High-Level Extracellular Expression of Collagenase ColH in Bacillus subtilis for Adipose-Derived Cells Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Construction and Transformation of Plasmids for Collagenase Expression
2.3. Gelatin Plate Screening
2.4. Quantification of Collagenase Protein and Enzyme Activity Assay
2.5. SDS-PAGE
2.6. Signal Peptide Engineering for Enzyme Expression
2.7. Promoter Engineering for Enzyme Expression
2.8. Purification
2.9. Optimization of Shaker Flask Culture Media and Fermenter Cultivation
2.10. Statistical Analysis
2.11. Adipose-Derived Cell Isolation
3. Results
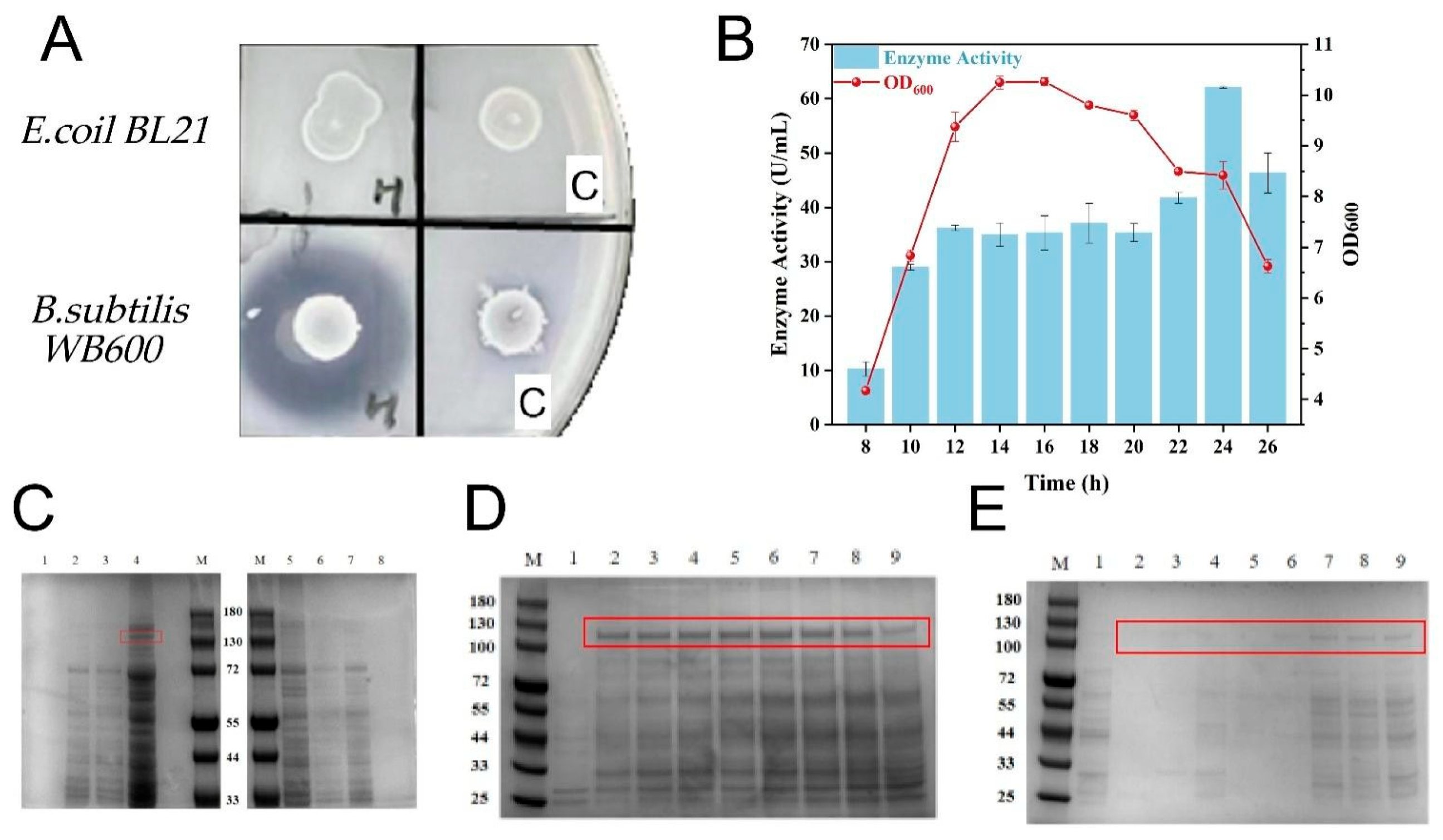
3.1. Expression of Recombinant Collagenase
3.2. Screening of Signal Peptide for Secretory Expression of Collagenase
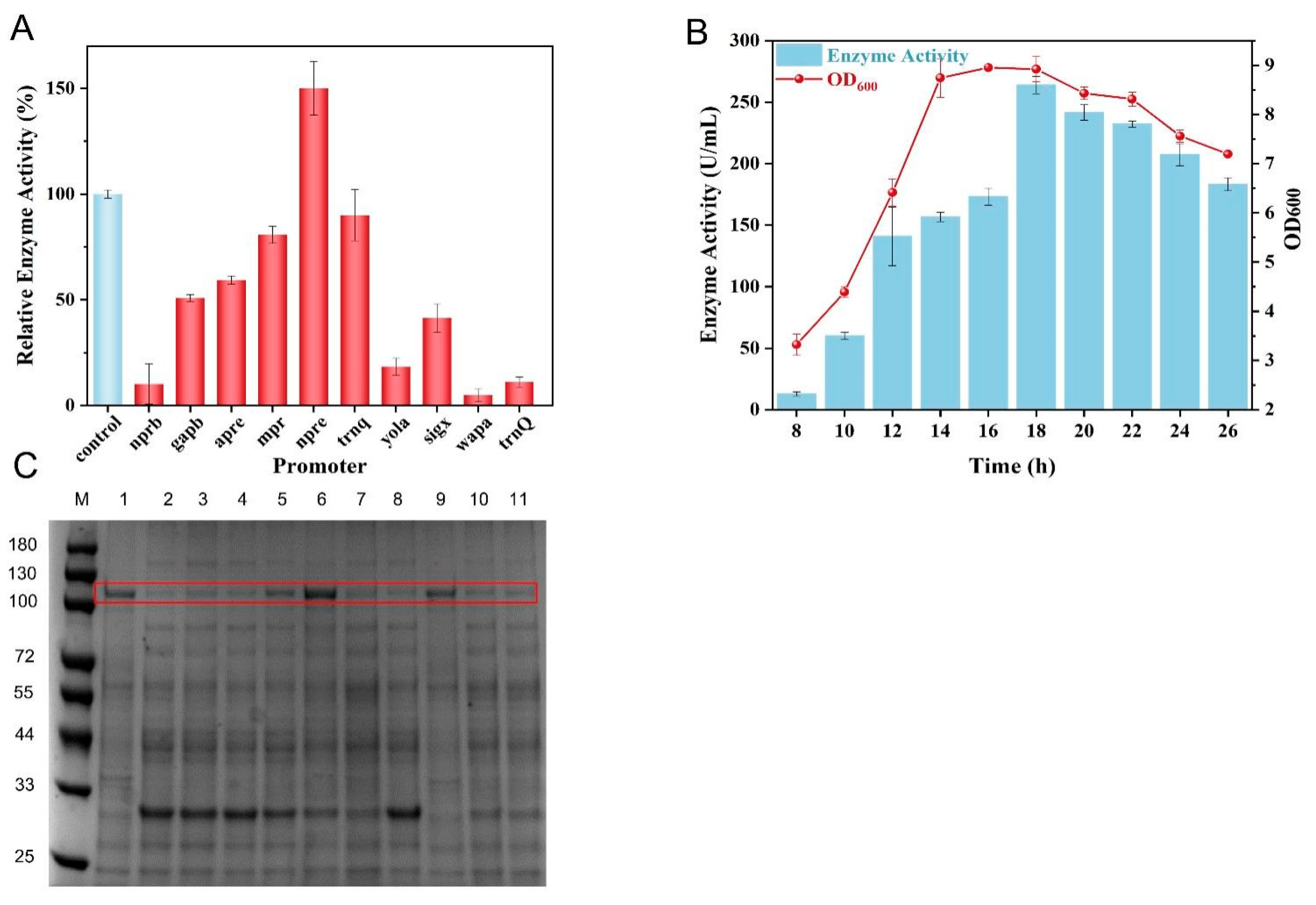
3.3. Optimization of Promoter to Facilitate Collagenase Expression
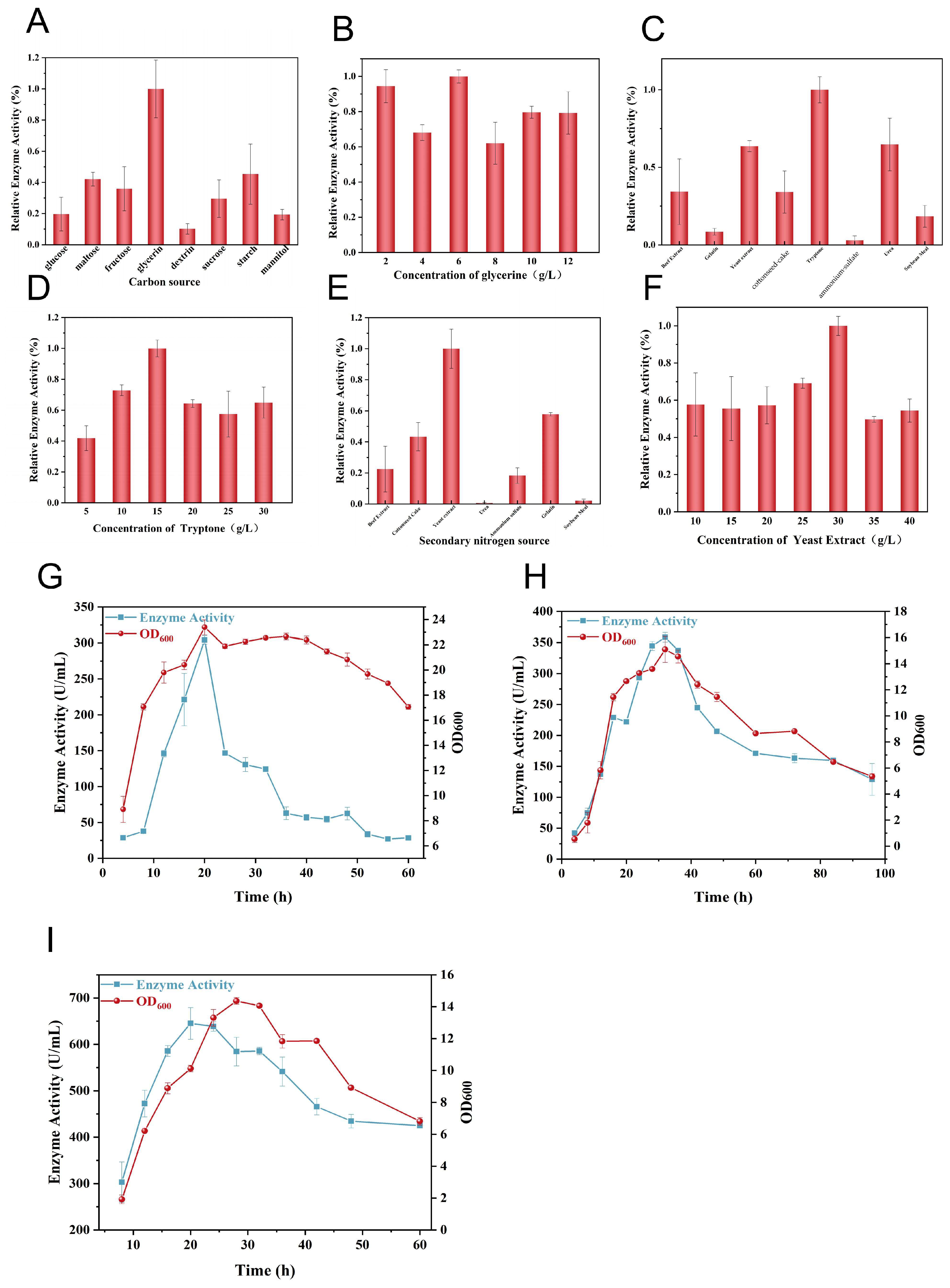
3.4. Fermentation Strategy for Collagenase Production
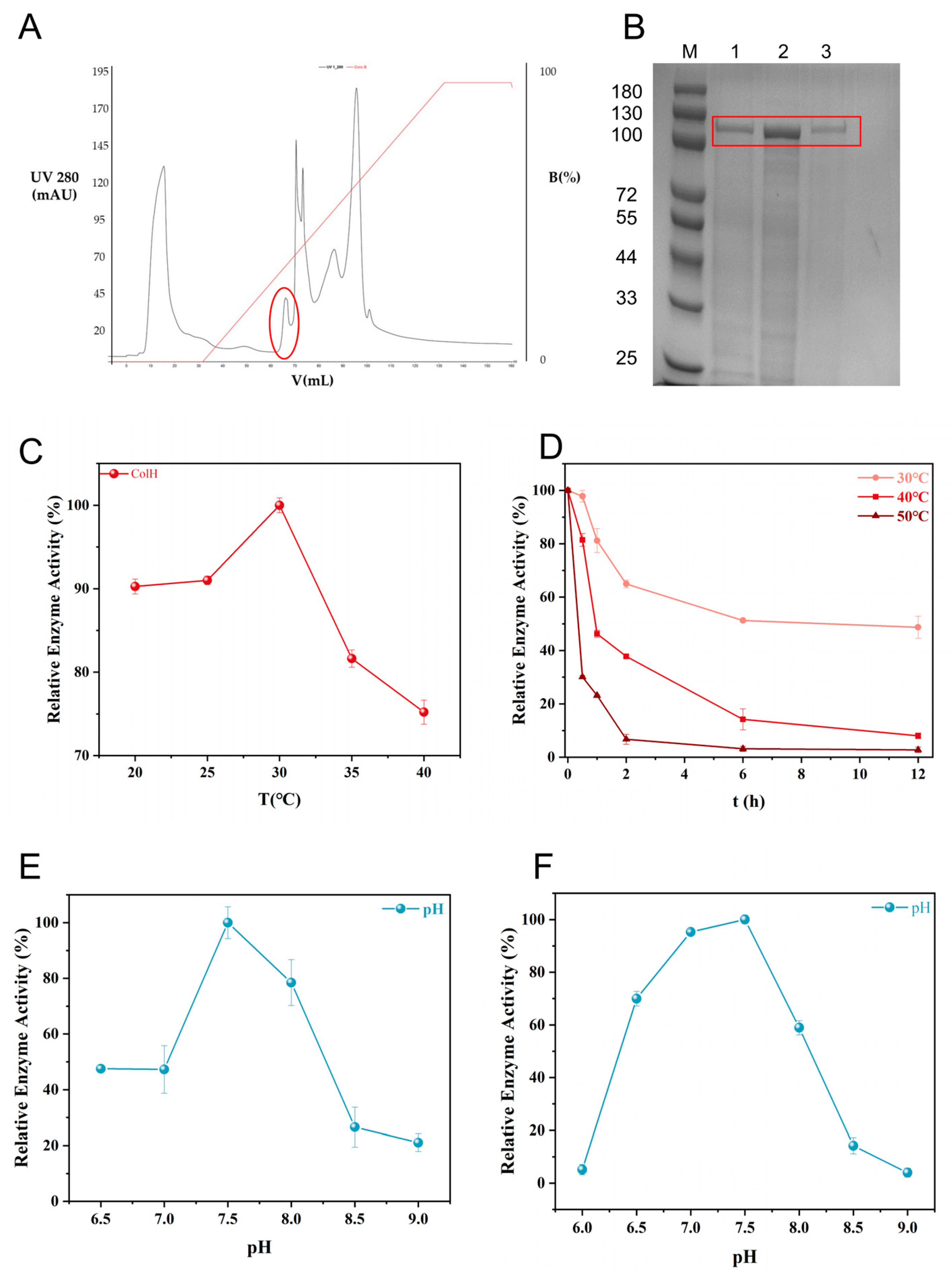
3.5. Purification and Characterization of Collagenase ColH
3.6. Adipose Tissue Isolation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Signal Peptide | Sequence |
|---|---|
| SPhrui | atgagaaaatggtattttattcttttggcgggtgttttaacgtctgtcatcctcgctttcgtttatgataagacaaaagcg |
| SPamye | atgtttgcaaaacgattcaaaacctctttactgccgttattcgctggatttttattgctgtttcatttggttctggcagga |
| SPbgls | atgccttatctgaaacgagtgttgctgcttcttgtcactggattgtttatgagtttgtttgcagtcactgctactgcctcagct |
| SPpelc | ttgaaaaaaatcgtgtctatcctatttatgttcggtttggttatgggtttcagccagtttcagccatcaaccgtttttgca |
| SPvpr | ttgaaaaaggggatcattcgctttctgcttgtaagtttcgtcttattttttgcgttatccacaggcattacgggcgttcaggca |
| SPmdh | atgggaaatactcgtaaaaaagtttctgttatcggagcaggttttaccggagctacaactgcatttttaatcgctcaaaaagagctggcagacgtt |
| SPnpre | gtgggtttaggtaagaaattgtctgttgctgtcgctgcttcgtttatgagtttatcaatcagcctgccaggtgttcaggct |
| SPdacb | atgcgcattttcaaaaaagcagtattcgtgatcatgatttcttttcttattgcaaccgtaaatgtgaatacagcacatgct |
| SPphob | ttgaaaaaattcccgaagaaattactgcctatcgcggttttatcatcaattgcgttcagcagcttagccagcggcagtgtgcctgaagccagcgcc |
| SPyhfm | atgaaaaaaatagtggcagccatcgtggtaatcggtcttgtgtttatcgcatttttttatctttacagccgatcaggcgatgtgtatcaatcggtagacgcg |
| SPesta | atgaaatttgtaaaaagaaggatcattgcacttgtaacaattttgatgctgtctgttacatcgctgtttgcgttgcagccgtcagcaaaagcc |
| Promoter | Sequence |
|---|---|
| Pgapb | gttggacaccccttatcaattatgccatcctaatttgttactatctaaaattagtataacacattaaaacaaattcgccagtacatattgataaaaatcaacgattataacgctaatttatgatgcatcttatgttttcttgcacagaatatcaggcgtaagaaccaaaaagcagccagcgctccgcactgactgccctcatgctttatgattcctgttttaacttatcattgagcacactttcttctagcacagaaaatgtatccccgtacactctgttaatatgattcgctccccaagcacacagcatatctaaaatcccttct |
| Pnprb | catatggagcaggggttattatttatgtcaataaaaaattagtagagtagaaaaagaagggcagagcgaaatatctctgcccttcttttttgggaaaataagaacgaagcaccacatacaagtttttgtatttttgataggatgaagaaaaatggtagattccaaaataggaaggatgtggtgtt |
| Pmpr | ttagcggattacactgttgaaggattggaaaacgcactggcagtcccaaagactcaagtgcgcgtctttggaaagccgataacaaaagccggacgtcgtatggcagttgcgctttctgctgctgattcagttgaaacggcaagagagaatgcaaagaaagcgttggaccagctaattttaaaatagagtttgaacaggtcttgtcatgggacaaggcctgtttttttctttctccgtaaaagttttatcataagaatcagaaacctgattataatgtaaaagtcttccatcgatacgggtggttgacactaaaggagggagatgacaaa |
| Papre | cgataatatccattgttctcacggaagcacacgcaggtcatttgaacgaattttttcgacaggaatttgccgggactcaggagcatttaacctaaaaaagcatgacatttcagcataatgaacatttactcatgtctattttcgttcttttctgtatgaaaatagttatttcgagtctctacggaaatagcgagagatgatatacctaaatagagataaaatcatctcaaaaaaatgggtctactaaaatattattccatctattacaataaattcacagaatagtcttttaagtaagtctactctgaatttttttaaaaggagagggtaaaga |
| Pyola | ttcctaaatcctccttggtacaagtttacatgttaaatatcgtcatttgaagggaattgtttaatatttgaaataaaaaaagaacctgcattaagcaagttcttttcatattgtcaatttattgtgaatttttaacgacaaggacatttatgtatagtataatatttcctgtacataaagtttgctcactcaagggagtcttgctcatcccctatgaaaggggtgggaaaatgactgtttacgaatcattaatgataatgatcaattttggcggattgatattaaataccgtcttgttgatcttcaatataatgatgattgtaacgtcaagccaaaagaaaaaatagaccttcccttgagtttggacacctgaagggttaggcctacgcagatttgacaacgagcaagcc |
| Psigx | caaagactccgggtctggcataccggaagaagatctgccatttatctttgagcggttttataaggcagataaagcgcggacaaggggcagagcaggaaccgggttagggctggctatcgttaaaaatatcgtggaagcccacaacggatcaattactgtgcacagccgaatagataaaggaacaacattttctttttatattccgacaaaacggtaaaatcgagtctgaatttgccgaagaatcttgttccataagaaacacccgctgactgagcgggtgtttttttaatagccaacattaataaaatttaaggatatgttaatataaattcccttccaaattccagttactcgtaatatagttgtaatgtaacttttcaagctattcatacgacaaaaaagtgaacggaggggtttcaa |
| Pnpre | attgaatcagcagggtgctttgtctgcttaatataaaataacgttcgaaatgcaatacataatgactgaataactccaacacgaacaacaatcctttacttcttattaaggcctcattcggttagacagcggacttttcaaaaagtttcaagatgaaacaaaaatatctcatcttccccttgatatgtaaaaaacataactcttgaatgaaccaccacatgacacttgactcatcttgatattattcaacaaaaacaaacacaggacaatactatcaattttgtctagttatgttagtttttgttgagtattccagaatgctagtttaatataacaatataaagttttcagtattttcaaaaagggggatttatt |
| Pp43 | agcattattgagtggatgattatattccttttgatagggtggtatgttttcgcttgaacttttaaatcagccattgaacatacggttgattttaataactgacaaacatcaccctcttgctaaagcggccaaggacgctgccgccggggctgtttgcgtttttgccgtgatttcgtgtatcattggtttacttatattttttgccaaagctgtaatggctgaaaattcttacatttattttacattttttagaaatgaggcgtgaaaaaaagcgcgcgattatgtaaaataaaaagtatagcgggtacca |
| Pwapa | atttcaatcattgtatttctcgaccccgctgtcgcgatcgtgctcgataccgtcttcacaggcttccgccctgacctctatcaaacgcttggcatcgtaatgatctttgcgggcatggccttgacgcttgtcaggaggcaggggaaggcgaatgtgacagctgagggtacggatattgaacaaatacaataaaaaatgtaaaaaggcctatgcggcctttttttgttttaggtcaattgactctcgctaatccttaaaataagataaattttctagaaaaatattgtaatgatatttcagtctagttaagattattgagtaaatattacttttattacaaaaggagagaggaa |
| Ptrnq | gagaataaatgtcatacgctctttccccgcggttcgtttgttcaatgactgtaggtattaaattcataatgctcctccttcaccttttaggtaagtatgtagttacatgatacatttttggtcaataaaggtcaaacaaaaagctggcctgatatgcaaagtcgtctctttttcccattttccccaaaaatacaggggttcaaaccatcgtatgtcagattgccaattaagatgctttgtctatttaaaaaacggcctctcgaaatagagggttgttatttgaaaggaattatcgtataattagttgtgctgacgttctcataacgcagtctatat |
References
- Frederick, R.E.; Bearden, R.; Jovanovic, A.; Jacobson, N.; Sood, R.; Dhall, S. Clostridium Collagenase Impact on Zone of Stasis Stabilization and Transition to Healthy Tissue in Burns. Int. J. Mol. Sci. 2021, 22, 8643. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Sims, H.; Trueheart, I.; Simpson, K.; Wang, K.C.; Patzkowsky, K.; Wegman, T.; Soma, J.-M.; Dixon, R.; Jayes, F.; et al. A Phase I Clinical Trial to Assess Safety and Tolerability of Injectable Collagenase in Women with Symptomatic Uterine Fibroids. Reprod. Sci. 2021, 28, 2699–2709. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.; Park, J.k.; Shim, J.; Ko, N.; Kim, H.J.; Lee, Y.; Kim, J.H.; Alexander, M.; Lakey, J.R.T.; Kim, H.; et al. Comparison of islet isolation result and clinical applicability according to GMP-grade collagenase enzyme blend in adult porcine islet isolation and culture. Xenotransplantation 2021, 28, 12703. [Google Scholar] [CrossRef]
- Narwani, K.; Stark, J.; Cortez, D.; Yang, I.; Au, C.; Diaz, A.; Guerra, C.; Niihara, Y.; Bardag-Gorce, F. cG-CAOMECS—Clinical-grade cultured autologous oral mucosal epithelial cell sheet. Cell Tissue Res. 2021, 386, 47–57. [Google Scholar] [CrossRef]
- Jin, Y.-Q.; Liu, W.; Hong, T.-H.; Cao, Y. Efficient Schwann cell purification by differential cell detachment using multiplex collagenase treatment. J. Neurosci. Methods 2008, 170, 140–148. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar] [CrossRef]
- Pal, G.K.; Pv, S. Microbial collagenases: Challenges and prospects in production and potential applications in food and nutrition. RSC Adv. 2016, 6, 33763–33780. [Google Scholar] [CrossRef]
- Xiao, H.; Liu, X.; Feng, Y.; Zheng, L.; Zhao, M.; Huang, M. Secretion of collagenases by Saccharomyces cerevisiae for collagen degradation. Biotechnol. Biofuels Bioprod. 2022, 15, 89. [Google Scholar] [CrossRef]
- Han, X.; Xiufang, L.; Lin, Z.; Mouming, Z.; Mingtao, H. Recent advances in collagenase from Clostridium histolyticum. Food Sci. 2022, 43, 316–325. [Google Scholar]
- Serwanja, J.; Wieland, A.C.; Haubenhofer, A.; Brandstetter, H.; Schoenauer, E. A conserved strategy to attack collagen: The activator domain in bacterial collagenases unwinds triple-helical collagen. Proc. Natl. Acad. Sci. USA 2024, 121, e2321002121. [Google Scholar] [CrossRef]
- Jung, C.M.; Matsushita, O.; Katayama, S.; Minami, J.; Ohhira, L.; Okabe, A. Expression of the colH Gene Encoding Clostridium histolyticum Collagenase in Bacillus subtilis and Its Application to Enzyme Purification. Microbiol. Immunol. 2013, 40, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, O.; Jung, C.M.; Minami, J.; Katayama, S.; Nishi, N.; Okabe, A. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J. Biol. Chem. 1998, 273, 3643–3648. [Google Scholar] [CrossRef] [PubMed]
- Tamai, E.; Miyata, S.; Tanaka, H.; Nariya, H.; Suzuki, M.; Matsushita, O.; Hatano, N.; Okabe, A. High-level expression of his-tagged clostridial collagenase in Clostridium perfringens. Appl. Microbiol. Biotechnol. 2008, 80, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, X.; Li, S. Secretory Expression of Clostridium histolyticum Collagenase H in Escherichia coli. J. East China Univ. Sci. Technoloy Nat. Sci. Ed. 2022, 48, 797–805. [Google Scholar]
- Zhu, Y.; Wang, L.; Zheng, K.; Liu, P.; Li, W.; Lin, J.; Liu, W.; Shan, S.; Sun, L.; Zhang, H. Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600. Fermentation 2022, 8, 449. [Google Scholar] [CrossRef]
- Hoppe, I.J.; Brandstetter, H.; Schonauer, E. Biochemical characterisation of a collagenase from Bacillus cereus strain Q1. Sci. Rep. 2021, 11, 4187. [Google Scholar] [CrossRef]
- Subramanian, S.R.; Singam, E.R.A.; Berinski, M.; Subramanian, V.; Wade, R.C. Identification of an Electrostatic Ruler Motif for Sequence-Specific Binding of Collagenase to Collagen. J. Phys. Chem. B 2016, 120, 8580–8589. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Y.; Yan, X.; Li, Z. A high-performance protein preparation approach in a single column-free step. Trends Biotechnol. 2025, 43, 476–487. [Google Scholar] [CrossRef]
- Boe, C.; Blazar, P.; Iannuzzi, N. Dupuytren Contractures: An Update of Recent Literature. J. Hand Surg. 2021, 46, 896–907. [Google Scholar] [CrossRef]
- Holzer, L.A.; Holzer, G. Collagenase Clostridum histolyticum in the Management of Dupuytren’s Contracture. Handchir. Mikrochir. Plast. Chir. 2011, 43, 269–274. [Google Scholar]
- Leafblad, N.D.; Wagner, E.; Wanderman, N.R.; Anderson, G.R.; Visscher, S.L.; Kremers, H.M.; Larson, D.R.; Rizzo, M. Outcomes and Direct Costs of Needle Aponeurotomy, Collagenase Injection, and Fasciectomy in the Treatment of Dupuytren Contracture. J. Hand Surg. 2019, 44, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xie, Y.; Wen, B.; Ye, M.; Liu, Y.; Imam, K.M.S.U.; Cai, H.; Zhang, C.; Wang, F.; Xin, F. Porcine bone collagen peptides promote osteoblast proliferation and differentiation by activating the PI3K/Akt signaling pathway. J. Funct. Foods 2020, 64, 103697. [Google Scholar] [CrossRef]
- Mei, F.; Liu, J.; Wu, J.; Duan, Z.; Chen, M.; Meng, K.; Chen, S.; Shen, X.; Xia, G.; Zhao, M. Collagen Peptides Isolated from Salmo salar and Tilapia nilotica Skin Accelerate Wound Healing by Altering Cutaneous Microbiome Colonization via Upregulated NOD2 and BD14. J. Agric. Food Chem. 2020, 68, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Swanson, J.W.; Watt, A.J.; Vedder, N.B. Collagenase for Recurrent Dupuytren Contracture with Skin Grafts Reply. J. Hand Surg. 2013, 38, 1264–1265. [Google Scholar] [CrossRef]
- Warwick, D.; Arandes-Renu, J.M.; Pajardi, G.; Witthaut, J.; Hurst, L.C. Collagenase Clostridium histolyticum: Emerging practice patterns and treatment advances. J. Plast. Surg. Hand Surg. 2016, 50, 251–261. [Google Scholar] [CrossRef]
- Xiong, L.; Cui, M.; Zhou, Z.; Wu, M.; Wang, Q.; Song, H.; Ding, L. Primary culture of chondrocytes after collagenase IA or II treatment of articular cartilage from elderly patients undergoing arthroplasty. Asian Biomed. 2021, 15, 91–99. [Google Scholar] [CrossRef]
- Sheets, A.R.; Demidova-Rice, T.N.; Shi, L.; Ronfard, V.; Grover, K.V.; Herman, I.M. Identification and Characterization of Novel Matrix-Derived Bioactive Peptides: A Role for Collagenase from Santyl® Ointment in Post-Debridement Wound Healing? PLoS ONE 2016, 11, e0159598. [Google Scholar] [CrossRef]


| Strain or Plasmid | Relevant Characteristics | Source/Reference |
|---|---|---|
| Strain | ||
| E. coli JM109 | Clone strain | Our laboratory |
| E. coli BL21(DE3) | Expression strain | Our laboratory |
| B. subtilis 168 | Wild type | Our laboratory |
| B. subtilis WB600 | B. subtilis 168 derivate, deficient in nprE, aprE, epr, bpr, mpr, nprB, expression strain | Our laboratory |
| BL-K | E. coli BL21(DE3) derivative, E. coli BL21(DE3) carrying PET3b | This study |
| WB-K | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5 | This study |
| BL-H | E. coli BL21(DE3) derivative, E. coli BL21(DE3) carrying PET3b-H | This study |
| WB-H | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H | This study |
| WB-H-SPhrui | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPhrui | This study |
| WB-H-SPamye | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPamye | This study |
| WB-H-SPbgls | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPbgls | This study |
| WB-H-SPpelc | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPpelc | This study |
| WB-H-SPvpr | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPvpr | This study |
| WB-H-SPmdh | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh | This study |
| WB-H-SPnpre | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPnpre | This study |
| WB-H-SPdacb | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPdacb | This study |
| WB-H-SPphob | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPphob | This study |
| WB-H-SPyhfm | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPyhfm | This study |
| WB-H-SPesta | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPesta | This study |
| WB-H-SPmdh-Pgapb | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pgapb | This study |
| WB-H-SPmdh-Pnprb | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pnprb | This study |
| WB-H-SPmdh-Pnpre | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pnpre | This study |
| WB-H-SPmdh-Pmpr | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pmpr | This study |
| WB-H-SPmdh-Papre | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Papre | This study |
| WB-H-SPmdh-Pyola | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pyola | This study |
| WB-H-SPmdh-Psigx | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Psigx | This study |
| WB-H-SPmdh-Pp43 | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pp43 | This study |
| WB-H-SPmdh-Pwapa | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Pwapa | This study |
| WB-H-SPmdh-Ptrnq | B. subtilis WB600 derivative, B. subtilis WB600 carrying PMA5-H-SPmdh-Ptrnq | This study |
| Plasmid | ||
| PMA5 | F1ori Ampr, RepB Kanr, E. coli-B. subtilis shuttle vector | Our laboratory |
| PET3b | T7 Ampr, AmpR | Our laboratory |
| PET3b-H | PET3b derivative, PET3b carrying ColH, AmpR | This study |
| PMA5-H | PMA5 derivative, PMA5 carrying ColH, KanR | This study |
| PMA5-H-SPhrui | PMA5 derivative, PMA5 carrying ColH-SPhrui, KanR | This study |
| PMA5-H-SPamye | PMA5 derivative, PMA5 carrying ColH-SPamye, KanR | This study |
| PMA5-H-SPbgls | PMA5 derivative, PMA5 carrying ColH-SPbgls, KanR | This study |
| PMA5-H-SPpelc | PMA5 derivative, PMA5 carrying ColH-SPpelc, KanR | This study |
| PMA5-H-SPvpr | PMA5 derivative, PMA5 carrying ColH-SPvpr, KanR | This study |
| PMA5-H-SPmdh | PMA5 derivative, PMA5 carrying ColH-SPmdh, KanR | This study |
| PMA5-H-SPnpre | PMA5 derivative, PMA5 carrying ColH-SPnpre, KanR | This study |
| PMA5-H-SPdacb | PMA5 derivative, PMA5 carrying ColH-SPdacb, KanR | This study |
| PMA5-H-SPphob | PMA5 derivative, PMA5 carrying ColH-SPphob, KanR | This study |
| PMA5-H-SPyhfm | PMA5 derivative, PMA5 carrying ColH-SPyhfm, KanR | This study |
| PMA5-H-SPesta | PMA5 derivative, PMA5 carrying ColH-SPesta, KanR | This study |
| PMA5-H-SPmdh-Pgapb | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pgapb, KanR | This study |
| PMA5-H-SPmdh-Pnprb | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pnprb, KanR | This study |
| PMA5-H-SPmdh-Pmpr | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pmpr, KanR | This study |
| PMA5-H-SPmdh-Pnpre | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pnpre, KanR | This study |
| PMA5-H-SPmdh-Papre | PMA5 derivative, PMA5 carrying ColH-SPmdh-Papre, KanR | This study |
| PMA5-H-SPmdh-Pyola | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pyola, KanR | This study |
| PMA5-H-SPmdh-Psigx | PMA5 derivative, PMA5 carrying ColH-SPmdh-Psigx, KanR | This study |
| PMA5-H-SPmdh-Pp43 | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pp43, KanR | This study |
| PMA5-H-SPmdh-Pwapa | PMA5 derivative, PMA5 carrying ColH-SPmdh-Pwapa, KanR | This study |
| PMA5-H-SPmdh-Ptrnq | PMA5 derivative, PMA5 carrying ColH-SPmdh-Ptrnq, KanR | This study |
| Step | Volume (mL) | Protein (mg) | Activity (U) | Specific Activity (U/mg) | Fold | Yield (%) |
|---|---|---|---|---|---|---|
| Supernatant | 150 | 1830 | 40,200 | 21.97 | 1 | 100 |
| Ammonium sulfate | 25 | 88 | 25,950 | 294.89 | 54 | 64.55 |
| Q | 5 | 23.6 | 13,340 | 565.25 | 81 | 33.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.-F.; Xue, D.; Chen, N.; Su, C.; Gong, J.-S.; Qian, J.-Y.; Wang, Z.-Z.; Ma, X.-D.; Xie, N.; Xu, Z.-H.; et al. High-Level Extracellular Expression of Collagenase ColH in Bacillus subtilis for Adipose-Derived Cells Extraction. Fermentation 2025, 11, 242. https://doi.org/10.3390/fermentation11050242
Xu L-F, Xue D, Chen N, Su C, Gong J-S, Qian J-Y, Wang Z-Z, Ma X-D, Xie N, Xu Z-H, et al. High-Level Extracellular Expression of Collagenase ColH in Bacillus subtilis for Adipose-Derived Cells Extraction. Fermentation. 2025; 11(5):242. https://doi.org/10.3390/fermentation11050242
Chicago/Turabian StyleXu, Ling-Feng, Dai Xue, Nuo Chen, Chang Su, Jin-Song Gong, Jian-Ying Qian, Zhen-Zhen Wang, Xu-Dong Ma, Nan Xie, Zheng-Hong Xu, and et al. 2025. "High-Level Extracellular Expression of Collagenase ColH in Bacillus subtilis for Adipose-Derived Cells Extraction" Fermentation 11, no. 5: 242. https://doi.org/10.3390/fermentation11050242
APA StyleXu, L.-F., Xue, D., Chen, N., Su, C., Gong, J.-S., Qian, J.-Y., Wang, Z.-Z., Ma, X.-D., Xie, N., Xu, Z.-H., & Shi, J.-S. (2025). High-Level Extracellular Expression of Collagenase ColH in Bacillus subtilis for Adipose-Derived Cells Extraction. Fermentation, 11(5), 242. https://doi.org/10.3390/fermentation11050242








