Screening and Engineering Yeast Transporters to Improve Cellobiose Fermentation by Recombinant Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Molecular Biology Techniques
2.3. Enzymatic and Transport Activity Assays
2.4. Analytical Methods
2.5. Prediction of Lysine Residues in MgCBT2 Transporter with Ubiquitinylation Potential
3. Results
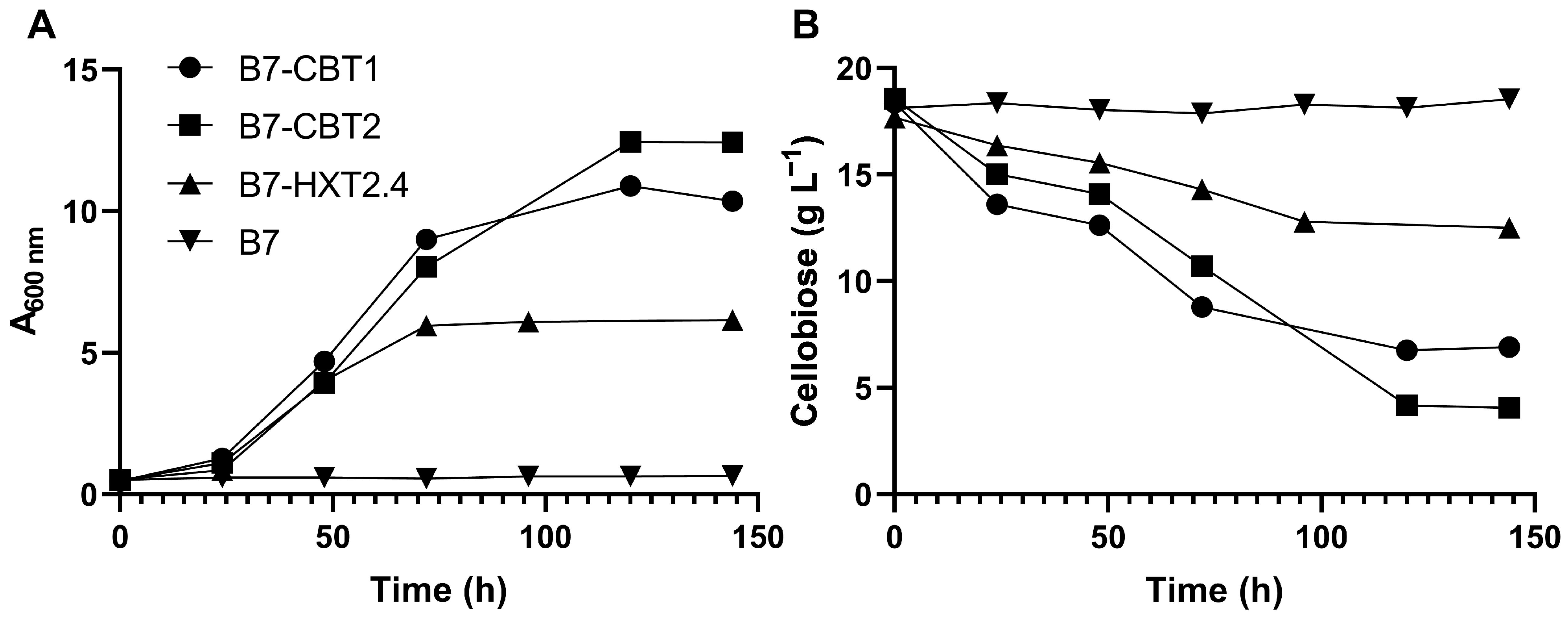
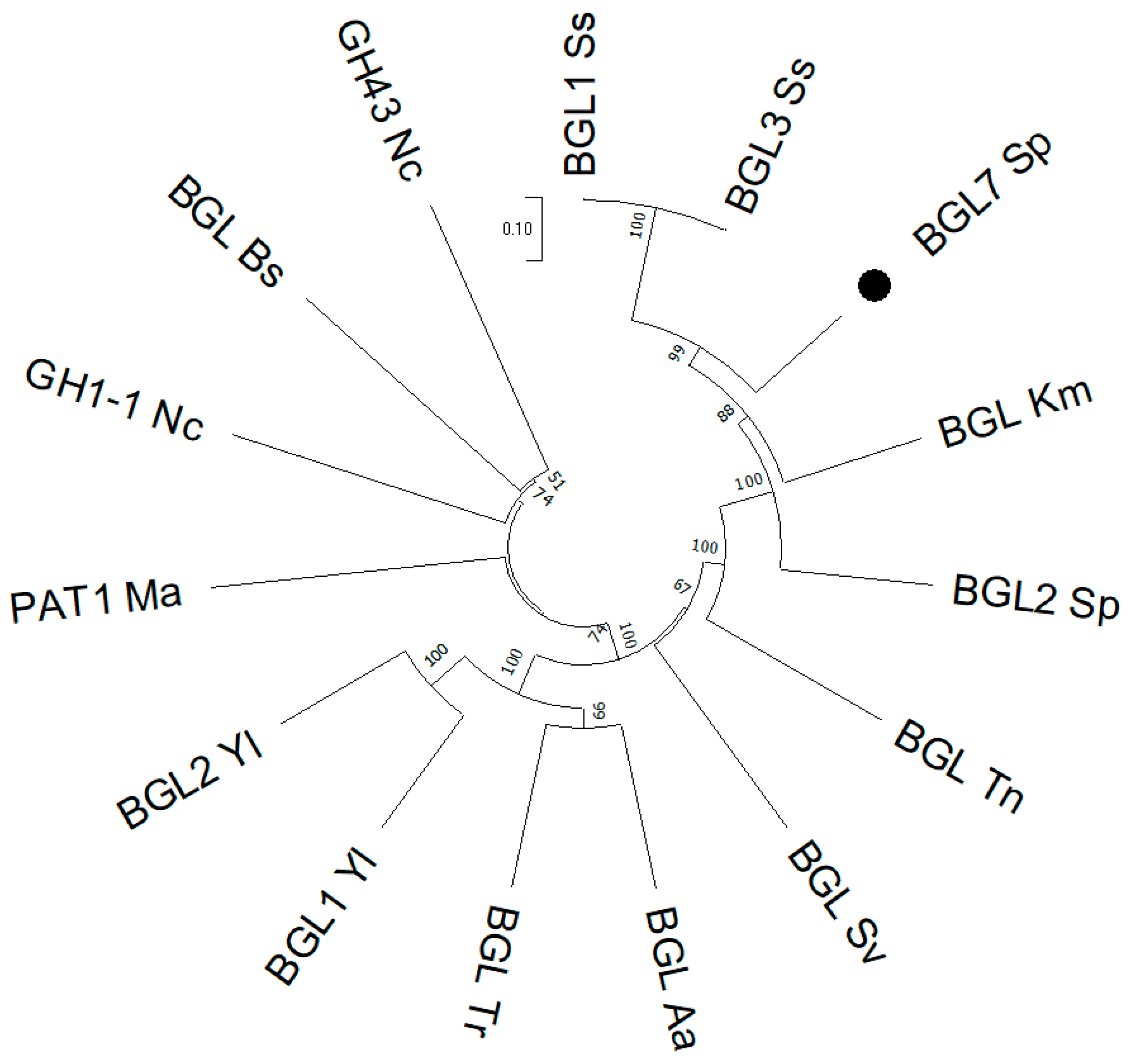
3.1. Cloning and Expression of an Intracellular Yeast β-Glucosidase in S. cerevisiae
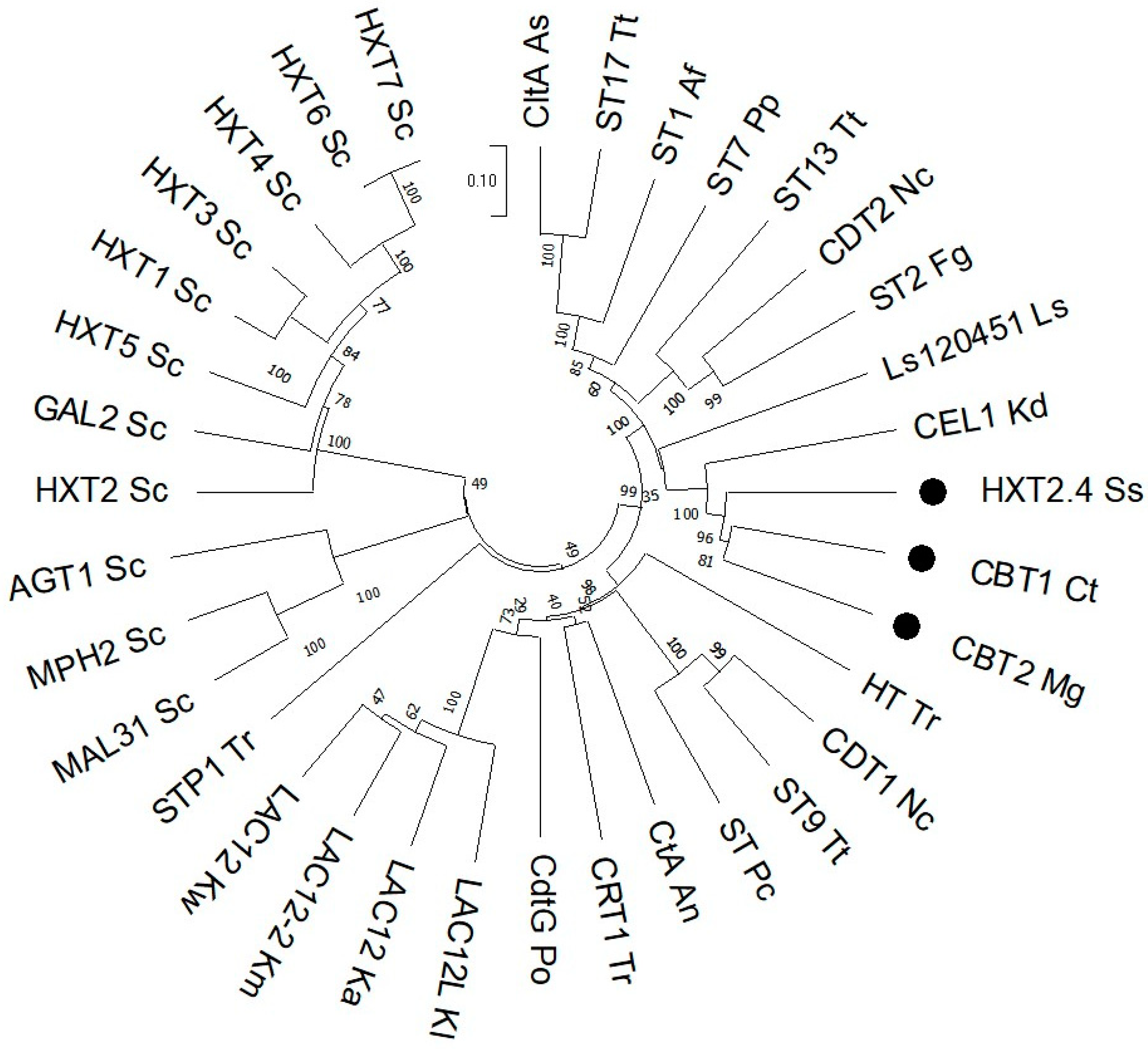
3.2. Cloning and Expression of Yeast Sugar (Cellobiose) Transporters in S. cerevisiae
3.3. Identification of Possible Lysine Residues Involved in Ubiquitinylation and Down-Regulation of the MgCBT2 Cellobiose Transporter Expressed in S. cerevisiae
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaiswal, D.; de Souza, A.; Larsen, S.; Lebauer, D.S.; Miguez, F.E.; Sparovek, G.; Bollero, G.; Buckeridge, M.S.; Long, S.P. Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nat. Clim. Chang. 2017, 7, 788–792. [Google Scholar] [CrossRef]
- Liu, Y.; Cruz-Morales, P.; Zargar, A.; Belcher, M.S.; Pang, B.; Englund, E.; Dan, Q.; Yin, K.; Keasling, J.D. Biofuels for a sustainable future. Cell 2021, 184, 1636–1647. [Google Scholar] [CrossRef] [PubMed]
- Jacobus, A.P.; Gross, J.; Evans, J.H.; Ceccato-Antonini, S.R.; Gombert, A.K. Saccharomyces cerevisiae strains used industrially for bioethanol production. Essays Biochem. 2021, 65, 147–161. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, L.V.; de Barros Grassi, M.C.; Gallardo, J.C.M.; Pirolla, R.A.S.; Calderón, L.L.; de Carvalho-Netto, O.V.; Parreiras, L.S.; Camargo, E.L.O.; Drezza, A.L.; Missawa, S.K.; et al. Second-generation ethanol: The need is becoming a reality. Ind. Biotechnol. 2016, 12, 40–57. [Google Scholar] [CrossRef]
- Igwebuike, C.M.; Awad, S.; Andrès, Y. Renewable energy potential: Second-generation biomass as feedstock for bioethanol production. Molecules 2024, 29, 1619. [Google Scholar] [CrossRef]
- Gao, M.; Ploessl, D.; Shao, Z. Enhancing the co-utilization of biomass-derived mixed sugars by yeasts. Front. Microbiol. 2019, 9, 3264. [Google Scholar] [CrossRef]
- Broda, M.; Yelle, D.J.; Serwańska, K. Bioethanol production from lignocellulosic biomass-Challenges and solutions. Molecules 2022, 27, 8717. [Google Scholar] [CrossRef]
- Yadav, A.; Sharma, V.; Tsai, M.L.; Chen, C.W.; Sun, P.P.; Nargotra, P.; Wang, J.X.; Dong, C.D. Development of lignocellulosic biorefineries for the sustainable production of biofuels: Towards circular bioeconomy. Bioresour. Technol. 2023, 381, 129145. [Google Scholar] [CrossRef]
- da Silva, A.S.; Espinheira, R.P.; Teixeira, R.S.S.; de Souza, M.F.; Ferreira-Leitão, V.; Bon, E.P.S. Constraints and advances in high-solids enzymatic hydrolysis of lignocellulosic biomass: A critical review. Biotechnol. Biofuels. 2020, 13, 58. [Google Scholar] [CrossRef]
- Østby, H.; Hansen, L.D.; Horn, S.J.; Eijsink, V.G.H.; Várnai, A. Enzymatic processing of lignocellulosic biomass: Principles, recent advances and perspectives. J. Ind. Microbiol. Biotechnol. 2020, 47, 623–657. [Google Scholar] [CrossRef]
- Sørensen, A.; Lübeck, M.; Lübeck, P.S.; Ahring, B.K. Fungal β-glucosidases: A bottleneck in industrial use of lignocellulosic materials. Biomolecules 2013, 3, 612–631. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and significance of β-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Pryor, S.W.; Nahar, N. β-glucosidase supplementation during biomass hydrolysis: How low can we go? Biomass Bioenergy 2015, 80, 298–302. [Google Scholar] [CrossRef]
- Ferreira, R.G.; Azzoni, A.R.; Freitas, S. On the production cost of lignocellulose-degrading enzymes. Biofuels Bioprod. Bioref. 2021, 15, 85–99. [Google Scholar] [CrossRef]
- Barbosa, A.M.; Giese, E.C.; Dekker, R.F.; Borsato, D.; Briones Pérez, A.I.; Ubeda Iranzo, J.F. Extracellular β-glucosidase production by the yeast Debaryomyces pseudopolymorphus UCLM-NS7A: Optimization using response surface methodology. N. Biotechnol. 2010, 27, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Z.L.; Weber, S.A.; Zhang, X. Two new native β-glucosidases from Clavispora NRRL Y-50464 confer its dual function as cellobiose fermenting ethanologenic yeast. PLoS ONE 2016, 11, e0151293. [Google Scholar] [CrossRef] [PubMed]
- Vaz, J.E.; Rabelo, L.; Zaiter, M.A.; Pereira, W.E.S.; Metzker, G.; Boscolo, M.; da Silva, R.; Gomes, E.; da Silva, R.R. Functional properties and potential application of ethanol tolerant β-glucosidases from Pichia ofunaensis and Trichosporon multisporum yeasts. 3 Biotech 2021, 11, 467. [Google Scholar] [CrossRef]
- Albarello, M.L.; Giehl, A.; Tadioto, V.; dos Santos, A.A.; Milani, L.M.; Bristot, J.C.; Tramontin, M.A.; Treichel, H.; Bernardi, O.; Treichel, H.; et al. Analysis of the holocellulolytic and fermentative potentials of yeasts isolated from the gut of Spodoptera frugiperda larvae. Bioener. Res. 2023, 16, 2046–2057. [Google Scholar] [CrossRef]
- Jeffries, T.W.; Van Vleet, J.R. Pichia stipitis genomics, transcriptomics, and gene clusters. FEMS Yeast Res. 2009, 9, 793–807. [Google Scholar] [CrossRef]
- Santos, R.O.; Cadete, R.M.; Badotti, F.; Mouro, A.; Wallheim, D.O.; Gomes, F.C.; Stambuk, B.U.; Lachance, M.A.; Rosa, C.A. Candida queiroziae sp. nov., a cellobiose-fermenting yeast species isolated from rotting wood in Atlantic Rain Forest. Antonie Van Leeuwenhoek 2011, 99, 635–642. [Google Scholar] [CrossRef]
- Lopes, M.R.; Lara, C.A.; Moura, M.E.F.; Uetanabaro, A.P.T.; Morais, P.B.; Vital, M.J.S.; Rosa, C.A. Characterization of the diversity and physiology of cellobiose-fermenting yeasts isolated from rotting wood in Brazilian ecosystems. Fungal Biol. 2018, 122, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Barrilli, É.T.; Tadioto, V.; Milani, L.M.; Deoti, J.R.; Fogolari, O.; Müller, C.; Barros, K.O.; Rosa, C.A.; dos Santos, A.A.; Stambuk, B.U.; et al. Biochemical analysis of cellobiose catabolism in Candida pseudointermedia strains isolated from rotten wood. Arch. Microbiol. 2020, 202, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Choi, H.J.; Lee, Y.R.; Kim, S.J.; Lee, S.; Lee, W.H. Comprehensive characterization of mutant Pichia stipitis co-fermenting cellobiose and xylose through genomic and transcriptomic analyses. J. Microbiol. Biotechnol. 2022, 32, 1485–1495. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Hipp, J.; Trinh, C.T. Activating and elucidating metabolism of complex sugars in Yarrowia lipolytica. Appl. Environ. Microbiol. 2015, 82, 1334–1345. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Duquesne, S.; Bozonnet, S.; Cioci, G.; Nicaud, J.M.; Marty, A.; O’Donohue, M.J. Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol. Biofuels 2015, 8, 109. [Google Scholar] [CrossRef]
- Galazka, J.M.; Tian, C.; Beeson, W.T.; Martinez, B.; Glass, N.L.; Cate, J.H. Cellodextrin transport in yeast for improved biofuel production. Science 2010, 330, 84–86. [Google Scholar] [CrossRef]
- Bae, Y.H.; Kang, K.H.; Jin, Y.S.; Seo, J.H. Molecular cloning and expression of fungal cellobiose transporters and β-glucosidases conferring efficient cellobiose fermentation in Saccharomyces cerevisiae. J. Biotechnol. 2014, 169, 34–41. [Google Scholar] [CrossRef]
- Gonçalves, D.L.; Matsushika, A.; de Sales, B.B.; Goshima, T.; Bon, E.P.S.; Stambuk, B.U. Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb. Technol. 2014, 63, 13–20. [Google Scholar] [CrossRef]
- Li, S.; Du, J.; Sun, J.; Galazka, J.M.; Glass, N.L.; Cate, J.H.; Yang, X.; Zhao, H. Overcoming glucose repression in mixed sugar fermentation by co-expressing a cellobiose transporter and a β-glucosidase in Saccharomyces cerevisiae. Mol. Biosyst. 2010, 6, 2129–2132. [Google Scholar] [CrossRef]
- Ha, S.J.; Galazka, J.M.; Kim, S.R.; Choi, J.H.; Yang, X.; Seo, J.H.; Glass, N.L.; Cate, J.H.; Jin, Y.S. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. USA 2011, 108, 504–509. [Google Scholar] [CrossRef]
- Ha, S.J.; Kim, S.R.; Kim, H.; Du, J.; Cate, J.H.; Jin, Y.S. Continuous co-fermentation of cellobiose and xylose by engineered Saccharomyces cerevisiae. Bioresour, Technol. 2013, 149, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Parisutham, V.; Chandran, S.P.; Mukhopadhyay, A.; Lee, S.K.; Keasling, J.D. Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries. Bioresour. Technol. 2017, 239, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.J.; Jin, Y.S. Engineering of Saccharomyces cerevisiae for efficient fermentation of cellulose. FEMS Yeast Res. 2020, 20, foz089. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Gu, R.; Wang, B.; Li, J.; Wan, L.; Tian, C.; Ma, Y. Evidence of a critical role for cellodextrin transporter 2 (CDT-2) in both cellulose and hemicellulose degradation and utilization in Neurospora crassa. PLoS ONE 2014, 9, e89330. [Google Scholar] [CrossRef]
- Znameroski, E.A.; Li, X.; Tsai, J.C.; Galazka, J.M.; Glass, N.L.; Cate, J.H. Evidence for transceptor function of cellodextrin transporters in Neurospora crassa. J. Biol. Chem. 2014, 289, 2610–2619. [Google Scholar] [CrossRef]
- Kim, H.; Lee, W.H.; Galazka, J.M.; Cate, J.H.; Jin, Y.S. Analysis of cellodextrin transporters from Neurospora crassa in Saccharomyces cerevisiae for cellobiose fermentation. Appl. Microbiol. Biotechnol. 2014, 98, 1087–1094. [Google Scholar] [CrossRef]
- Ha, S.J.; Kim, H.; Lin, Y.; Jang, M.U.; Galazka, J.M.; Kim, T.J.; Cate, J.H.; Jin, Y.S. Single amino acid substitutions in HXT2.4 from Scheffersomyces stipitis lead to improved cellobiose fermentation by engineered Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2013, 79, 1500–1507. [Google Scholar] [CrossRef]
- de Ruijter, J.C.; Igarashi, K.; Penttilä, M. The Lipomyces starkeyi gene Ls120451 encodes a cellobiose transporter that enables cellobiose fermentation in Saccharomyces cerevisiae. FEMS Yeast Res. 2020, 20, foaa019. [Google Scholar] [CrossRef]
- Long, T.M.; Su, Y.K.; Headman, J.; Higbee, A.; Willis, L.B.; Jeffries, T.W. Cofermentation of glucose, xylose, and cellobiose by the beetle-associated yeast Spathaspora passalidarum. Appl. Environ. Microbiol. 2012, 78, 5492–5500. [Google Scholar] [CrossRef]
- Lauwers, E.; Erpapazoglou, Z.; Haguenauer-Tsapis, R.; André, B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010, 20, 196–204. [Google Scholar] [CrossRef]
- Barata-Antunes, C.; Alves, R.; Talaia, G.; Casal, M.; Gerós, H.; Mans, R.; Paiva, S. Endocytosis of nutrient transporters in fungi: The ART of connecting signaling and trafficking. Comput. Struct. Biotechnol. J. 2021, 19, 1713–1737. [Google Scholar] [CrossRef] [PubMed]
- Meselson, M.; Yuan, R. DNA restriction enzyme from E. coli. Nature 1968, 217, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Lara, C.A.; Santos, R.O.; Cadete, R.M.; Ferreira, C.; Marques, S.; Gírio, F.; Oliveira, E.S.; Rosa, C.A.; Fonseca, C. Identification and characterisation of xylanolytic yeasts isolated from decaying wood and sugarcane bagasse in Brazil. Antonie Van Leeuwenhoek 2014, 105, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Cadete, R.M.; Melo-Cheab, M.A.; Dussán, K.J.; Rodrigues, R.C.L.B.; da Silva, S.S.; Gomes, F.C.O.; Rosa, C.A. Production of bioethanol in sugarcane bagasse hemicellulosic hydrolysate by Scheffersomyces parashehatae, Scheffersomyces illinoinensis and Spathaspora arborariae isolated from Brazilian ecosystems. J. Appl. Microbiol. 2017, 123, 1203–1213. [Google Scholar] [CrossRef]
- Mouro, A.; dos Santos, A.A.; Agnolo, D.D.; Gubert, G.F.; Bon, E.P.S.; Rosa, C.A.; Fonseca, C.; Stambuk, B.U. Combining xylose reductase from Spathaspora arborariae with xylitol dehydrogenase from Spathaspora passalidarum to promote xylose consumption and fermentation into xylitol by Saccharomyces cerevisiae. Fermentation 2020, 6, 72. [Google Scholar] [CrossRef]
- Entian, K.; Kotter, P. Yeast genetic strain and plasmid collections. Methods Microbiol. 2007, 36, 629–666. [Google Scholar] [CrossRef]
- Babrzadeh, F.; Jalili, R.; Wang, C.; Shokralla, S.; Pierce, S.; Robinson-Mosher, A.; Nyren, P.; Shafer, R.W.; Basso, L.C.; de Amorim, H.V.; et al. Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1. Mol. Genet. Genomics 2012, 287, 485–494. [Google Scholar] [CrossRef]
- Muller, G.; de Godoy, V.R.; Dário, M.G.; Duval, E.H.; Alves-Jr, S.L.; Bücker, A.; Rosa, C.A.; Dunn, B.; Sherlock, G.; Stambuk, B.U. Improved sugarcane-based fermentation processes by an industrial fuel-ethanol yeast strain. J. Fungi 2023, 9, 803. [Google Scholar] [CrossRef]
- Pereira, I.O.; dos Santos, Â.A.; Gonçalves, D.L.; Purificação, M.; Guimarães, N.C.; Tramontina, R.; Coutouné, N.; Zanella, E.; Matsushika, A.; Stambuk, B.U.; et al. Comparison of Spathaspora passalidarum and recombinant Saccharomyces cerevisiae for integration of first- and second-generation ethanol production. FEMS Yeast Res. 2021, 21, foab048. [Google Scholar] [CrossRef]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Vyas, V.K.; Bushkin, G.G.; Bernstein, D.A.; Getz, M.A.; Sewastianik, M.; Barrasa, M.I.; Bartel, D.P.; Fink, G.R. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. mSphere 2018, 3, e00154-18. [Google Scholar] [CrossRef]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology, 3rd ed.; John Wiley & Sons: New York, NY, USA, 1995. [Google Scholar]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.W.; Grigoriev, I.V.; Grimwood, J.; Laplaza, J.M.; Aerts, A.; Salamov, A.; Schmutz, J.; Lindquist, E.; Dehal, P.; Shapiro, H.; et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007, 25, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Wohlbach, D.J.; Kuo, A.; Sato, T.K.; Potts, K.M.; Salamov, A.A.; Labutti, K.M.; Sun, H.; Clum, A.; Pangilinan, J.L.; Lindquist, E.A.; et al. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc. Natl. Acad. Sci. USA 2011, 108, 13212–13217. [Google Scholar] [CrossRef]
- Reider Apel, A.; d’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.J.; et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 496–508. [Google Scholar] [CrossRef]
- Stambuk, B.U. A simple experiment illustrating metabolic regulation: Induction versus repression of yeast α-glucosidase. Biochem. Educ. 1999, 27, 177–180. [Google Scholar] [CrossRef]
- Hollatz, C.; Stambuk, B.U. Colorimetric determination of active α-glucoside transport in Saccharomyces cerevisiae. J. Microbiol. Methods 2001, 46, 253–259. [Google Scholar] [CrossRef]
- Pirovano, W.; Feenstra, K.A.; Heringa, J. PRALINETM: A strategy for improved multiple alignment of transmembrane proteins. Bioinformatics 2008, 24, 492–497. [Google Scholar] [CrossRef]
- Li, X.; Gao, X.; Ren, J.; Jin, C.; Xue, Y. BDM-PUB: Computational Prediction of Protein Ubiquitination Sites with a Bayesian Discriminant Method, 2009. BDM-PUB: Prediction of Ubiquitination Website. Available online: https://www.scienceopen.com/document?vid=cdb6582a-8d17-4b0a-aae0-7044b4d6363a (accessed on 30 November 2021).
- Radivojac, P.; Vacic, V.; Haynes, C.; Cocklin, R.R.; Mohan, A.; Heyen, J.W.; Goebl, M.G.; Iakoucheva, L.M. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010, 78, 365–380. [Google Scholar] [CrossRef]
- Urbina, H.; Blackwel, L.M. Multilocus phylogenetic study of the Scheffersomyces yeast clade and characterization of the N-terminal region of xylose reductase gene. PLoS ONE 2012, 7, e39128. [Google Scholar] [CrossRef] [PubMed]
- Stambuk, B.U. Transcriptional and posttranslational regulation of a membrane nutrient transporter. Biochem. Mol. Biol. Educ. 2002, 30, 388–393. [Google Scholar] [CrossRef]
- Horák, J. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: Insights from yeast. Biochim. Biophys. Acta 2003, 1614, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Skory, C.D.; Freer, S.N.; Bothast, R.J. Properties of an intracellular β-glucosidase purified from the cellobiose-fermenting yeast Candida wickerhamii. Appl. Microbiol. Biotechnol. 1996, 46, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Chemardin, P.; Arnaud, A. Purification and characterization of an intracellular β-glucosidase from a Candida sake strain isolated from fruit juices. Appl. Biochem. Biotechnol. 2001, 95, 151–162. [Google Scholar] [CrossRef]
- Varela, J.A.; Puricelli, M.; Ortiz-Merino, R.A.; Giacomobono, R.; Braun-Galleani, S.; Wolfe, K.H.; Morrissey, J.P. Origin of lactose fermentation in Kluyveromyces lactis by interspecies transfer of a neo-functionalized gene cluster during domestication. Curr. Biol. 2019, 29, 4284–4290.e2. [Google Scholar] [CrossRef]
- Sen, A.; Acosta-Sampson, L.; Alvaro, C.G.; Ahn, J.S.; Cate, J.H.; Thorner, J. Internalization of heterologous sugar transporters by endogenous α-arrestins in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2016, 82, 7074–7085. [Google Scholar] [CrossRef]
- Knychala, M.M.; dos Santos, A.A.; Kretzer, L.G.; Gelsleichter, F.; Leandro, M.J.; Fonseca, C.; Stambuk, B.U. Strategies for efficient expression of heterologous monosaccharide transporters in Saccharomyces cerevisiae. J. Fungi 2022, 8, 84. [Google Scholar] [CrossRef]
- Li, H.; Schmitz, O.; Alper, H.S. Enabling glucose/xylose co-transport in yeast through the directed evolution of a sugar transporter. Appl. Microbiol. Biotechnol. 2016, 100, 10215–10223. [Google Scholar] [CrossRef]
- Luo, X.; Tao, X.; Ran, G.; Deng, Y.; Wang, H.; Tan, L.; Pang, Z. Molecular modification enhances xylose uptake by the sugar transporter KM_SUT5 of Kluyveromyces marxianus. Int. J. Mol. Sci. 2024, 25, 8322. [Google Scholar] [CrossRef]
- Tamayo Rojas, S.A.; Schmidl, S.; Boles, E.; Oreb, M. Glucose-induced internalization of the S. cerevisiae galactose permease Gal2 is dependent on phosphorylation and ubiquitination of its aminoterminal cytoplasmic tail. FEMS Yeast Res. 2021, 21, foab019. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kim, Y.B.; Cho, K.H.; Kim, J.H. Glucose starvation-induced turnover of the yeast glucose transporter Hxt1. Biochim. Biophys. Acta 2014, 1840, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jimenez, F.D.; Pereira, I.O.; Ribeiro, M.P.A.; Sargo, C.R.; dos Santos, A.A.; Zanella, E.; Stambuk, B.U.; Ienczak, J.; Morais, E.; Costa, A. Integration of first- and second-generation ethanol production: Evaluation of a mathematical model to describe sucrose and xylose co-fermentation by recombinant Saccharomyces cerevisiae. Renew. Energy 2022, 192, 326–339. [Google Scholar] [CrossRef]
- de Oliveira Pereira, I.; dos Santos, Â.A.; Guimarães, N.C.; Lima, C.S.; Zanella, E.; Matsushika, A.; Rabelo, S.C.; Stambuk, B.U.; Ienczak, J.L. First- and second-generation integrated process for bioethanol production: Fermentation of molasses diluted with hemicellulose hydrolysate by recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 2024, 121, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Sousa, M.J.; Cardoso, H.; Inácio, J.; Silva, S.; Spencer-Martins, I.; Leão, C. Ethanol tolerance of sugar transport, and the rectification of stuck wine fermentations. Microbiology 2008, 154, 422–430. [Google Scholar] [CrossRef]
- Swinnen, S.; Goovaerts, A.; Schaerlaekens, K.; Dumortier, F.; Verdyck, P.; Souvereyns, K.; Van Zeebroeck, G.; Foulquié-Moreno, M.R.; Thevelein, J.M. Auxotrophic mutations reduce tolerance of Saccharomyces cerevisiae to very high levels of ethanol stress. Eukaryot. Cell 2015, 14, 884–897. [Google Scholar] [CrossRef]
- Eriksen, D.T.; Hsieh, P.C.; Lynn, P.; Zhao, H. Directed evolution of a cellobiose utilization pathway in Saccharomyces cerevisiae by simultaneously engineering multiple proteins. Microb. Cell Fact. 2013, 12, 61. [Google Scholar] [CrossRef]
- Lian, J.; Li, Y.; HamediRad, M.; Zhao, H. Directed evolution of a cellodextrin transporter for improved biofuel production under anaerobic conditions in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2014, 111, 1521–1531. [Google Scholar] [CrossRef]
- Hu, M.L.; Zha, J.; He, L.W.; Lv, Y.J.; Shen, M.H.; Zhong, C.; Li, B.Z.; Yuan, Y.J. Enhanced bioconversion of cellobiose by industrial Saccharomyces cerevisiae used for cellulose utilization. Front. Microbiol. 2016, 7, 241. [Google Scholar] [CrossRef]
- Oh, E.J.; Kwak, S.; Kim, H.; Jin, Y.S. Transporter engineering for cellobiose fermentation under lower pH conditions by engineered Saccharomyces cerevisiae. Bioresour. Technol. 2017, 245, 1469–1475. [Google Scholar] [CrossRef]
- Kim, H.; Oh, E.J.; Lane, S.T.; Lee, W.H.; Cate, J.H.D.; Jin, Y.S. Enhanced cellobiose fermentation by engineered Saccharomyces cerevisiae expressing a mutant cellodextrin facilitator and cellobiose phosphorylase. J. Biotechnol. 2018, 275, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.J.; Skerker, J.M.; Kim, S.R.; Wei, N.; Turner, T.L.; Maurer, M.J.; Arkin, A.P.; Jin, Y.S. Gene amplification on demand accelerates cellobiose utilization in engineered Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2016, 82, 3631–3639. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, W.H.; Turner, T.L.; Kwak, S.; Jin, Y.S. An extra copy of the β-glucosidase gene improved the cellobiose fermentation capability of an engineered Saccharomyces cerevisiae strain. 3 Biotech 2019, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Chomvong, K.; Benjamin, D.I.; Nomura, D.K.; Cate, J.H.D. Cellobiose consumption uncouples extracellular glucose sensing and glucose metabolism in Saccharomyces cerevisiae. mBio 2017, 8, e00855-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chomvong, K.; Acosta-Sampson, L.; Estrela, R.; Galazka, J.M.; Kim, S.R.; Jin, Y.S.; Cate, J.H. Leveraging transcription factors to speed cellobiose fermentation by Saccharomyces cerevisiae. Biotechnol. Biofuels 2014, 7, 126. [Google Scholar] [CrossRef]
- Sullivan, S.F.; Shetty, A.; Bharadwaj, T.; Krishna, N.; Trivedi, V.D.; Endalur Gopinarayanan, V.; Chappell, T.C.; Sellers, D.M.; Kumar, R.P.; Nair, N.U. Towards universal synthetic heterotrophy using a metabolic coordinator. Metab. Eng. 2023, 79, 14–26. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Lu, M.; Ding, B.; Chen, S.; Wen, Z.; Yu, Y.; Zhou, L.; Jin, M. Rapid evolution and mechanism elucidation for efficient cellobiose-utilizing Saccharomyces cerevisiae through synthetic chromosome rearrangement and modification by LoxPsym-mediated evolution. Bioresour. Technol. 2022, 356, 127268. [Google Scholar] [CrossRef]
- Blount, B.A.; Gowers, G.F.; Ho, J.C.H.; Ledesma-Amaro, R.; Jovicevic, D.; McKiernan, R.M.; Xie, Z.X.; Li, B.Z.; Yuan, Y.J.; Ellis, T. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome. Nat. Commun. 2018, 9, 1932. [Google Scholar] [CrossRef]





| Strains and Plasmids | Relevant Features or Genotype | Source |
|---|---|---|
| Yeast strains: | ||
| C. tropicalis UFMG-HB-93a | Isolated from decaying sugarcane bagasse in São Paulo, Brazil | [43] |
| M. guilliermondii NRRL Y-27844 | Clinical isolate | USDA-ARC Culture Collection |
| Sc. illinoinensis UFMG-CM-Y512 | Isolated from rotting wood in Rio de Janeiro, Brazil | [44] |
| Sp. passalidarum UFMG-CM-Y474 | Isolated from rotting wood in Roraima, Brazil | [45] |
| S. cerevisiae CEN.PK2-1C | MATa leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2 | [46] |
| S. cerevisiae B7 | CEN.PK2-1C + pGPD-424-SpBGL7 | This work |
| S. cerevisiae B7-HXT2.4 | CEN.PK2-1C + pGPD-424-SpBGL7 + pGPD-426-SsHXT2.4 | This work |
| S. cerevisiae B7-CBT1 | CEN.PK2-1C + pGPD-424-SpBGL7 + pGPD-426-CtCBT1 | This work |
| S. cerevisiae B7-CBT2 | CEN.PK2-1C + pGPD-424-SpBGL7 + pGPD-426-MgCBT2 | This work |
| S. cerevisiae CAT-1 | Industrial strain isolated from Usina VO Catanduva, São Paulo, Brazil | [47,48] |
| S. cerevisiae MP-C5H1 | Isogenic to CAT-1, but AUR1::pAUR-XKXDHXR loxP-KanMX-loxP-PADH1::[4- 59∆]HXT1 | [49] |
| S. cerevisiae MP-B7 | Isogenic to MP-C5H1, but ARS208::PTEF1-SpBGL7-TPGK1 | This work |
| S. cerevisiae MP-B7-CBT2 | Isogenic to MP-B7, but ARS1309::PTDH3-MgCBT2-TCYC1 | This work |
| S. cerevisiae MP-B7-CBT2ΔC | Isogenic to MP-B7, but ARS1309::PTDH3-MgCBT2ΔC-TCYC1 | This work |
| S. cerevisiae MP-B7-CBT2ΔNΔC | Isogenic to MP-B7, but ARS1309::PTDH3-MgCBT2ΔNΔC-TCYC1 | This work |
| Plasmids: | ||
| pGPD-424 | AmpR ori 2µ TRP1 PTDH3-TCYC1 | ATCC® 87357TM [50] |
| pGPD-426 | AmpR ori 2µ URA3 PTDH3-TCYC1 | ATCC® 87361TM [50] |
| pGPD-424-SpBGL7 | AmpR ori 2µ TRP1 PTDH3-SpBGL7-TCYC1 | This work |
| pGPD-426-SsHXT2.4 | AmpR ori 2µ URA3 PTDH3-SsHXT2.4-TCYC1 | This work |
| pGPD-426-CtCBT1 | AmpR ori 2µ URA3 PTDH3-CtCBT1-TCYC1 | This work |
| pGPD-426-MgCBT2 | AmpR ori 2µ URA3 PTDH3-MgCBT2-TCYC1 | This work |
| pGPD-426-MgCBT2ΔC | AmpR ori 2µ URA3 PTDH3-MgCBT2ΔC-TCYC1 | This work |
| pGPD-426-MgCBT2ΔNΔC | AmpR ori 2µ URA3 PTDH3-MgCBT2ΔNΔC-TCYC1 | This work |
| pV1382 | AmpR ori CEN ARS URA3 NATR PTEF1-CaCas9-TCYC1 PSNR52-sgRNA-TSNR52 | [51] |
| pV1382-ARS1309 | pV1382 PSNR52-sgRNA(ARS1309)-TSNR52 | This work |
| pV1382-ARS208 | pV1382 PSNR52-sgRNA(ARS208)-TSNR52 | This work |
| pMV | AmpR ori (pBR322 derivative) | BGI Group |
| pMV-SpBGL7 | AmpR ori [5’ARS208-PTEF1-SpBGL2-TPGK1-3’ARS208] | This work |
| Primer 1 | Sequence 2 |
|---|---|
| Cloning: | |
| SpBGL7-F | GAATTCATGACCGTGTCTGATTTCGATGTTG |
| SpBGL7-R | CTCGAGCTAATTACCTTTCCAGAAGAAACTTTGATC |
| HXT2.4-F | GGCGGATCCAAAATGTCTGACAAACTTCACAACATCAAG |
| HXT2.4-R | GGCCTCGAGGTCGACATAATCAGGTATAATTTATTGACTAAAGCTTAG |
| CtCBT1-F | GGCGAATTCAAAATGTCATCCAAAGATAATATCATCATCACTGAAG |
| CtCBT1-R | GGCCTCGAGGTCGACCTAGGCCAATTTTTCAACGTGATCAACC |
| MgCBT2-F | GGCGGATCCATGGTTTCCAATTCGTCTTCATACTGG |
| MgCBT2-R | GGCAAGCTTTCATACTTTTTCAGCATGTTCAAGCG |
| MgCBT2ΔC-F | CATGGATCCATGGGTTTCCATTCGTCTTC |
| MgCBT2ΔC-R | TGAAAGCTTTCACGGAGTGGCAAGAATATGGA |
| MgCBT2ΔNΔC-F | CATGGATCCATGCACCAGGATATCGCTACTCA |
| CRISPR-Cas9: | |
| sgRNA.ARS1309-F | 5P-GATCGCCTGTGGTGACTACGTATCCG |
| sgRNA.ARS1309-R | 5P-AAAACGGATACGTAGTCACCACAGGC |
| sgRNA.ARS208-F | 5P-GATCGGTCCGCTAAACAAAAGATCTG |
| sgRNA.ARS208-R | 5P-AAAACAGATCTTTTGTTTAGCGGACC |
| ARS208-F | CCGCAGTGTCTTGCGTCTCTGATCTTACCTGGTGAATTGG |
| ARS208-R | TTGGCAGTGACTCCGTCTCTAGTAGGTGCCAGTTGAATAG |
| Sequencing: | |
| seq.p1382.sgRNA-F | GCTGTAGAAGTGAAAGTTGG |
| seq.p1382.sgRNA-R | CAAGTTGATAACGGACTAGC |
| Strain a | Transporter | β-Glucosidase | Cellobiose (g/L) | Cellobiose Consumption (%) | Incubation Time (h) | Ethanol Yield/g Cellobiose b | Reference |
|---|---|---|---|---|---|---|---|
| YPH499 | Nc_CDT-1 | Nc_gh1-1 | 20 | 100 | 150 | 0.44 | [26] |
| BY4741 | Nc_CDT-2 | Nc_gh1-1 | 80-90 | 38–89 | >170 | 0.16–0.35 | [69] |
| Nc_CDT-2 c | Nc_gh1-1 | 80 | 78 | 170 | 0.23 | ||
| INVSc1 | Nc_CDT-2 | Nc_gh1-1 | 22 | 91 | 30 | 0.35 | [80] |
| Nc_CDT-2 d | Nc_gh1-1 | 22 | 98 | 10 | 0.45 | ||
| D452-2 | Nc_CDT-2 d | Nc_gh1-1 | 80 | 100 | 36 | 0.35 | [82] |
| D452-2 | Nc_CDT-1 | Pc_BG | 40 | 100 | 36 | 0.30 | [27] |
| Nc_CDT-1 | Tt_BG | 40 | 100 | 48 | 0.29 | ||
| Pc_ST | Tt_BG | 40 | 100 | 38 | 0.38 | ||
| CEN.PK2-1D | Ss_HXT2.4 | Nc_gh1-1 | 80 | 90 | 54 | 0.40 | [37] |
| Ss_HXT2.4 d | Nc_gh1-1 | 80 | 94 | 36 | 0.45 | ||
| D452-2 | Ss_HXT2.4 | Nc_gh1-1 | 80 | 70 | 60 | 0.16 | |
| Ss_HXT2.4 d | Nc_gh1-1 | 80 | 100 | 24 | 0.45 | ||
| YSS1 | Ls_120451 | Nc_gh1-1 | 21 | 64 | 96 | 0.18 | [38] |
| Still Spirits Turbo | Nc_CDT-1 | Nc_gh1-1 | 86 | 100 | 56 | 0.40 | [79] |
| Nc_CDT-1 d | Nc_gh1-1 d | 86 | 100 | 30 | 0.44 | ||
| Angel Yeast | Nc_CDT-1 | Nc_gh1-1 | 45 | 92 | 96 | 0.34 | [81] |
| Angel Yeast e | Nc_CDT-1 | Nc_gh1-1 | 45 | 100 | 18 | 0.50 | |
| MP-C5H1 | Mg_CBT2 | Sp_BGL7 | 20 | 55 | 192 | 0.47 | This work |
| Mg_CBT2ΔNΔC c | Sp_BGL7 | 20 | 94 | 144 | 0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kretzer, L.G.; Knychala, M.M.; da Silva, L.C.; da Fontoura, I.C.C.; Leandro, M.J.; Fonseca, C.; Verstrepen, K.J.; Stambuk, B.U. Screening and Engineering Yeast Transporters to Improve Cellobiose Fermentation by Recombinant Saccharomyces cerevisiae. Fermentation 2024, 10, 490. https://doi.org/10.3390/fermentation10090490
Kretzer LG, Knychala MM, da Silva LC, da Fontoura ICC, Leandro MJ, Fonseca C, Verstrepen KJ, Stambuk BU. Screening and Engineering Yeast Transporters to Improve Cellobiose Fermentation by Recombinant Saccharomyces cerevisiae. Fermentation. 2024; 10(9):490. https://doi.org/10.3390/fermentation10090490
Chicago/Turabian StyleKretzer, Leonardo G., Marilia M. Knychala, Lucca C. da Silva, Isadora C. C. da Fontoura, Maria José Leandro, César Fonseca, Kevin J. Verstrepen, and Boris U. Stambuk. 2024. "Screening and Engineering Yeast Transporters to Improve Cellobiose Fermentation by Recombinant Saccharomyces cerevisiae" Fermentation 10, no. 9: 490. https://doi.org/10.3390/fermentation10090490
APA StyleKretzer, L. G., Knychala, M. M., da Silva, L. C., da Fontoura, I. C. C., Leandro, M. J., Fonseca, C., Verstrepen, K. J., & Stambuk, B. U. (2024). Screening and Engineering Yeast Transporters to Improve Cellobiose Fermentation by Recombinant Saccharomyces cerevisiae. Fermentation, 10(9), 490. https://doi.org/10.3390/fermentation10090490









