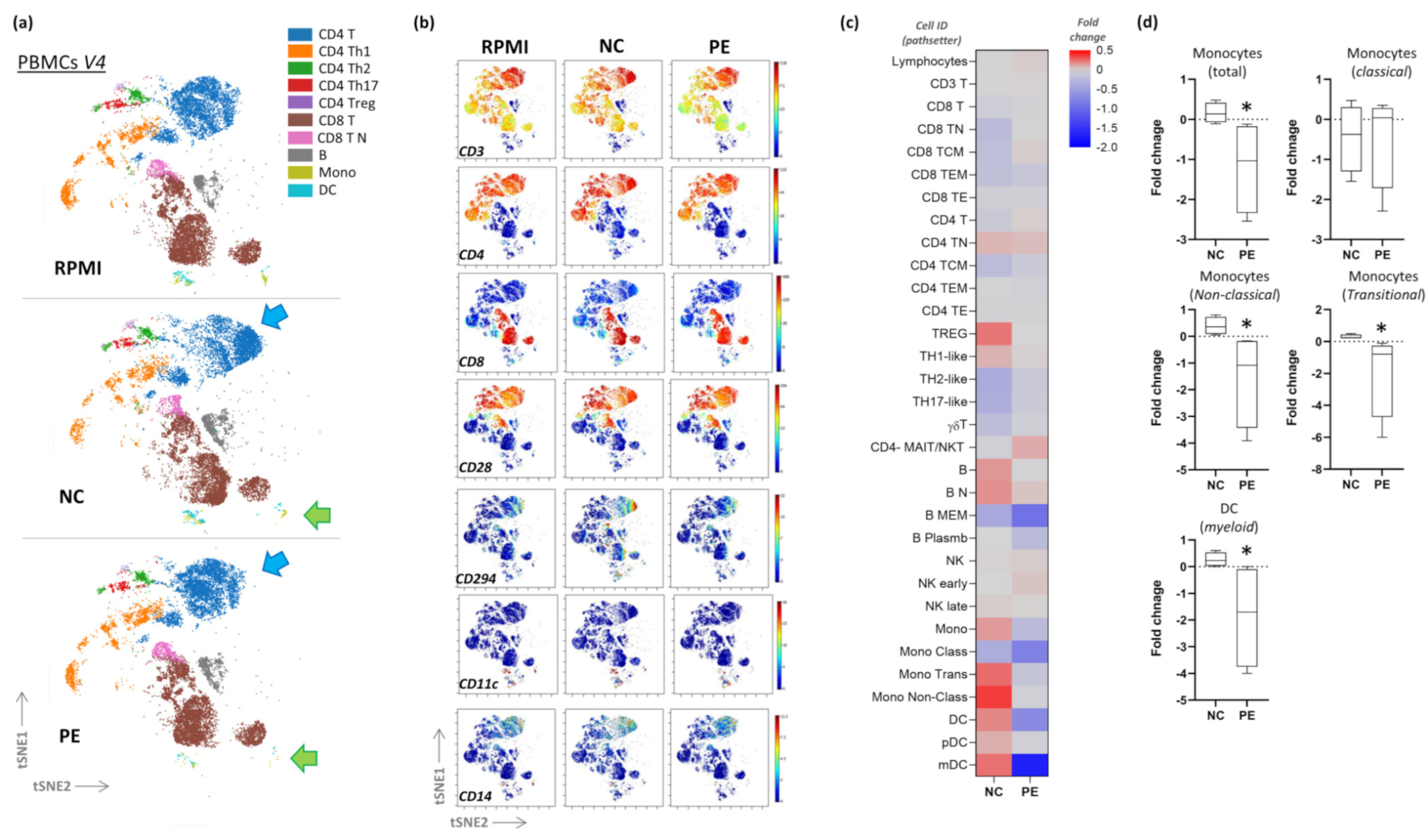
2.6. CyTOF
2.7. Statistical Analysis
3. Results
3.1. Cytokine Expression and Secretion by U937-Derived Macrophages Treated with FSs
3.1.1. TNF-α Gene Expression and Protein Secretion
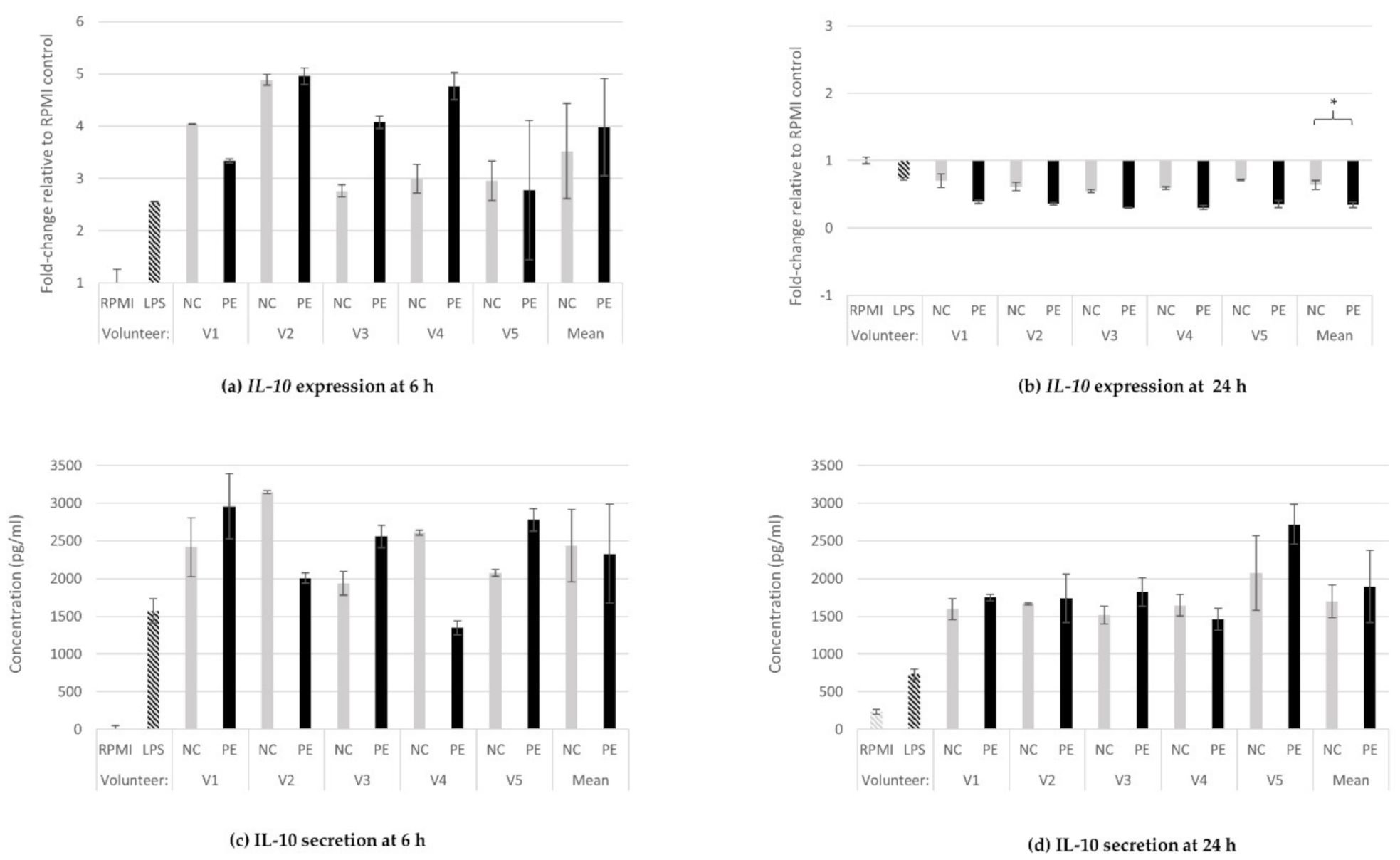
3.1.2. IL-10 Gene Expression and Protein Secretion
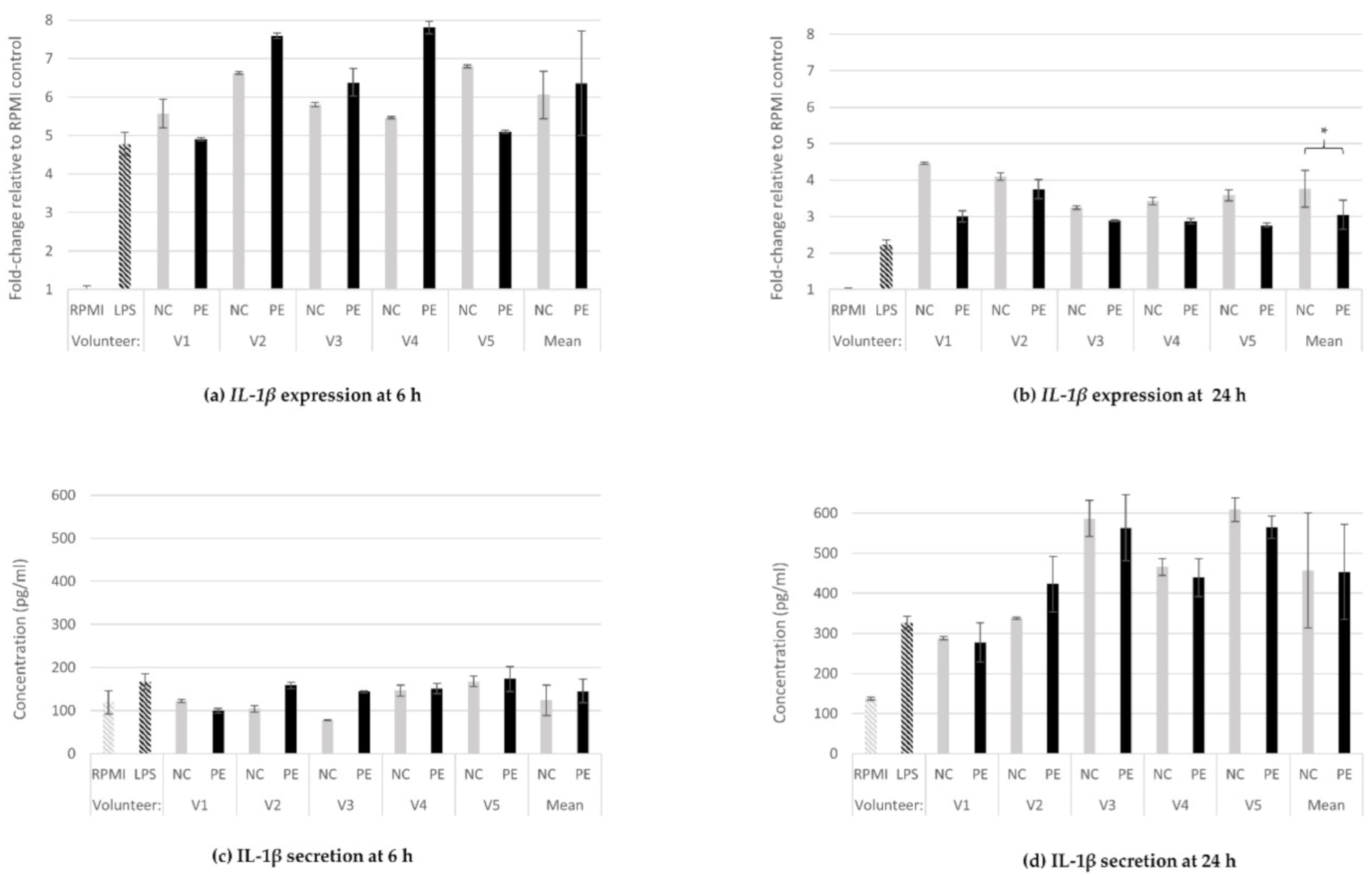
3.1.3. IL-1β Gene Expression and Protein Secretion
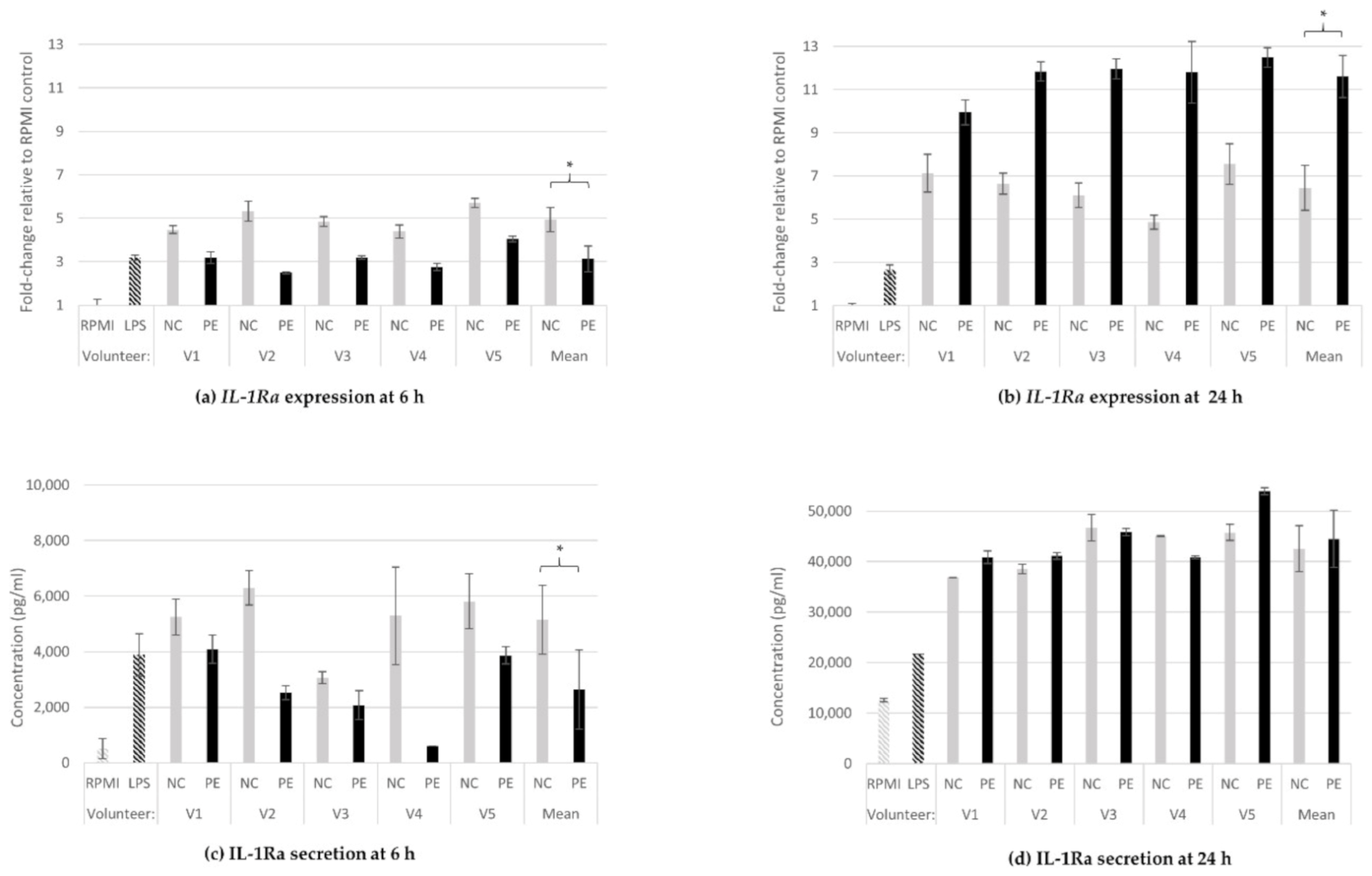
3.1.4. IL-1Ra Gene Expression and Protein Release
3.2. CyTOF Pilot Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giavasis, I. Bioactive fungal polysaccharides as potential functional ingredients in food and nutraceuticals. Curr. Opin. Biotechnol. 2014, 26, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liang, H.; Luo, L. Antitumor polysaccharides from mushrooms: A review on the structural characteristics, antitumor mechanisms and immunomodulating activities. Carbohydr. Res. 2016, 424, 30–41. [Google Scholar] [CrossRef]
- Roncero-Ramos, I.; Delgado-Andrade, C. The beneficial role of edible mushrooms in human health. Curr. Opin. Food Sci. 2017, 14, 122–128. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, M.; Wang, X.; Ren, Y.; Yue, T.; Wang, Z.; Gao, Z. Edible fungal polysaccharides, the gut microbiota, and host health. Carbohydr. Polym. 2021, 273, 118558. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.M.I.; Choi, J.S. Clinical and Physiological Perspectives of β-Glucans: The Past, Present, and Future. Int. J. Mol. Sci. 2017, 18, 1906. [Google Scholar] [CrossRef]
- Ooi, V.E.; Liu, F. Immunomodulation and anti-cancer activity of polysaccharide-protein complexes. Curr. Med. Chem. 2000, 7, 715–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasser, S.P.; Weis, A.L. Therapeutic effects of substances occurring in higher Basidiomycetes mushrooms: A modern perspective. Crit. Rev. Immunol. 1999, 19, 65–96. [Google Scholar] [CrossRef]
- Jayachandran, M.; Xiao, J.; Xu, B. A Critical Review on Health Promoting Benefits of Edible Mushrooms through Gut Microbiota. Int. J. Mol. Sci. 2017, 18, 1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laroche, C.; Michaud, P. New developments and prospective applications for β (1,3) glucans. Recent Pat. Biotechnol. 2007, 1, 59–73. [Google Scholar] [CrossRef]
- Koutrotsios, G.; Kalogeropoulos, N.; Kaliora, A.C.; Zervakis, G.I. Toward an Increased Functionality in Oyster (Pleurotus) Mushrooms Produced on Grape Marc or Olive Mill Wastes Serving as Sources of Bioactive Compounds. J. Agric. Food Chem. 2018, 66, 5971–5983. [Google Scholar] [CrossRef]
- Vetvicka, V.; Vannucci, L.; Sima, P.; Richter, J. Beta Glucan: Supplement or Drug? From Laboratory to Clinical Trials. Molecules 2019, 24, 1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjbar, R.; Vahdati, S.N.; Tavakoli, S.; Khodaie, R.; Behboudi, H. Immunomodulatory roles of microbiota-derived short-chain fatty acids in bacterial infections. Biomed. Pharmacother. 2021, 141, 111817. [Google Scholar] [CrossRef]
- Mitsou, E.K.; Panopoulou, N.; Turunen, K.; Spiliotis, V.; Kyriacou, A. Prebiotic potential of barley derived β-glucan at low intake levels: A randomised, double-blinded, placebo-controlled clinical study. Food Res. Int. 2010, 43, 1086–1092. [Google Scholar] [CrossRef]
- Jayachandran, M.; Chen, J.; Chung, S.S.M.; Xu, B. A critical review on the impacts of β-glucans on gut microbiota and human health. J. Nutr. Biochem. 2018, 61, 101–110. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, C.E.; Cruz, I.B.; Cadoná, F.C.; Machado, A.K.; Assmann, C.; Schlemmer, K.B.; Zanette, R.A.; Leal, D.B.; Santurio, J.M. Cytoprotective and genoprotective effects of β-glucans against aflatoxin B1-induced DNA damage in broiler chicken lymphocytes. Toxicol. In Vitro 2015, 29, 538–543. [Google Scholar] [CrossRef]
- Kerche-Silva, L.E.; Cólus, I.M.S.; Malini, M.; Mori, M.P.; Dekker, R.F.H.; Barbosa-Dekker, A.M. In vitro protective effects of botryosphaeran, a (1 → 3;1 → 6)-β-D-glucan, against mutagens in normal and tumor rodent cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2017, 814, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Madrigal-Bujaidar, E.; Morales-González, J.A.; Sánchez-Gutiérrez, M.; Izquierdo-Vega, J.A.; Reyes-Arellano, A.; Álvarez-González, I.; Pérez-Pasten, R.; Madrigal-Santillán, E. Prevention of Aflatoxin B1-Induced DNA Breaks by β-D-Glucan. Toxins 2015, 7, 2145–2158. [Google Scholar] [CrossRef] [Green Version]
- Tohamy, A.A.; El-Ghor, A.A.; El-Nahas, S.M.; Noshy, M.M. β-glucan inhibits the genotoxicity of cyclophosphamide, adriamycin and cisplatin. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2003, 541, 45–53. [Google Scholar] [CrossRef]
- Silva-Sena, G.G.; Malini, M.; Delarmelina, J.M.; Dutra, J.C.V.; Gervásio, S.V.; Leal, M.A.S.; Costa Pereira, T.M.; Barbosa-Dekker, A.M.; Dekker, R.F.H.; de Paula, F.; et al. In vivo antimutagenic and antiatherogenic effects of the (1 → 3)(1 → 6)-β-d- glucan botryosphaeran. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 826, 6–14. [Google Scholar] [CrossRef] [PubMed]
- de Souza Silva, P.M.; de Sousa, R.V.; Simão, A.A.; Cesar, P.H.S.; Trento, M.V.C.; Marcussi, S. Protective effect of β-D-glucan and glutamine on the genomic instability induced by Cytarabine/Ara-C in BALB/c mice. Int. J. Biol. Macromol. 2018, 117, 559–564. [Google Scholar] [CrossRef]
- Turunen, K.T.; Pletsa, V.; Georgiadis, P.; Triantafillidis, J.K.; Karamanolis, D.; Kyriacou, A. Impact of β-glucan on the Fecal Water Genotoxicity of Polypectomized Patients. Nutr. Cancer 2016, 68, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Śliżewska, K.; Otlewska, A. Antigenotoxic activity of lactic acid bacteria, prebiotics, and products of their fermentation against selected mutagens. Regul. Toxicol. Pharmacol. 2015, 73, 938–946. [Google Scholar] [CrossRef]
- Christophersen, C.T.; Petersen, A.; Licht, T.R.; Conlon, M.A. Xylo-oligosaccharides and inulin affect genotoxicity and bacterial populations differently in a human colonic simulator challenged with soy protein. Nutrients 2013, 5, 3740–3756. [Google Scholar] [CrossRef] [Green Version]
- Allsopp, P.; Possemiers, S.; Campbell, D.; Oyarzábal, I.S.; Gill, C.; Rowland, I. An exploratory study into the putative prebiotic activity of fructans isolated from Agave angustifolia and the associated anticancer activity. Anaerobe 2013, 22, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.J.; Rowland, I.R. Antigenotoxicity of probiotics and prebiotics on faecal water-induced DNA damage in human colon adenocarcinoma cells. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2004, 551, 233–243. [Google Scholar] [CrossRef]
- Munjal, U.; Scharlau, D.; Glei, M. Gut fermentation products of inulin-type fructans modulate the expression of xenobiotic-metabolising enzymes in human colonic tumour cells. Anticancer Res. 2012, 32, 5379–5386. [Google Scholar]
- Boulaka, A.; Christodoulou, P.; Vlassopoulou, M.; Koutrotsios, G.; Bekiaris, G.; Zervakis, G.I.; Mitsou, E.K.; Saxami, G.; Kyriacou, A.; Zervou, M.; et al. Genoprotective Properties and Metabolites of β-Glucan-Rich Edible Mushrooms Following Their In Vitro Fermentation by Human Faecal Microbiota. Molecules 2020, 25, 3554. [Google Scholar] [CrossRef]
- Polemis, E.; Zervakis, G.I. Mushrooms in Greece: Present status and threats. In Proceedings of the Workshop on Mushrooms (including Truffles) Regulating Policies, Ioannina, Greece, 20 April 2016; p. 11. [Google Scholar] [CrossRef]
- Ntougias, S.; Baldrian, P.; Ehaliotis, C.; Nerud, F.; Merhautová, V.; Zervakis, G.I. Olive mill wastewater biodegradation potential of white-rot fungi—Mode of action of fungal culture extracts and effects of ligninolytic enzymes. Bioresour. Technol. 2015, 189, 121–130. [Google Scholar] [CrossRef]
- Koutrotsios, G.; Patsou, M.; Mitsou, E.K.; Bekiaris, G.; Kotsou, M.; Tarantilis, P.A.; Pletsa, V.; Kyriacou, A.; Zervakis, G.I. Valorization of Olive By-Products as Substrates for the Cultivation of Ganoderma lucidum and Pleurotus ostreatus Mushrooms with Enhanced Functional and Prebiotic Properties. Catalysts 2019, 9, 537. [Google Scholar] [CrossRef] [Green Version]
- Koutrotsios, G.; Kalogeropoulos, N.; Stathopoulos, P.; Kaliora, A.C.; Zervakis, G.I. Bioactive compounds and antioxidant activity exhibit high intraspecific variability in Pleurotus ostreatus mushrooms and correlate well with cultivation performance parameters. World J. Microbiol. Biotechnol. 2017, 33, 98. [Google Scholar] [CrossRef] [PubMed]
- Mitsou, E.K.; Saxami, G.; Stamoulou, E.; Kerezoudi, E.; Terzi, E.; Koutrotsios, G.; Bekiaris, G.; Zervakis, G.I.; Mountzouris, K.C.; Pletsa, V.; et al. Effects of Rich in Β-Glucans Edible Mushrooms on Aging Gut Microbiota Characteristics: An In Vitro Study. Molecules 2020, 25, 2806. [Google Scholar] [CrossRef] [PubMed]
- Abreu, H.; Zavadinack, M.; Smiderle, F.R.; Cipriani, T.R.; Cordeiro, L.M.C.; Iacomini, M. Polysaccharides from Pleurotus eryngii: Selective extraction methodologies and their modulatory effects on THP-1 macrophages. Carbohydr. Polym. 2021, 252, 117177. [Google Scholar] [CrossRef] [PubMed]
- Minato, K.I.; Laan, L.C.; van Die, I.; Mizuno, M. Pleurotus citrinopileatus polysaccharide stimulates anti-inflammatory properties during monocyte-to-macrophage differentiation. Int. J. Biol. Macromol. 2019, 122, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Lewis, J.S.; Lee, J.A.; Underwood, J.C.; Harris, A.L.; Lewis, C.E. Macrophage responses to hypoxia: Relevance to disease mechanisms. J. Leukoc. Biol. 1999, 66, 889–900. [Google Scholar] [CrossRef]
- Benoit, M.; Desnues, B.; Mege, J.L. Macrophage polarization in bacterial infections. J. Immunol. 2008, 181, 3733–3739. [Google Scholar] [CrossRef] [Green Version]
- Shiratori, H.; Feinweber, C.; Luckhardt, S.; Linke, B.; Resch, E.; Geisslinger, G.; Weigert, A.; Parnham, M.J. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Mol. Immunol. 2017, 88, 58–68. [Google Scholar] [CrossRef]
- Chanput, W.; Peters, V.; Wichers, H.J.P.S. THP-1 and U937 Cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 147–159. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–534, 536–537. [Google Scholar]
- Bagwell, C.B.; Hunsberger, B.; Hill, B.; Herbert, D.; Bray, C.; Selvanantham, T.; Li, S.; Villasboas, J.C.; Pavelko, K.; Strausbauch, M.; et al. Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system. Cytom. B Clin. Cytom. 2020, 98, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Kotecha, N.; Krutzik, P.O.; Irish, J.M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010, 10, cy1017s53. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.; Araújo, J.R.; Di Santo, J.P. A Cross-Talk Between Microbiota-Derived Short-Chain Fatty Acids and the Host Mucosal Immune System Regulates Intestinal Homeostasis and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2018, 24, 558–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Cai, X.; Fei, W.; Ye, Y.; Zhao, M.; Zheng, C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 2022, 62, 1–12. [Google Scholar] [CrossRef]
- Chikina, A.S.; Nadalin, F.; Maurin, M.; San-Roman, M.; Thomas-Bonafos, T.; Li, X.V.; Lameiras, S.; Baulande, S.; Henri, S.; Malissen, B.; et al. Macrophages Maintain Epithelium Integrity by Limiting Fungal Product Absorption. Cell 2020, 183, 411–428.e16. [Google Scholar] [CrossRef]
- Desai, C. Meyler’s side effects of drugs: The international encyclopedia of adverse drug reactions and interactions. Indian J. Pharmacol. 2016, 48, 224. [Google Scholar]
- Gane, J.M.; Stockley, R.A.; Sapey, E. TNF-α Autocrine Feedback Loops in Human Monocytes: The Pro- and Anti-Inflammatory Roles of the TNF-α Receptors Support the Concept of Selective TNFR1 Blockade In Vivo. J. Immunol. Res. 2016, 2016, 1079851. [Google Scholar] [CrossRef] [Green Version]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [Green Version]
- Llauradó Maury, G.; Morris-Quevedo, H.J.; Heykers, A.; Lanckacker, E.; Cappoen, D.; Delputte, P.; Vanden Berghe, W.; Salgueiro, Z.; Cos, P. Differential Induction Pattern Towards Classically Activated Macrophages in Response to an Immunomodulatory Extract from Pleurotus ostreatus Mycelium. J. Fungi 2021, 7, 206. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Zhang, F.; Linhardt, R.J.; Zeng, G.; Zhang, A. Extraction, structure and bioactivities of the polysaccharides from Pleurotus eryngii: A review. Int. J. Biol. Macromol. 2020, 150, 1342–1347. [Google Scholar] [CrossRef]
- Saraiva, M.; Vieira, P.; O’Garra, A. Biology and therapeutic potential of interleukin-10. J. Exp. Med. 2020, 217, e20190418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef]
- Pattison, M.J.; Mackenzie, K.F.; Arthur, J.S. Inhibition of JAKs in macrophages increases lipopolysaccharide-induced cytokine production by blocking IL-10-mediated feedback. J. Immunol. 2012, 189, 2784–2792. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Diez, D.; Miranda-Saavedra, D. The IL-10/STAT3-mediated anti-inflammatory response: Recent developments and future challenges. Brief. Funct. Genom. 2013, 12, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baseler, W.A.; Davies, L.C.; Quigley, L.; Ridnour, L.A.; Weiss, J.M.; Hussain, S.P.; Wink, D.A.; McVicar, D.W. Autocrine IL-10 functions as a rheostat for M1 macrophage glycolytic commitment by tuning nitric oxide production. Redox Biol. 2016, 10, 12–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koelink, P.J.; Bloemendaal, F.M.; Li, B.; Westera, L.; Vogels, E.W.M.; van Roest, M.; Gloudemans, A.K.; van ‘t Wout, A.B.; Korf, H.; Vermeire, S.; et al. Anti-TNF therapy in IBD exerts its therapeutic effect through macrophage IL-10 signalling. Gut 2020, 69, 1053–1063. [Google Scholar] [CrossRef] [Green Version]
- Shokryazdan, P.; Faseleh Jahromi, M.; Navidshad, B.; Liang, J.B. Effects of prebiotics on immune system and cytokine expression. Med. Microbiol. Immunol. 2017, 206, 1–9. [Google Scholar] [CrossRef]
- Mantovani, A.; Barajon, I.; Garlanda, C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol. Rev. 2018, 281, 57–61. [Google Scholar] [CrossRef]
- Dinarello, C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018, 281, 8–27. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, D.; Dong, M.; Dai, C.; Wu, S. Inflammation and Inflammatory Cytokine Contribute to the Initiation and Development of Ulcerative Colitis and Its Associated Cancer. Inflamm. Bowel Dis. 2019, 25, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, D.; Nan, C.; Mori, K.; Hanayama, M.; Kikuchi, H.; Hirai, S.; Egashira, Y. Effect of mushroom polysaccharides from Pleurotus eryngii on obesity and gut microbiota in mice fed a high-fat diet. Eur. J. Nutr. 2020, 59, 3231–3244. [Google Scholar] [CrossRef]
- Oguma, T.; Asano, K.; Ishizaka, A. Role of prostaglandin D2 and its receptors in the pathophysiology of asthma. Allergol. Int. 2008, 57, 307–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, T.; Hirahara, K.; Onodera, A.; Endo, Y.; Hosokawa, H.; Shinoda, K.; Tumes, D.J.; Okamoto, Y. Th2 Cells in Health and Disease. Annu. Rev. Immunol. 2017, 35, 53–84. [Google Scholar] [CrossRef] [PubMed]

| Gene | RefSeqNumber | Sequence (5′ -> 3′) |
|---|---|---|
| GAPDH | NM_002046 | Primer 1: 5′-ACATCGCTCAGACACCATG-3′ Primer 2: 5′-TGTAGTTGAGGTCAATGAAGGG-3′ |
| IL-1β | NM_000576 | Primer 1: 5′-CAGCCAATCTTCATTGCTCAAG-3′ Primer 2: 5′-GAACAAGTCATCCTCATTGCC-3′ |
| IL-1RN | NM_173843 | Primer 1: 5′-CTGTCCTGTGTCAAGTCTGG-3′ Primer 2: 5′- TTGTCCTGCTTTCTGTTCTCG-3′ |
| IL-10 | NM_000572 | Primer 1: 5′-GCGCTGTCATCGATTTCTTC-3′ Primer 2: 5′-TCACTCATGGCTTTGTAGATGC-3′ |
| TNF | NM_000594 | Primer 1: 5′-TGCACTTTGGAGTGATCGG-3′ Primer 2: 5′-TCAGCTTGAGGGTTTGCTAC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlassopoulou, M.; Paschalidis, N.; Savvides, A.L.; Saxami, G.; Mitsou, E.K.; Kerezoudi, E.N.; Koutrotsios, G.; Zervakis, G.I.; Georgiadis, P.; Kyriacou, A.; et al. Immunomodulating Activity of Pleurotus eryngii Mushrooms Following Their In Vitro Fermentation by Human Fecal Microbiota. J. Fungi 2022, 8, 329. https://doi.org/10.3390/jof8040329
Vlassopoulou M, Paschalidis N, Savvides AL, Saxami G, Mitsou EK, Kerezoudi EN, Koutrotsios G, Zervakis GI, Georgiadis P, Kyriacou A, et al. Immunomodulating Activity of Pleurotus eryngii Mushrooms Following Their In Vitro Fermentation by Human Fecal Microbiota. Journal of Fungi. 2022; 8(4):329. https://doi.org/10.3390/jof8040329
Chicago/Turabian StyleVlassopoulou, Marigoula, Nikolaos Paschalidis, Alexandros L. Savvides, Georgia Saxami, Evdokia K. Mitsou, Evangelia N. Kerezoudi, Georgios Koutrotsios, Georgios I. Zervakis, Panagiotis Georgiadis, Adamantini Kyriacou, and et al. 2022. "Immunomodulating Activity of Pleurotus eryngii Mushrooms Following Their In Vitro Fermentation by Human Fecal Microbiota" Journal of Fungi 8, no. 4: 329. https://doi.org/10.3390/jof8040329
APA StyleVlassopoulou, M., Paschalidis, N., Savvides, A. L., Saxami, G., Mitsou, E. K., Kerezoudi, E. N., Koutrotsios, G., Zervakis, G. I., Georgiadis, P., Kyriacou, A., & Pletsa, V. (2022). Immunomodulating Activity of Pleurotus eryngii Mushrooms Following Their In Vitro Fermentation by Human Fecal Microbiota. Journal of Fungi, 8(4), 329. https://doi.org/10.3390/jof8040329