Bioinformatics Analysis of Diadenylate Cyclase Regulation on Cyclic Diadenosine Monophosphate Biosynthesis in Exopolysaccharide Production by Leuconostoc mesenteroides DRP105
Abstract
1. Introduction
2. Materials and Methods
2.1. Dac Sequence Analysis
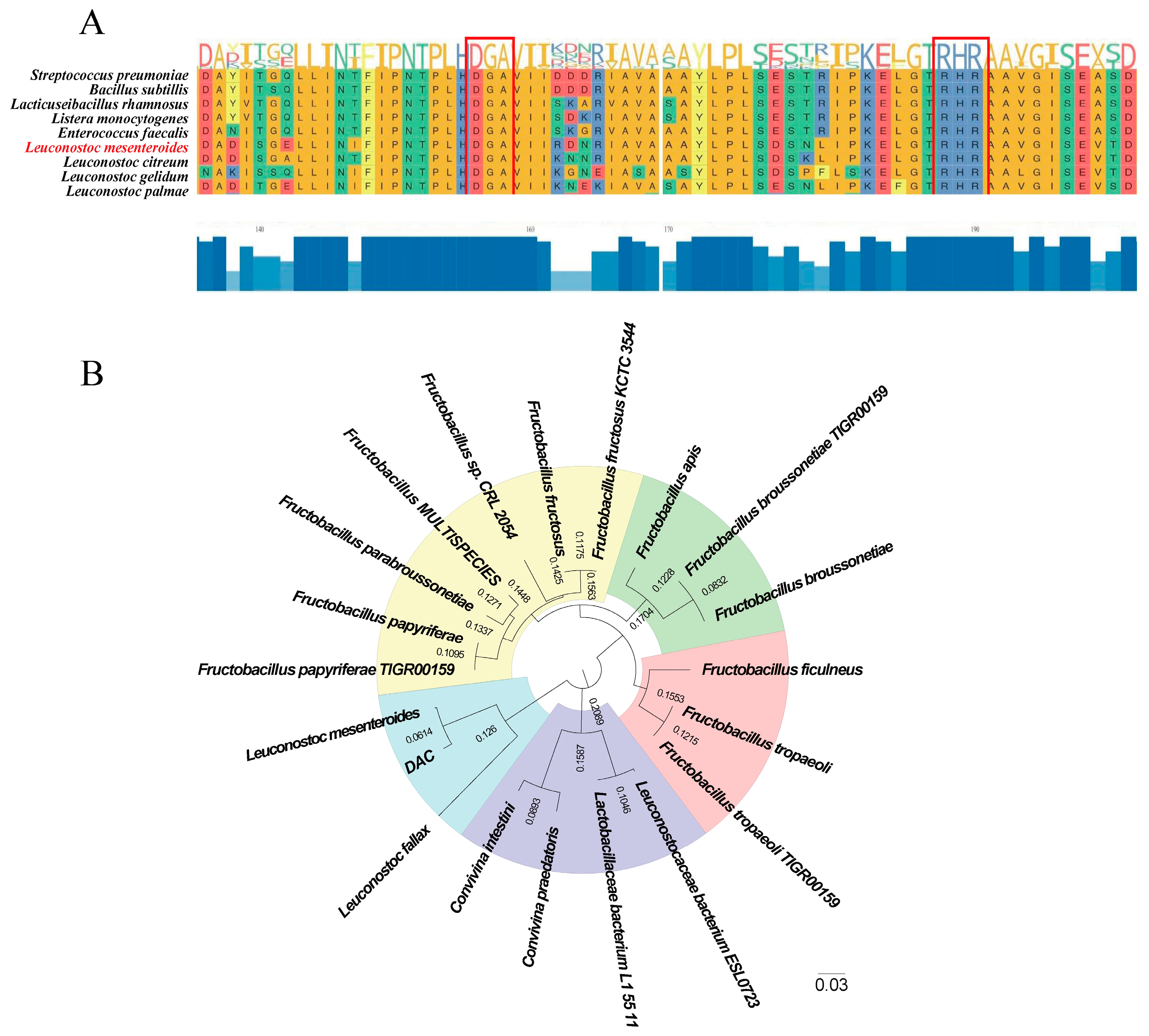
2.2. DAC Protein Evolutionary Tree Analysis
2.3. DAC Secondary Structure and Tertiary Structure Prediction
2.4. Evaluation of DAC Protein Conformation
2.5. DAC-Targeted Mutagenesis and ATP Docking
2.6. Prediction of Interaction Between DAC and Glucansucrase Proteins
2.7. Molecular Dynamics (MD) Simulation of DAC and ATP
2.8. Determination of EPS Content
2.9. Determination of Glucansucrase Activity
2.10. Detection of c-di-AMP Content and Related Enzyme Activities
2.11. Determination of Relative Gene Expression
2.12. Data Statistical Methods
3. Results
3.1. Dac Sequence Analysis and Multiple Sequence Alignment
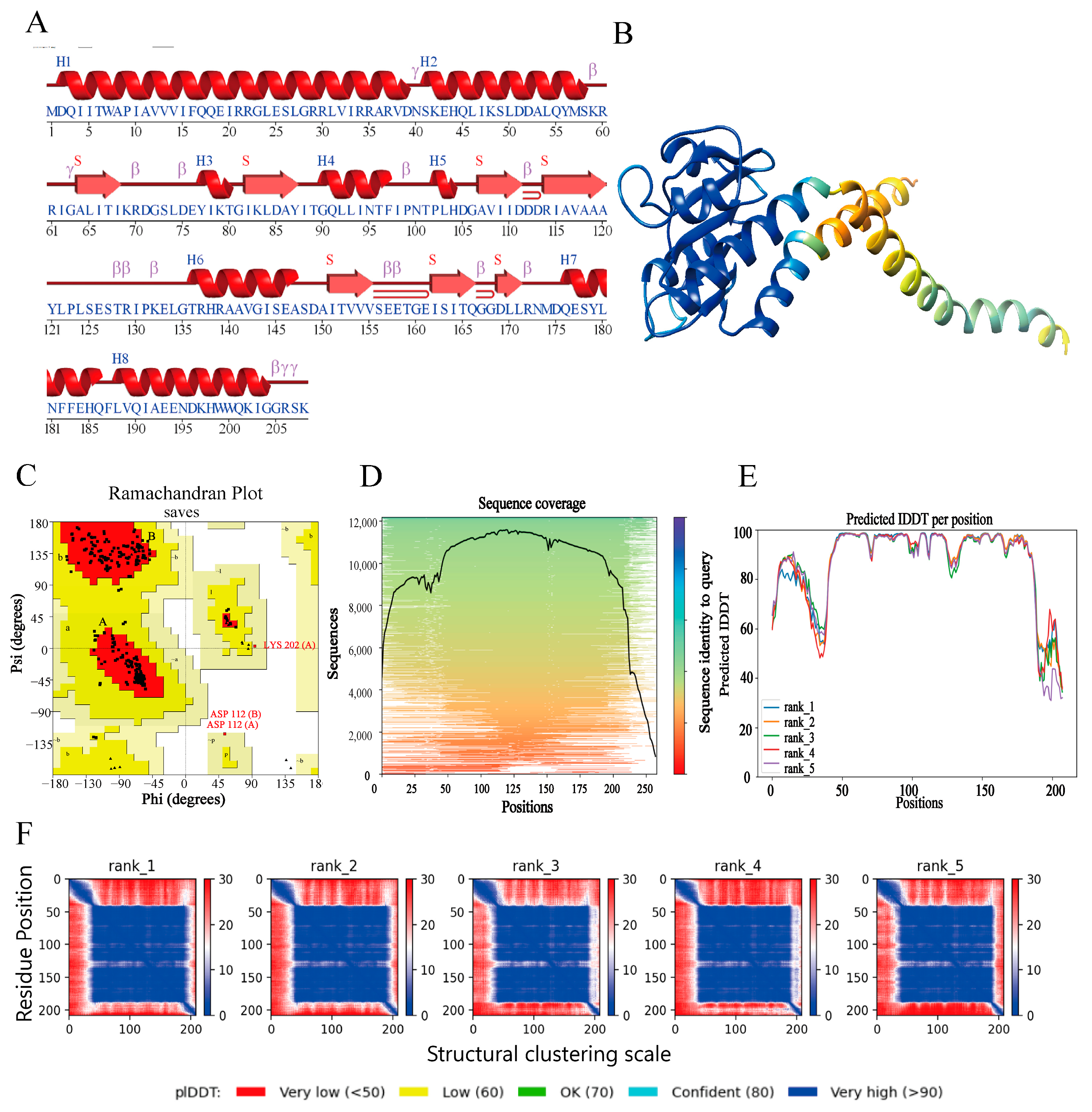
3.2. Prediction of DAC Secondary and Tertiary Structure
3.3. Saves Server for DAC Protein Evaluation
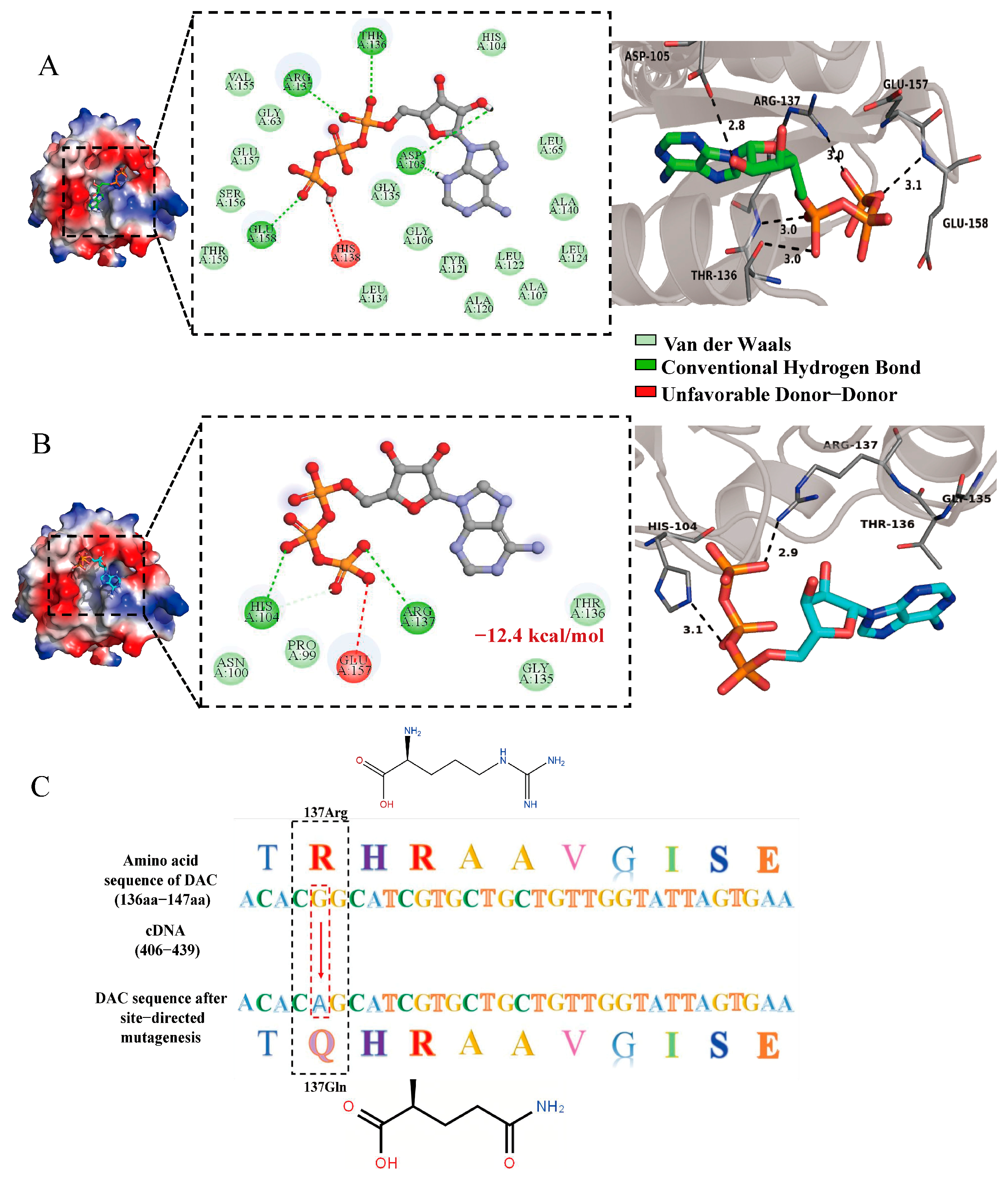
3.4. Docking Analysis of DAC and Two ATP Molecules
3.5. Targeted Mutagenesis of DAC and Docking with ATP
3.6. Analysis of Interaction Between DAC and Glucansucrase
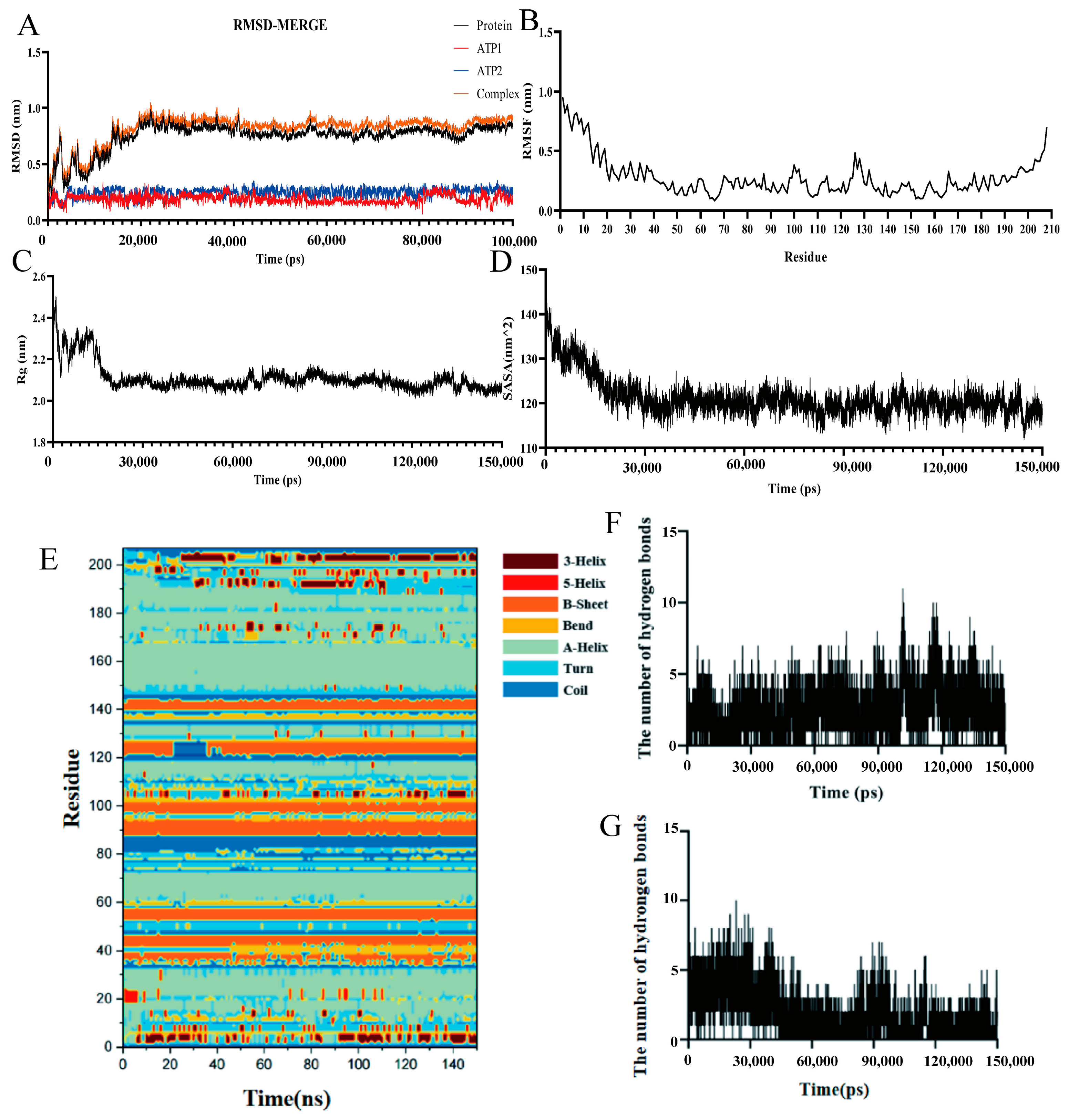
3.7. MD Simulation of DAC and ATP
3.7.1. RMSD Analysis of DAC and ATP
3.7.2. RMSF Analysis of DAC
3.7.3. Rg, SASA and DSSP Simulation, and Hydrogen Bond Analysis Between DAC and ATP
3.7.4. Free Energy Landscape Analysis of DAC and ATP Free Energy
3.7.5. Free Energy Analysis of Binding ATP to DAC
3.8. Fermentation Process Genes and Metabolites and Correlation Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, R.; Pei, F.; Kang, J.; Zhang, W.; Wang, S.; Ping, W.; Ling, H.; Ge, J. Analysis of the structure and properties of dextran produced by Weissella confusa. Int. J. Biol. Macromol. 2022, 204, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Devi, P.V.; Islam, J.; Narzary, P.; Sharma, D.; Sultana, F. Bioactive compounds, nutraceutical values and its application in food product development of oyster mushroom. J. Future Foods 2024, 4, 335–342. [Google Scholar] [CrossRef]
- Ning, E.; Sun, C.; Wang, X.; Chen, L.; Li, F.; Zhang, L.; Wang, L.; Ma, Y.; Zhu, J.; Li, X.; et al. Artemisia argyi polysaccharide alleviates intestinal inflammation and intestinal flora dysbiosis in lipopolysaccharide-treated mice. Food Med. Homol. 2024, 1, 9420008. [Google Scholar] [CrossRef]
- Gao, X.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Modulation of gut health using probiotics: The role of probiotic effector molecules. J. Future Foods 2022, 2, 1–12. [Google Scholar] [CrossRef]
- Suo, K.; Li, X.; Liu, X.; Zhu, J.; Shi, Y.; Yi, J.; Lu, J. From environment to environmental adaptation: Environmental perspectives on the study of food and medicine homology. Food Med. Homol. 2025, 3, 9420082. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, K.; Luo, W.; Chen, A.; Liang, Z.; Ji, P.; Wang, Y.; Wang, X.; Homology, M. Anti-virulence potential of carvone against Serratia marcescens. Food Med. Homol. 2024, 1, 9420001. [Google Scholar] [CrossRef]
- Molina, M.; Cioci, G.; Moulis, C.; Severac, E.; Remaud-Simeon, M. Bacterial α-Glucan and branching sucrases from GH70 family: Discovery, structure-function relationship studies and engineering. Microorganisms 2021, 9, 1607. [Google Scholar] [CrossRef] [PubMed]
- Arskold, E.; Svensson, M.; Grage, H.; Roos, S.; Radstrom, P.; van Niel, E.W.J. Environmental influences on exopolysaccharide formation in Lactobacillus reuteri ATCC 55730. Int. J. Food Microbiol. 2007, 116, 159–167. [Google Scholar] [CrossRef]
- Lv, L.; Wei, Z.; Li, W.; Chen, J.; Tian, Y.; Gao, W.; Wang, P.; Sun, L.; Ren, Z.; Zhang, G.; et al. Regulation of extracellular polymers based on quorum sensing in wastewater biological treatment from mechanisms to applications: A critical review. Water Res. 2024, 250, 121057. [Google Scholar] [CrossRef]
- Yan, Z.; Meng, H.; Yang, X.; Zhu, Y.; Li, X.; Xu, J.; Sheng, G. Insights into the interactions between triclosan (TCS) and extracellular polymeric substance (EPS) of activated sludge. J. Environ. Manag. 2019, 232, 219–225. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Xu, X.; Zhao, C.; Yang, F.; Wang, D. Potential coupling effects of ammonia-oxidizing and anaerobic ammonium-oxidizing bacteria on completely autotrophic nitrogen removal over nitrite biofilm formation induced by the second messenger cyclic diguanylate. Appl. Microbiol. Biotechnol. 2017, 101, 3821–3828. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Blanco-Toral, C.; Larrouy-Maumus, G. The role of cyclic nucleotides in bacterial antimicrobial resistance and tolerance. Trends Microbiol. 2024, 33, 164–183. [Google Scholar] [CrossRef]
- Agostoni, M.; Logan-Jackson, A.R.; Heinz, E.R.; Severin, G.B.; Bruger, E.L.; Waters, C.M.; Montgomery, B.L. Homeostasis of second messenger cyclic-di-AMP is critical for cyanobacterial fitness and acclimation to abiotic stress. Front. Microbiol. 2018, 9, 1121. [Google Scholar] [CrossRef]
- Tang, Q.; Luo, Y.; Zheng, C.; Yin, K.; Ali, M.K.; Li, X.; He, J. Functional Analysis of a c-di-AMP-specific Phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 2015, 11, 813–824. [Google Scholar] [CrossRef]
- Commichau, F.M.; Heidemann, J.L.; Ficner, R.; Stuelke, J. Making and breaking of an essential poison: The cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in Bacteria. J. Bacteriol. 2019, 201, 1110–1128. [Google Scholar] [CrossRef]
- Galperin, M.Y. All DACs in a row: Domain architectures of bacterial and archaeal diadenylate cyclases. J. Bacteriol. 2023, 205, e00023-23. [Google Scholar] [CrossRef]
- Zhu, Y.; Pham, T.H.; Nhiep, T.H.; Vu, N.M.; Marcellin, E.; Chakrabortti, A.; Wang, Y.; Waanders, J.; Lo, R.; Huston, W.M.; et al. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol. Microbiol. 2016, 99, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, C.; Meißner, J.; Warneke, R.; Stülke, J. The many roles of cyclic di-AMP to control the physiology of Bacillus subtilis. MicroLife 2023, 4, uqad043. [Google Scholar] [CrossRef]
- Wright, M.J.; Bai, G. Bacterial second messenger cyclic di-AMP in Streptococci. Mol. Microbiol. 2023, 120, 791–804. [Google Scholar] [CrossRef]
- Schwedt, I.; Wang, M.; Gibhardt, J.; Commichau, F.M. Cyclic di-AMP, a multifaceted regulator of central metabolism and osmolyte homeostasis in Listeria monocytogenes. MicroLife 2023, 4, uqad005. [Google Scholar] [CrossRef]
- Du, B.; Sun, J.H. Diadenylate cyclase evaluation of ssDacA (SSU98_1483) in Streptococcus suis serotype 2. Genet. Mol. Res. 2015, 14, 6917–6924. [Google Scholar] [CrossRef]
- Bai, Y.; Yang, J.; Zhou, X.; Ding, X.; Eisele, L.E.; Bai, G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS ONE 2012, 7, e35206. [Google Scholar] [CrossRef]
- Xiong, Z.; Fan, Y.; Song, X.; Liu, X.; Xia, Y.; Ai, L. The second messenger c-di-AMP mediates bacterial exopolysaccharide biosynthesis: A review. Mol. Biol. Rep. 2020, 47, 9149–9157. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Zhou, Z.; Han, Y. Functional identification of the dextransucrase gene of Leuconostoc mesenteroides DRP105. Int. J. Mol. Sci. 2020, 21, 6596. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Lanave, C.; Attimonelli, M.; De Robertis, M.; Licciulli, F.; Liuni, S.; Sbisa, E.; Saccone, C. Update of AMmtDB: A database of multi-aligned metazoa mitochondrial DNA sequences. Nucleic Acids Res. 1999, 27, 134–137. [Google Scholar] [CrossRef][Green Version]
- Du, R.; Yu, L.; Yu, N.; Ping, W.; Song, G.; Ge, J. Characterization of exopolysaccharide produced by Levilactobacillus brevis HDE-9 and evaluation of its potential use in dairy products. Int. J. Biol. Macromol. 2022, 217, 303–311. [Google Scholar] [CrossRef]
- Kolb, A.W.; Adams, M.; Cabot, E.L.; Craven, M.; Brandt, C.R. Multiplex sequencing of seven ocular herpes simplex virus type-1 genomes: Phylogeny, sequence variability, and SNP distribution. Investig. Ophthalmol. Vis. Sci. 2011, 52, 9061–9073. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhao, J.; Chen, Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: A case of oligopeptide binding protein. Comput. Math. Methods Med. 2018, 2018, 3502514. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E.J.S. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softw. X 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Gorelov, S.; Titov, A.; Tolicheva, O.; Konevega, A.; Shvetsov, A. DSSP in GROMACS: Tool for defining secondary structures of proteins in trajectories. J. Chem. Inf. Model. 2024, 64, 3593–3598. [Google Scholar] [CrossRef]
- Bojovschi, A.; Liu, M.S.; Sadus, R.J. Conformational dynamics of ATP/Mg:ATP in motor proteins via data mining and molecular simulation. J. Chem. Phys. 2012, 137, 075101. [Google Scholar] [CrossRef]
- Pillsbury, M.; Orland, H.; Zee, A. Steepest descent calculation of RNA pseudoknots. Phys. Rev. E 2005, 72, 011911. [Google Scholar] [CrossRef]
- Yue, F.; Zhang, J.; Xu, J.; Niu, T.; Lu, X.; Liu, M. Effects of monosaccharide composition on quantitative analysis of total sugar content by phenol-sulfuric acid method. Front. Nutr. 2022, 9, 963318. [Google Scholar] [CrossRef]
- Wang, L.; Shan, T.; Xie, B.; Ling, C.; Shao, S.; Jin, P.; Zheng, Y. Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chem. 2019, 272, 530–538. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90. [Google Scholar] [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.J.N.l. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. Imeta 2023, 2, e85. [Google Scholar] [CrossRef] [PubMed]
- Zarrella, T.M.; Metzger, D.W.; Bai, G. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a trk family effector protein in Streptococcus pneumoniae. J. Bacteriol. 2018, 200, 1110–1128. [Google Scholar] [CrossRef]
- Oppenheimer-Shaanan, Y.; Wexselblatt, E.; Katzhendler, J.; Yavin, E.; Ben-Yehuda, S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. Embo Rep. 2011, 12, 594–601. [Google Scholar] [CrossRef]
- Kwon, Y.; Park, C.; Lee, J.; Park, D.H.; Jeong, S.; Yun, C.H.; Park, O.J.; Han, S.H. Regulation of bone cell differentiation and activation by microbe-associated molecular patterns. Int. J. Mol. Sci. 2021, 22, 5805. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, J.M.; Faguy, D.M. Thermotoga heats up lateral gene transfer. Curr. Biol. 1999, 9, R747–R751. [Google Scholar] [CrossRef] [PubMed]
- Bruley, A.; Mornon, J.-P.; Duprat, E.; Callebaut, I. Digging into the 3D structure predictions of AlphaFold2 with low confidence: Disorder and beyond. Biomolecules 2022, 12, 1467. [Google Scholar] [CrossRef]
- Hung, L.; Samudrala, R. PROTINFO: Secondary and tertiary protein structure prediction. Nucleic Acids Res. 2003, 31, 3296–3299. [Google Scholar] [CrossRef]
- Gogoi, C.R.; Rahman, A.; Saikia, B.; Baruah, A. Protein dihedral angle prediction: The state of the art. Chem. Select 2023, 8, e202203427. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Manjula, M.; Pampa, K.J.; Kumar, S.M.; Mukherjee, S.; Kunishima, N.; Rangappa, K.S.; Lokanath, N.K. Crystal structure of ATP-binding subunit of an ABC transporter from Geobacillus kaustophilus. Biochem. Biophys. Res. Commun. 2015, 459, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Domanski, T.L.; Halpert, J.R. Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr. Drug Metab. 2001, 2, 117–137. [Google Scholar] [CrossRef]
- Rosenberg, J.; Dickmanns, A.; Neumann, P.; Gunka, K.; Arens, J.; Kaever, V.; Stuelke, J.; Ficner, R.; Commichau, F.M. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J. Biol. Chem. 2015, 290, 6596–6606. [Google Scholar] [CrossRef]
- Chandel, T.I.; Zaman, M.; Khan, M.V.; Ali, M.; Rabbani, G.; Ishtikhar, M.; Khan, R.H. A mechanistic insight into protein-ligand interaction, folding, misfolding, aggregation and inhibition of protein aggregates: An overview. Int. J. Biol. Macromol. 2018, 106, 1115–1129. [Google Scholar] [CrossRef]
- Peng, X.; Michalek, S.; Wu, H. Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of Streptococcus mutans revisit. Environ. Microbiol. 2016, 18, 3612–3619. [Google Scholar] [CrossRef]
- Agarwal, V.; McShan, A.C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat. Chem. Biol. 2024, 20, 950–959. [Google Scholar] [CrossRef]
- Pijning, T.; te Poele, E.M.; de Leeuw, T.C.; Guskov, A.; Dijkhuizen, L. Crystal structure of 4,6-α-Glucanotransferase GtfC-ΔC from Thermophilic Geobacillus 12AMOR1: Starch transglycosylation in non-permuted GH70 enzymes. J. Agric. Food Chem. 2022, 70, 15283–15295. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, Y.; Bai, G.; Zhou, X.; Wu, H. Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 2016, 99, 945–959. [Google Scholar] [CrossRef]
- Holcomb, M.; Chang, Y.-T.; Goodsell, D.S.; Forli, S. Evaluation of AlphaFold2 structures as docking targets. Protein Sci. 2023, 32, e4530. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, L.; Li, E.; Dong, G. Molecular dynamics simulation study on the structures of fascin mutants. J. Mol. Recognit. 2023, 36, e2998. [Google Scholar] [CrossRef]
- Fuglebakk, E.; Echave, J.; Reuter, N. Measuring and comparing structural fluctuation patterns in large protein datasets. Bioinformatics 2012, 28, 2431–2440. [Google Scholar] [CrossRef]
- Dong, W.; Zhu, W.; Wu, Q.; Li, W.; Li, X. Improvement the thermostability and specific activity of acidic xylanase PjxA from Penicillium janthinellum via rigid flexible sites. Int. J. Biol. Macromol. 2024, 279, 135399. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Islam, R.; Parves, M.R.; Paul, A.S.; Uddin, N.; Rahman, M.S.; Al Mamun, A.; Hossain, M.N.; Ali, M.A.; Halim, M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 3213–3224. [Google Scholar] [CrossRef]
- Shibata, M.; Zielinski, T.J. Computer graphics presentations and analysis of hydrogen bonds from molecular dynamics simulation. J. Mol. Graph. 1992, 10, 88–95. [Google Scholar] [CrossRef]
- Duong, T.; Devi, A.P.; Tran, N.; Phan, H.; Huynh, N.; Sichaem, J.; Tran, H.; Alam, M.; Nguyen, T.; Nguyen, H.; et al. Synthesis, α-glucosidase inhibition, and molecular docking studies of novel N-substituted hydrazide derivatives of atranorin as antidiabetic agents. Bioorg. Med. Chem. Lett. 2020, 30, 127359. [Google Scholar] [CrossRef] [PubMed]
- Pietrucci, F.J.R.i.P. Strategies for the exploration of free energy landscapes: Unity in diversity and challenges ahead. Rev. Phys. 2017, 2, 32–45. [Google Scholar] [CrossRef]
- Tavernelli, I.; Cotesta, S.; Di Iorio, E.E. Protein dynamics, thermal stability, and free-energy landscapes: A molecular dynamics investigation. Biophys. J. 2003, 85, 2641–2649. [Google Scholar] [CrossRef] [PubMed]
- Alrouji, M.; DasGupta, D.; Ashraf, G.M.; Bilgrami, A.L.; Alhumaydhi, F.A.; Al Abdulmonem, W.; Shahwan, M.; Alsayari, A.; Atiya, A.; Shamsi, A. Inhibition of microtubule affinity regulating kinase 4 by an acetylcholinesterase inhibitor, Huperzine A: Computational and experimental approaches. Int. J. Biol. Macromol. 2023, 235, 123831. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, G.; Qi, X.; Zhou, B.; Yu, L.; Song, G.; Du, R. Exploration of exopolysaccharide from Leuconostoc mesenteroides HDE-8: Unveiling structure, bioactivity, and food industry applications. Polymers 2024, 16, 954. [Google Scholar] [CrossRef]
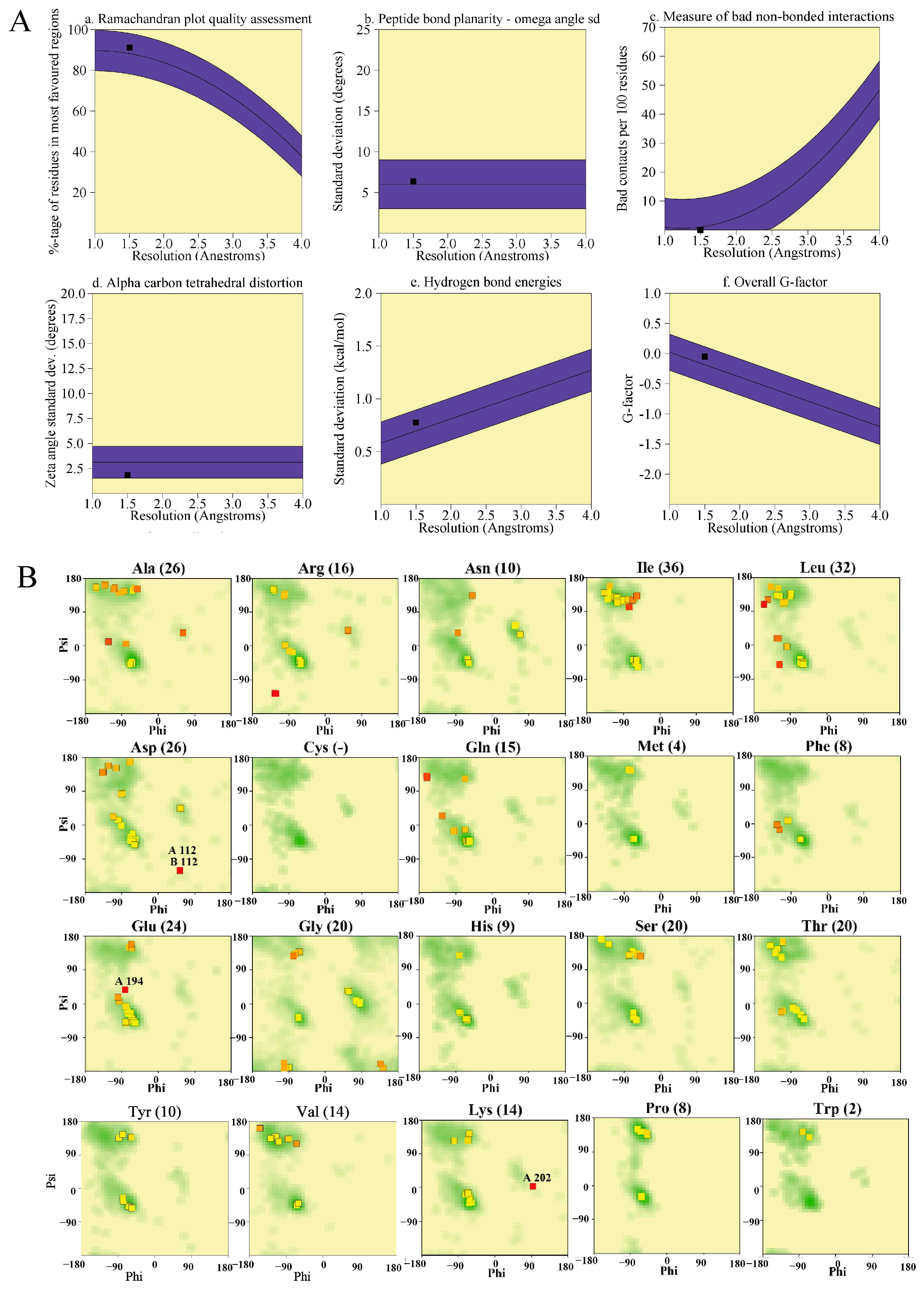




| Main Chain Parameter | Number of Data Points | Parameter Values | Typical Comparison Value | Wideband Data Peak Frequency Deviation | The Amount of Bandwidth That Deviates from the Average | Evaluation |
|---|---|---|---|---|---|---|
| a. Percentage of amino acid residues in A, B, and L | 286 | 91.3 | 88.2 | 10.0 | 0.3 | Qualified |
| b. Ω angle | 316 | 6.4 | 6.0 | 3.0 | 0.1 | Qualified |
| b. Error residue/100 residues | 0 | 0.0 | 1.0 | 10.0 | −0.1 | Qualified |
| c. Zeta angle | 298 | 1.8 | 3.1 | 1.6 | −0.8 | Qualified |
| d. Hydrogen bond energy | 198 | 0.8 | 0.7 | 0.2 | 0.4 | Qualified |
| e. Overall G-coefficient | 318 | −0.0 | −0.2 | 0.3 | 0.5 | Qualified |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Yu, L.; Li, T.; Wang, Z.; Du, R.; Ping, W. Bioinformatics Analysis of Diadenylate Cyclase Regulation on Cyclic Diadenosine Monophosphate Biosynthesis in Exopolysaccharide Production by Leuconostoc mesenteroides DRP105. Fermentation 2025, 11, 196. https://doi.org/10.3390/fermentation11040196
Yu W, Yu L, Li T, Wang Z, Du R, Ping W. Bioinformatics Analysis of Diadenylate Cyclase Regulation on Cyclic Diadenosine Monophosphate Biosynthesis in Exopolysaccharide Production by Leuconostoc mesenteroides DRP105. Fermentation. 2025; 11(4):196. https://doi.org/10.3390/fermentation11040196
Chicago/Turabian StyleYu, Wenna, Liansheng Yu, Tengxin Li, Ziwen Wang, Renpeng Du, and Wenxiang Ping. 2025. "Bioinformatics Analysis of Diadenylate Cyclase Regulation on Cyclic Diadenosine Monophosphate Biosynthesis in Exopolysaccharide Production by Leuconostoc mesenteroides DRP105" Fermentation 11, no. 4: 196. https://doi.org/10.3390/fermentation11040196
APA StyleYu, W., Yu, L., Li, T., Wang, Z., Du, R., & Ping, W. (2025). Bioinformatics Analysis of Diadenylate Cyclase Regulation on Cyclic Diadenosine Monophosphate Biosynthesis in Exopolysaccharide Production by Leuconostoc mesenteroides DRP105. Fermentation, 11(4), 196. https://doi.org/10.3390/fermentation11040196






