Development of Marker Recycling Systems for Sequential Genetic Manipulation in Marine-Derived Fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Culture Conditions
2.2. DNA Manipulation
2.3. Plasmid Construction
2.4. 5-FOA Sensitivity Assay
2.5. Acetamide Utilization Assay
2.6. Protoplast Transformation
2.7. Secondary Metabolite Analysis by HPLC
3. Results and Discussion
3.1. Construction of CRISPR-Cas9 Vectors Containing pyrG/amdS Marker Gene
3.2. Construction of Uracil Auxotroph Mutant by Disruption of the pyrG Gene in Aspergillus sp. SCSIO SX7S7
3.3. Gene Disruption Using the CRISPR-Cas9-pyrG System in Aspergillus sp. SCSIO SX7S7
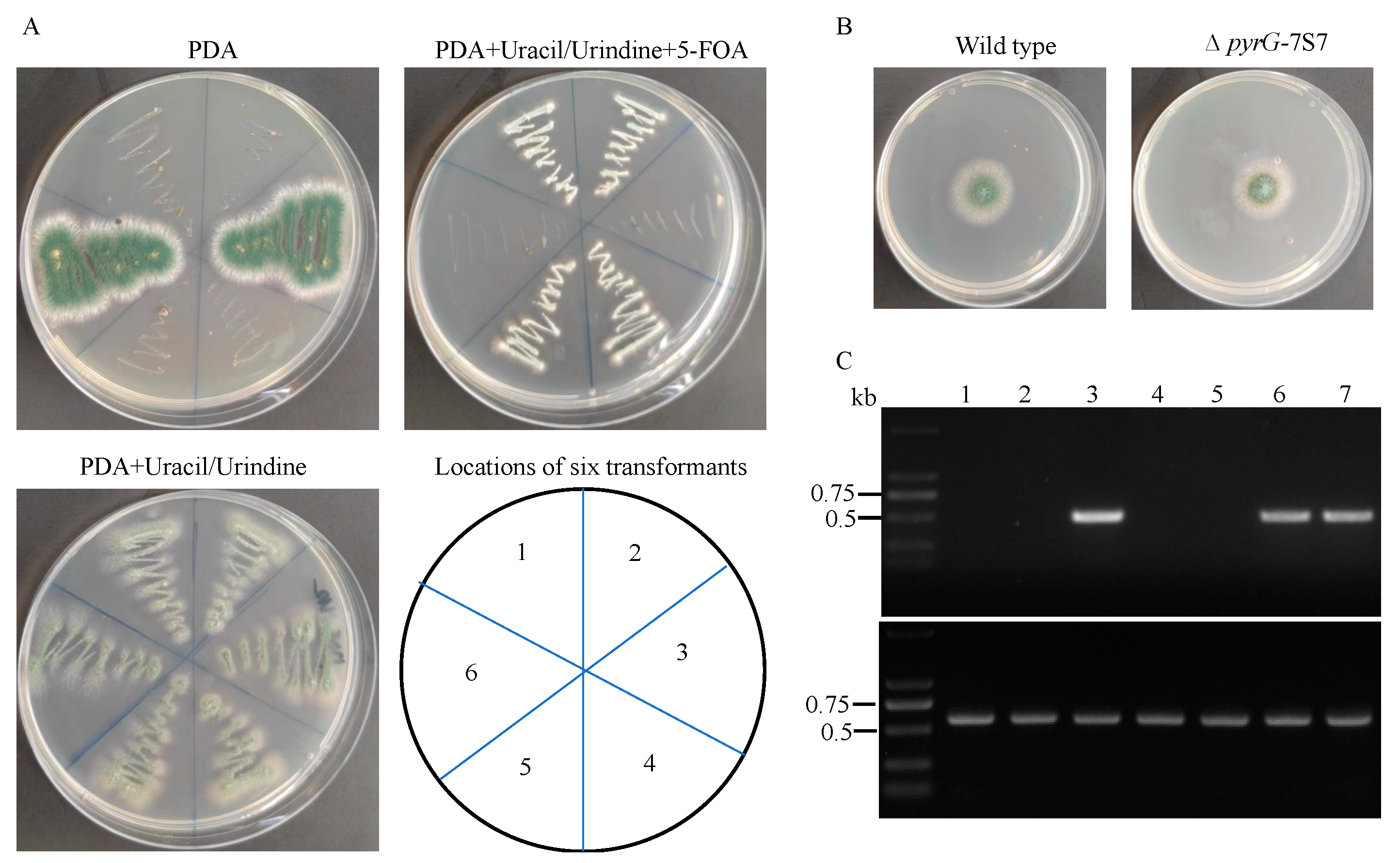
3.4. Remove of pyrG Marker by Counter Selection
3.5. Gene Disruptions Using CRISPR-Cas9-amdS System in Spiromastix sp. SCSIO F190
3.6. Remove of amdS Marker by Counter Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasan, S.; Ansari, M.I.; Ahmad, A.; Mishra, M. Major bioactive metabolites from marine fungi: A Review. Bioinformation 2015, 11, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Frank, M.; Yu, X.; Yu, H.; Tran-Cong, N.M.; Gao, Y.; Proksch, P. Secondary metabolites from marine-derived fungi from China. Prog. Chem. Org. Nat. Prod. 2020, 111, 81–153. [Google Scholar] [PubMed]
- Shin, H.J. Natural Products from Marine Fungi. Mar. Drugs 2020, 18, 230. [Google Scholar] [CrossRef] [PubMed]
- Shabana, S.; Lakshmi, K.R.; Satya, A.K. An Updated Review of Secondary Metabolites from Marine Fungi. Mini. Rev. Med. Chem. 2021, 21, 602–642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, C.; Li, Q.; Ma, J.; Ju, J. Metabolic blockade-based genome mining reveals lipochain-linked dihydro-beta-alanine synthetases involved in autucedine biosynthesis. Org. Lett. 2022, 24, 5535–5540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ding, W.; Qin, X.; Ju, J. Genome sequencing of Streptomyces olivaceus SCSIO T05 and activated production of lobophorin CR4 via metabolic engineering and genome mining. Mar. Drugs 2019, 17, 593. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Boeke, J.D.; LaCroute, F.; Fink, G.R. A positive selection for mutants lacking orotidine-5’-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984, 197, 345–346. [Google Scholar] [CrossRef]
- Delmas, S.; Llanos, A.; Parrou, J.L.; Kokolski, M.; Pullan, S.T.; Shunburne, L.; Archer, D.B. Development of an unmarked gene deletion system for the filamentous fungi Aspergillus niger and Talaromyces versatilis. Appl. Environ. Microbiol. 2014, 80, 3484–3487. [Google Scholar] [CrossRef]
- D’Enfert, C.; Diaquin, M.; Delit, A.; Wuscher, N.; Debeaupuis, J.P.; Huerre, M.; Latge, J.P. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 1996, 64, 4401–4405. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.O.; Storms, R.; Zheng, Y.; Rodzi, M.R.; Mahadi, N.M.; Illias, R.M.; Abdul Murad, A.M.; Abu Bakar, F.D. Development of a pyrG mutant of Aspergillus oryzae strain S1 as a host for the production of heterologous proteins. Sci. World J. 2013, 2013, 634317. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.L.; Albertsen, L.; Lettier, G.; Nielsen, J.B.; Mortensen, U.H. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 2006, 43, 54–64. [Google Scholar] [CrossRef]
- Sun, R.; Xu, H.; Feng, Y.; Hou, X.; Zhu, T.; Che, Q.; Pfeifer, B.; Zhang, G.; Li, D. An efficient marker recycling system for sequential gene deletion in a deep sea-derived fungus Acremonium sp. HDN16-126. Synth. Syst. Biotechnol. 2021, 6, 127–133. [Google Scholar] [CrossRef]
- Jorgensen, M.S.; Skovlund, D.A.; Johannesen, P.F.; Mortensen, U.H. A novel platform for heterologous gene expression in Trichoderma reesei (Teleomorph Hypocrea jecorina). Microb. Cell Fact. 2014, 13, 33. [Google Scholar] [CrossRef]
- Erpf, P.E.; Stephenson, C.J.; Fraser, J.A. amdS as a dominant recyclable marker in Cryptococcus neoformans. Fungal Genet. Biol. 2019, 131, 103241. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M.J.; Pateman, J.A. The genetic analysis of regulation of amidase synthesis in Aspergillus nidulans. II. Mutants resistant to fluoroacetamide. Mol. Gen. Genet. 1970, 108, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.D. Fluoroacetate and fluorocitrate: Mechanism of action. Neurochem. Res. 1991, 16, 1055–1058. [Google Scholar] [CrossRef]
- Apirion, D. The two-way selection of mutants and revertants in respect of acetate utilization and resistance to fluoro-acetate in Aspergillus nidulans. Genet. Res. 1965, 6, 317–329. [Google Scholar] [CrossRef]
- Nan, Y.; Ouyang, L.; Chu, J. In vitro CRISPR/Cas9 system for genome editing of Aspergillus niger based on removable bidirectional selection marker AmdS. Biotechnol. Appl. Biochem. 2021, 68, 964–970. [Google Scholar] [CrossRef]
- Fu, J.; Blaylock, M.; Wickes, C.F.; Welte, W.; Mehrtash, A.; Wiederhold, N.; Wickes, B.L. Development of a Candida glabrata dominant nutritional transformation marker utilizing the Aspergillus nidulans acetamidase gene (amdS). FEMS. Yeast. Res. 2016, 16, fow023. [Google Scholar] [CrossRef]
- Beri, R.K.; Turner, G. Transformation of Penicillium chrysogenum using the Aspergillus nidulans amdS gene as a dominant selective marker. Curr. Genet. 1987, 11, 639–641. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, C.; Yang, J.; Shi, J.; Song, Y.; Hu, D.; Ma, J.; Ju, J. Development of the CRISPR-Cas9 system for the marine-derived fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7. J. Fungi 2022, 8, 715. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, L.; Zhou, Z.; Song, Y.; Ju, J. Anti-pathogenic depsidones and its derivatives from a coral-derived fungus Aspergillus sp. SCSIO SX7S7. Biochem. Syst. Ecol. 2022, 102, 104415. [Google Scholar] [CrossRef]
- Shao, M.; Sun, C.; Liu, X.; Wang, X.; Li, W.; Wei, X.; Li, Q.; Ju, J. Upregulation of a marine fungal biosynthetic gene cluster by an endobacterial symbiont. Commun. Biol. 2020, 3, 527. [Google Scholar] [CrossRef]
- Cai, C.; Chen, Y.; Zhou, L.; Gong, N.; Zhang, H.; Sun, C.; Ma, J.; Ju, J. Antimicrobial polyketides from the marine-derived fungus Spiromastix sp. SCSIO F190. J. Nat. Prod. 2022. [Google Scholar] [CrossRef]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, A.; Tan, K.; Guo, S.; Chen, Y.; Wang, F.; Wong, K.H. Universal plasmids to facilitate gene deletion and gene tagging in filamentous fungi. Fungal Genet. Biol. 2019, 125, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.M.; Lin, F.L.; Gao, H.; Zou, G.; Zhang, J.W.; Wang, G.Q.; Chen, G.D.; Zhou, Z.H.; Yao, X.S.; Hu, D. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci. Rep. 2017, 7, 9250. [Google Scholar] [CrossRef]
- Nodvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Zhai, Q.; Sun, L.; Huang, E.; Zhang, Y.; Zhu, Y.; Guo, Q.; Tian, Y.; Zhao, B.; Lu, H. CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 2019, 103, 6919–6932. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zheng, J.; Yu, D.; Wang, B.; Pan, L. Efficient genome editing in Aspergillus niger with an improved recyclable CRISPR-HDR toolbox and its application in introducing multiple copies of heterologous genes. J. Microbiol. Methods 2019, 163, 105655. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Ram, A.F.; van den Hondel, C.A. The Aspergillus nidulans amdS gene as a marker for the identification of multicopy T-DNA integration events in Agrobacterium-mediated transformation of Aspergillus awamori. Curr. Genet. 2004, 45, 399–403. [Google Scholar] [CrossRef]
- Penttila, M.; Nevalainen, H.; Ratto, M.; Salminen, E.; Knowles, J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 1987, 61, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Alani, E.; Cao, L.; Kleckner, N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 1987, 116, 541–545. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids. Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Liu, X.; Homma, A.; Sayadi, J.; Yang, S.; Ohashi, J.; Takumi, T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 2016, 6, 19675. [Google Scholar] [CrossRef] [PubMed]
- Tilburn, J.; Scazzocchio, C.; Taylor, G.G.; Zabicky-Zissman, J.H.; Lockington, R.A.; Davies, R.W. Transformation by integration in Aspergillus nidulans. Gene 1983, 26, 205–221. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yang, J.; Cai, C.; Shi, J.; Song, Y.; Ma, J.; Ju, J. Development of Marker Recycling Systems for Sequential Genetic Manipulation in Marine-Derived Fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7. J. Fungi 2023, 9, 302. https://doi.org/10.3390/jof9030302
Chen Y, Yang J, Cai C, Shi J, Song Y, Ma J, Ju J. Development of Marker Recycling Systems for Sequential Genetic Manipulation in Marine-Derived Fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7. Journal of Fungi. 2023; 9(3):302. https://doi.org/10.3390/jof9030302
Chicago/Turabian StyleChen, Yingying, Jiafan Yang, Cunlei Cai, Junjie Shi, Yongxiang Song, Junying Ma, and Jianhua Ju. 2023. "Development of Marker Recycling Systems for Sequential Genetic Manipulation in Marine-Derived Fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7" Journal of Fungi 9, no. 3: 302. https://doi.org/10.3390/jof9030302
APA StyleChen, Y., Yang, J., Cai, C., Shi, J., Song, Y., Ma, J., & Ju, J. (2023). Development of Marker Recycling Systems for Sequential Genetic Manipulation in Marine-Derived Fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7. Journal of Fungi, 9(3), 302. https://doi.org/10.3390/jof9030302






