High-Level Expression of β-Glucosidase in Aspergillus niger ATCC 20611 Using the Trichoderma reesei Promoter Pcdna1 to Enhance Cellulose Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Construction of Expression Strains
2.3. Fluorescence and Light Microscopy
2.4. RNA Extraction and RT-qPCR Analysis
2.5. Enzyme Assay and SDS PAGE
2.6. Enzymatic Characterization
2.7. Saccharification of Different Pretreated Corncob Residues
3. Results
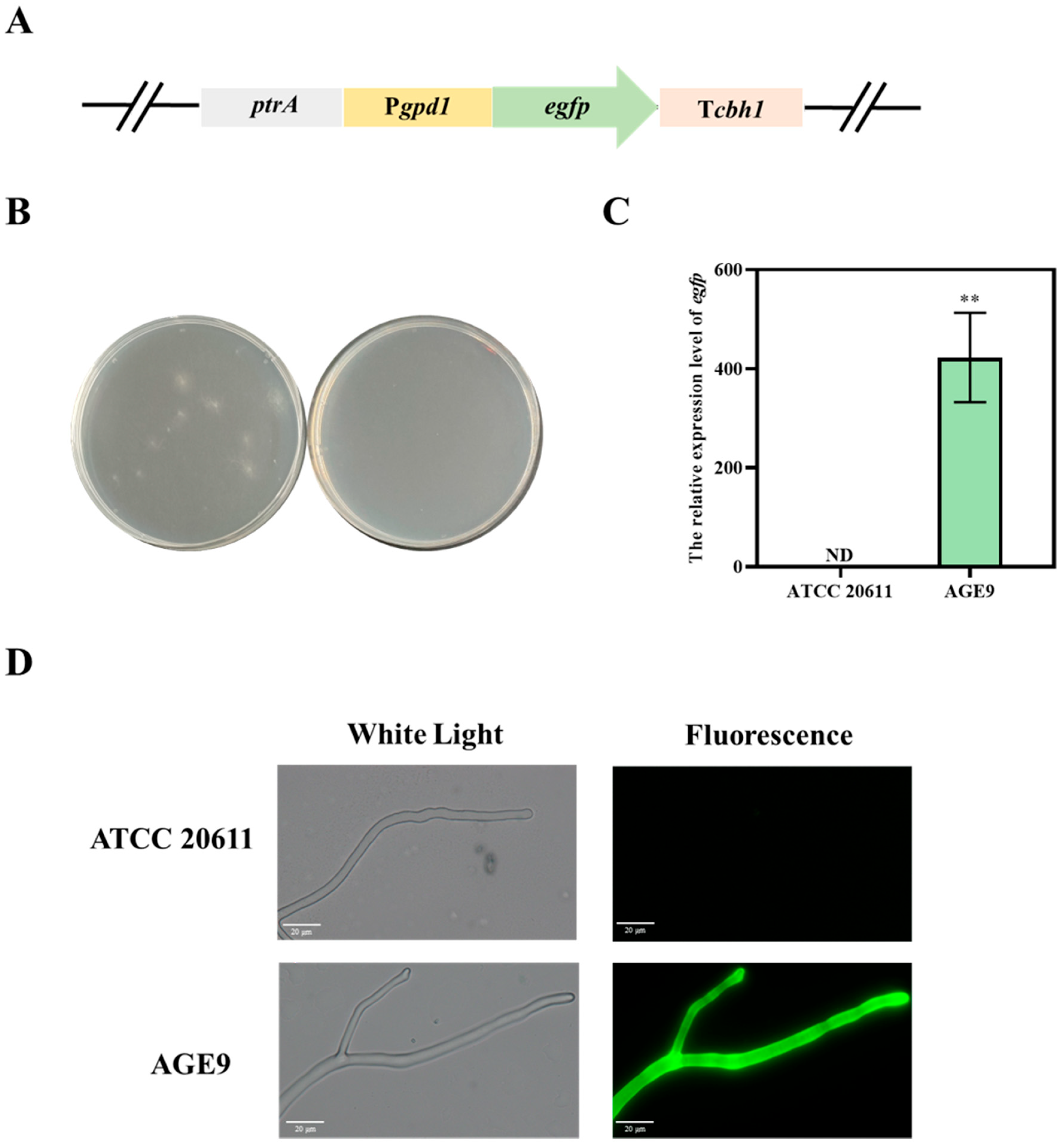
3.1. The T. reesei gpd1 Promoter-Driven EGFP Expression in A. niger ATCC 20611
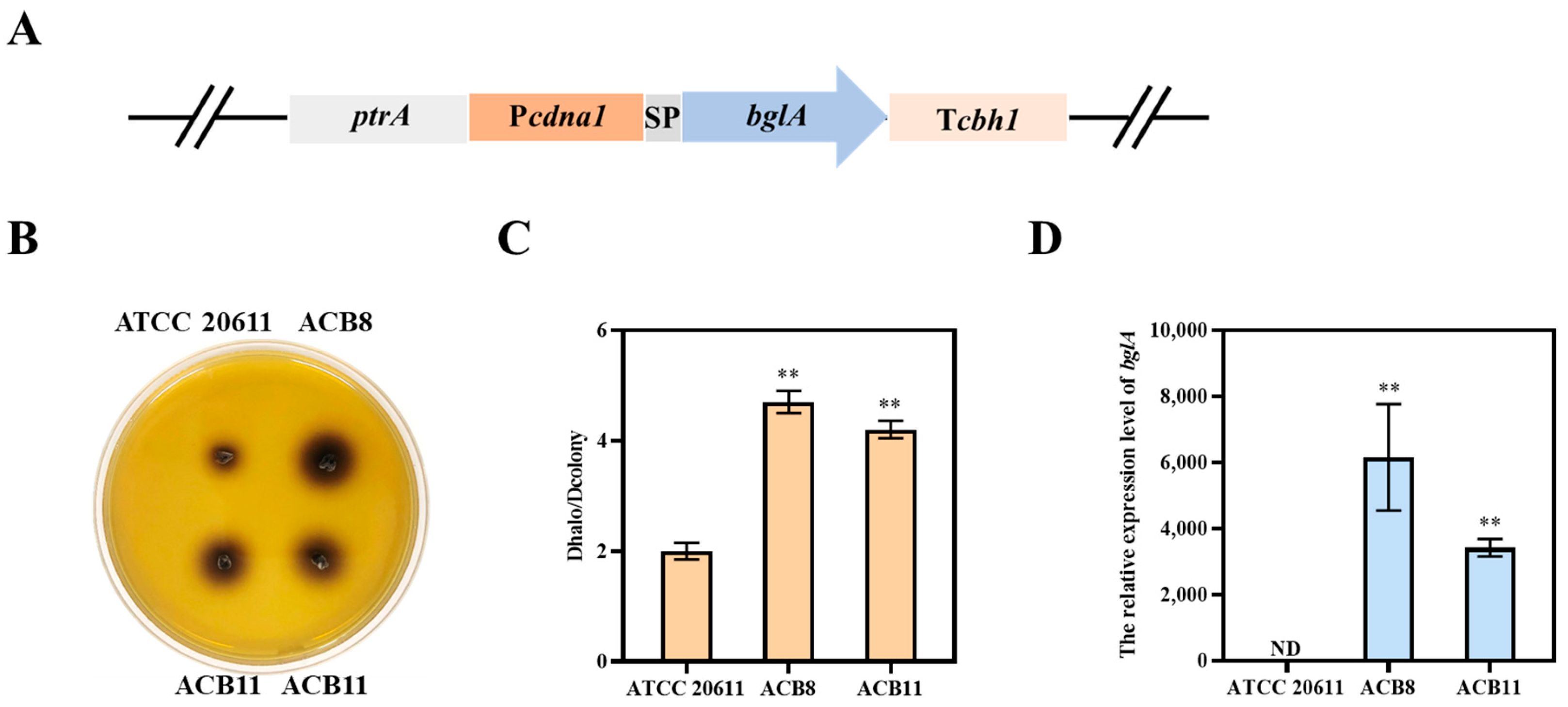
3.2. The gpd1 Promoter-Driven β-Glucosidase Expression in A. niger ATCC 20611
3.3. Detection of the ER Pressure in the gpd1 Promoter-Driven β-glucosidase Expression Strains
3.4. The cdna1 Promoter-Driven β-Glucosidase Expression in A. niger ATCC 20611
3.5. Enzymatic Property of β-Glucosidase in A. niger ACB8
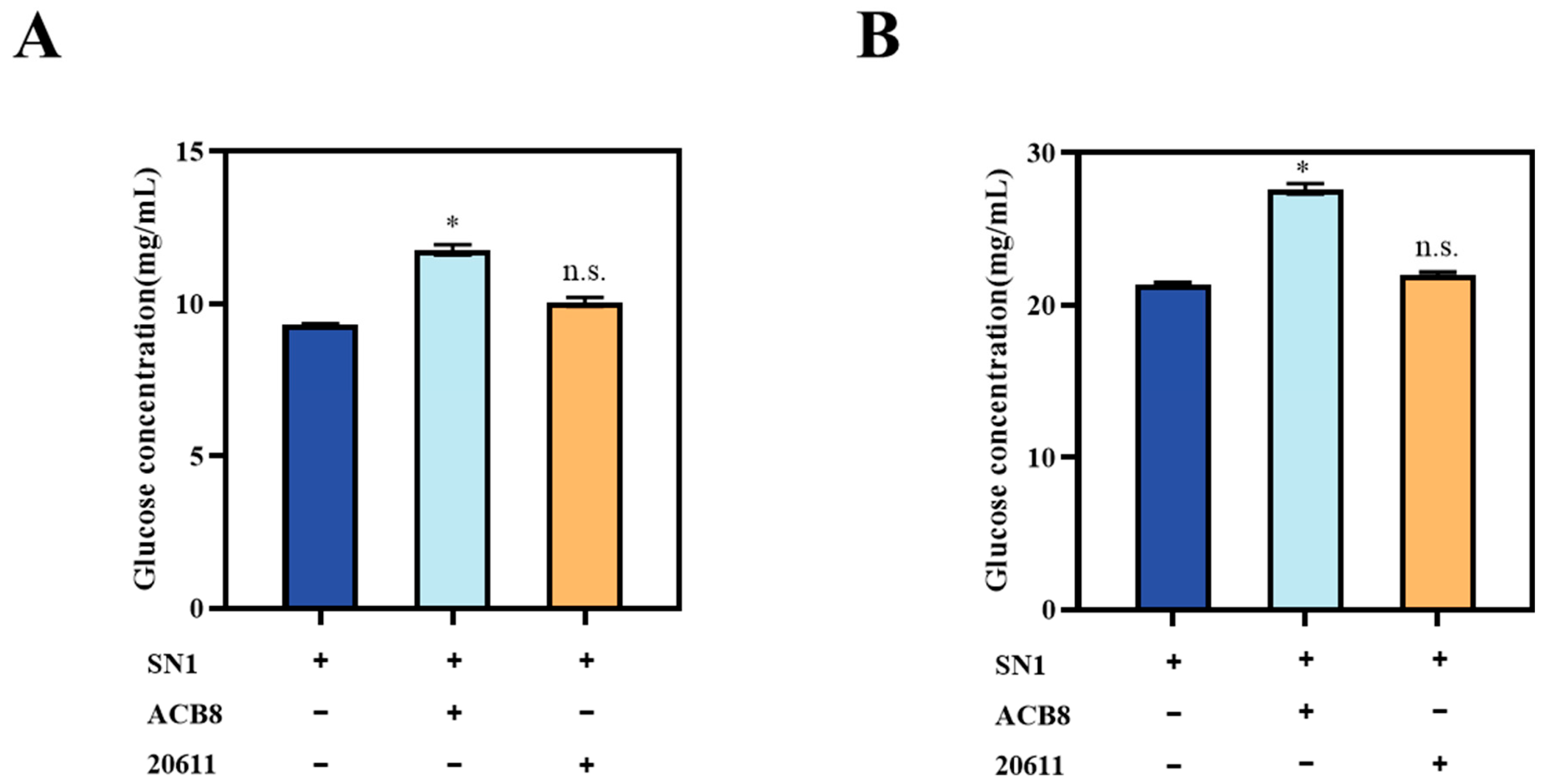
3.6. Saccharification of the Corncob Residues by Supplementing the β-glucosidase to the Cellulase Mixture of T. reesei
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sangeetha, P.T.; Ramesh, M.N.; Prapulla, S.G. Recent trends in the microbial production, analysis and application of fructooligosaccharides. Trends Food. Sci. Technol. 2005, 16, 442–457. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, C.; Xie, Y.; Li, N.; Ning, Z.; Du, N.; Huang, X.; Zhong, Y. Enhancing fructooligosaccharides production by genetic improvement of the industrial fungus Aspergillus niger ATCC 20611. J. Biotechnol. 2017, 249, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Kluge, J.; Terfehr, D.; Kück, U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 6357–6372. [Google Scholar] [CrossRef]
- Wan, X.; Wang, L.; Chang, J.; Zhang, J.; Zhang, Z.; Li, K.; Sun, G.; Liu, C.; Zhong, Y. Effective synthesis of high-content fructooligosaccharides in engineered Aspergillus niger. Microb. Cell Fact. 2024, 23, 76. [Google Scholar] [CrossRef]
- Wang, L.; Xie, Y.; Chang, J.; Wang, J.; Liu, H.; Shi, M.; Zhong, Y. A novel sucrose-inducible expression system and its application for production of biomass-degrading enzymes in Aspergillus niger. Biotechnol. Biofuels Bioprod. 2023, 16, 23. [Google Scholar] [CrossRef]
- Adnan, M.; Ma, X.; Olsson, S.; Wang, J.; Liu, G. Promoter regulation and genetic engineering strategies for enhanced cellulase expression in Trichoderma reesei. Microbiol. Res. 2022, 259, 127011. [Google Scholar] [CrossRef]
- Nakari-Set, L.T.; Penttil, M. Production of Trichoderma reesei cellulases on glucose-containing media. Appl. Environ. Microb. 1995, 61, 3650–3655. [Google Scholar] [CrossRef]
- Uzbas, F.; Sezerman, U.; Hartl, L.; Kubicek, C.P.; Seiboth, B. A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl. Microbiol. Biotechnol. 2012, 93, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Qin, Y.; Liu, Z.; Liu, K.; Wang, F.; Qu, Y. Isolation and characterization of a β-glucosidase from Penicillium decumbens and improving hydrolysis of corncob residue by using it as cellulase supplementation. Enzym. Microb. Technol. 2010, 46, 444–449. [Google Scholar] [CrossRef]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 2016, 15, 106. [Google Scholar] [CrossRef]
- Dashtban, M.; Qin, W. Overexpression of an exotic thermotolerant β-glucosidase in Trichoderma reesei and its signifcant increase in cellulolytic activity and saccharifcation of barley straw. Microb. Cell Fact. 2012, 11, 63. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, J.; Xia, L. Enzymatic hydrolysis of maize straw polysaccharides for the production of reducing sugars. Carbohyd. Polym. 2008, 71, 411–415. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, J.; Zou, G.; Wang, C.; Zhou, Z. Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzym. Microb. Techol. 2011, 49, 366–371. [Google Scholar] [CrossRef]
- Qian, Y.; Zhong, L.; Hou, Y.; Qu, Y.; Zhong, Y. Characterization and Strain Improvement of a Hypercellulytic Variant, Trichoderma reesei SN1, by Genetic Engineering for Optimized Cellulase Production in Biomass Conversion Improvement. Front. Microbiol. 2016, 7, 1349. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, S.; Schuster, M.; Cannon, S.; Steinberg, G. Optimised red- and green-fluorescent proteins for live cell imaging in the industrial enzyme-producing fungus Trichoderma reesei. Fungal Genet. Biol. 2020, 138, 103366. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Lin, X.; Yue, J.; Li, X.; Fang, X.; Zhu, M.; Lin, J.; Qu, Y.; Xiao, L. High concentration ethanol production from corncob residues by fed-batch strategy. Bioresour. Technol. 2010, 101, 4952–4958. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Pandey, A.; Sukumaran, R.K. Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis. Bioresour. Technol. 2017, 245, 1334–1342. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Wang, S.; Xing, M.; Yu, S.; Liu, G. Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters. Microb. Cell. Fact. 2012, 11, 84. [Google Scholar] [CrossRef]
- Sørensen, A.; Lübeck, M.; Lübeck, P.S.; Ahring, B.K. Fungal beta-glucosidases: A bottleneck in industrial use of lignocellulosic materials. Biomolecules 2013, 3, 612–631. [Google Scholar] [CrossRef]
- Sun, Y.; Qian, Y.; Zhang, J.; Yao, C.; Wang, Y.; Liu, H.; Zhong, Y. Development of a novel expression platform for heterologous protein production via deleting the p53-like regulator Vib1 in Trichoderma reesei. Enzym. Microb. Technol. 2022, 155, 109993. [Google Scholar] [CrossRef]
- Yang, S.; Song, L.; Wang, J.; Zhao, J.; Tang, H.; Bao, X. Engineering Saccharomyces cerevisiae for efficient production of recombinant proteins. Eng. Microb. 2024, 4, 100122. [Google Scholar] [CrossRef]
- Zhou, S.; Du, G.; Kang, Z.; Li, J.; Chen, J.; Li, H.; Zhou, J. The application of powerful promoters to enhance gene expression in industrial microorganisms. World J. Microbiol. Biotechnol. 2017, 33, 23. [Google Scholar] [CrossRef] [PubMed]
- Paasikallio, T.; Huuskonen, A.; Wiebe, M.G. Scaling up and scaling down the production of galactaric acid from pectin using Trichoderma reesei. Microb. Cell Fact. 2017, 16, 119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, Y.; Liu, Q.; Zhao, Q.; Gao, L.; Song, X.; Li, X.; Qu, Y.; Liu, G. Genetic engineering and raising temperature enhance recombinant protein production with the cdna1 promoter in Trichoderma reesei. Bioresour. Bioprocess 2022, 9, 113. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Gao, L.; Waghmare, P.R.; Qu, J.; Liu, G. Expression of a SARS-CoV-2 neutralizing nanobody in Trichoderma reesei. Sheng Wu Gong Cheng Xue Bao 2022, 38, 2250–2258. [Google Scholar] [CrossRef]
- Keren, L.; Zackay, O.; Lotan Pompan, M.; Barenholz, U.; Dekel, E.; Sasson, V.; Aidelberg, G.; Bren, A.; Zeevi, D.; Weinberger, A.; et al. Promoters maintain their relative activity levels under different growth conditions. Mol. Syst. Biol. 2013, 9, 701. [Google Scholar] [CrossRef]
- Fitz, E.; Wanka, F.; Seiboth, B. The Promoter Toolbox for Recombinant Gene Expression in Trichoderma reesei. Front. Bioeng. Biotech. 2018, 6, 135. [Google Scholar] [CrossRef]
- Erden-Karaoğlan, F.; Karaoğlan, M. Applicability of the heterologous yeast promoters for recombinant protein production in Pichia pastoris. Appl. Microbiol. Biot. 2022, 106, 7073–7083. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Lu, L.; Zhang, C.; Cai, F.; Lin, Y.; Huang, Y. Isolation and evaluation of strong endogenous promoters for the heterologous expression of proteins in Pichia pastoris. World J. Microbiol. Biotechnol. 2022, 38, 226. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef]
- Gusakov, A.V. Cellulases and hemicellulases in the 21st century race for cellulosic ethanol. Biofuels 2013, 4, 567–569. [Google Scholar] [CrossRef]
- Li, J.; Gao, K.; Secundo, F.; Mao, X. Biochemical characterization of two β-N-acetylglucosaminidases from Streptomyces violascens for efficient production of N-acetyl-d-glucosamine. Food Chem. 2021, 364, 130393. [Google Scholar] [CrossRef]
- Kamaruddin, N.; Storms, R.; Mahadi, N.M.; Illias, R.M.; Bakar, F.D.A.; Murad, A.M.A. Reduction of extracellular proteases increased activity and stability of heterologous protein in Aspergillus niger. Arab. J. Sci. Eng. 2018, 43, 3327–3338. [Google Scholar] [CrossRef]
- Yang, W.; Su, Y.; Wang, R.; Zhang, H.; Jing, H.; Meng, J.; Zhang, G.; Huang, L.; Guo, L.; Wang, J.; et al. Microbial production and applications of beta-glucosidase—A review. Int. J. Biol. Macromol. 2024, 256, 127915. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, J.; Wang, J.; Li, Z.; Wang, L.; Lu, P.; Zhong, Y.; Liu, H. High-Level Expression of β-Glucosidase in Aspergillus niger ATCC 20611 Using the Trichoderma reesei Promoter Pcdna1 to Enhance Cellulose Degradation. Fermentation 2024, 10, 461. https://doi.org/10.3390/fermentation10090461
Chang J, Wang J, Li Z, Wang L, Lu P, Zhong Y, Liu H. High-Level Expression of β-Glucosidase in Aspergillus niger ATCC 20611 Using the Trichoderma reesei Promoter Pcdna1 to Enhance Cellulose Degradation. Fermentation. 2024; 10(9):461. https://doi.org/10.3390/fermentation10090461
Chicago/Turabian StyleChang, Jingjing, Juan Wang, Zhihong Li, Lu Wang, Peng Lu, Yaohua Zhong, and Hong Liu. 2024. "High-Level Expression of β-Glucosidase in Aspergillus niger ATCC 20611 Using the Trichoderma reesei Promoter Pcdna1 to Enhance Cellulose Degradation" Fermentation 10, no. 9: 461. https://doi.org/10.3390/fermentation10090461




