A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
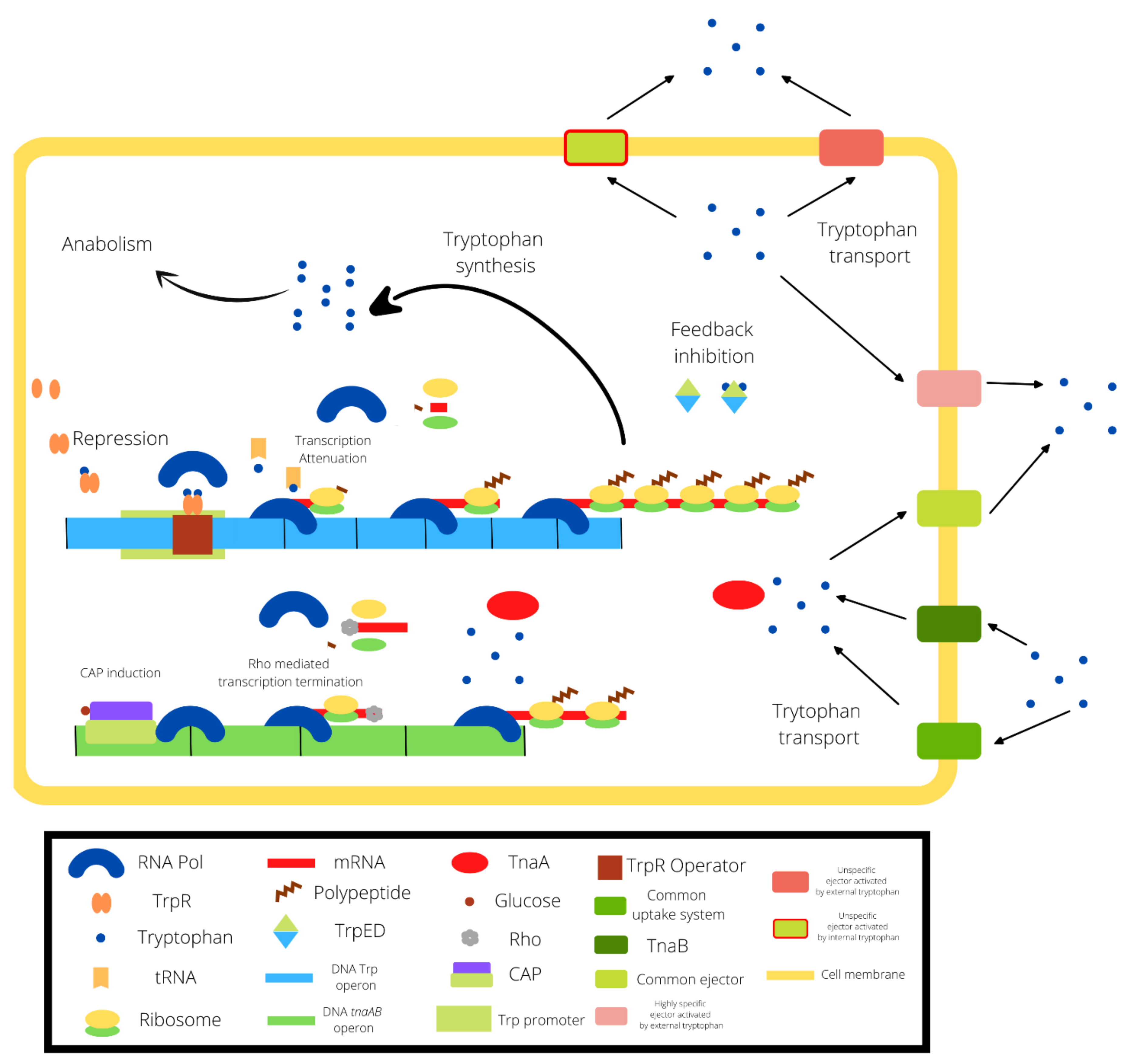
2.1. The Tryptophan Operon
2.2. The Tryptophan Biosynthesis Mathematical Models
 |
2.3. Model Implementation
2.4. Inclusion of TnaAB Enzymes and Transmembranal Transporters in the Model
2.5. Adjusting Parameters of Trp Transmembranal Transport
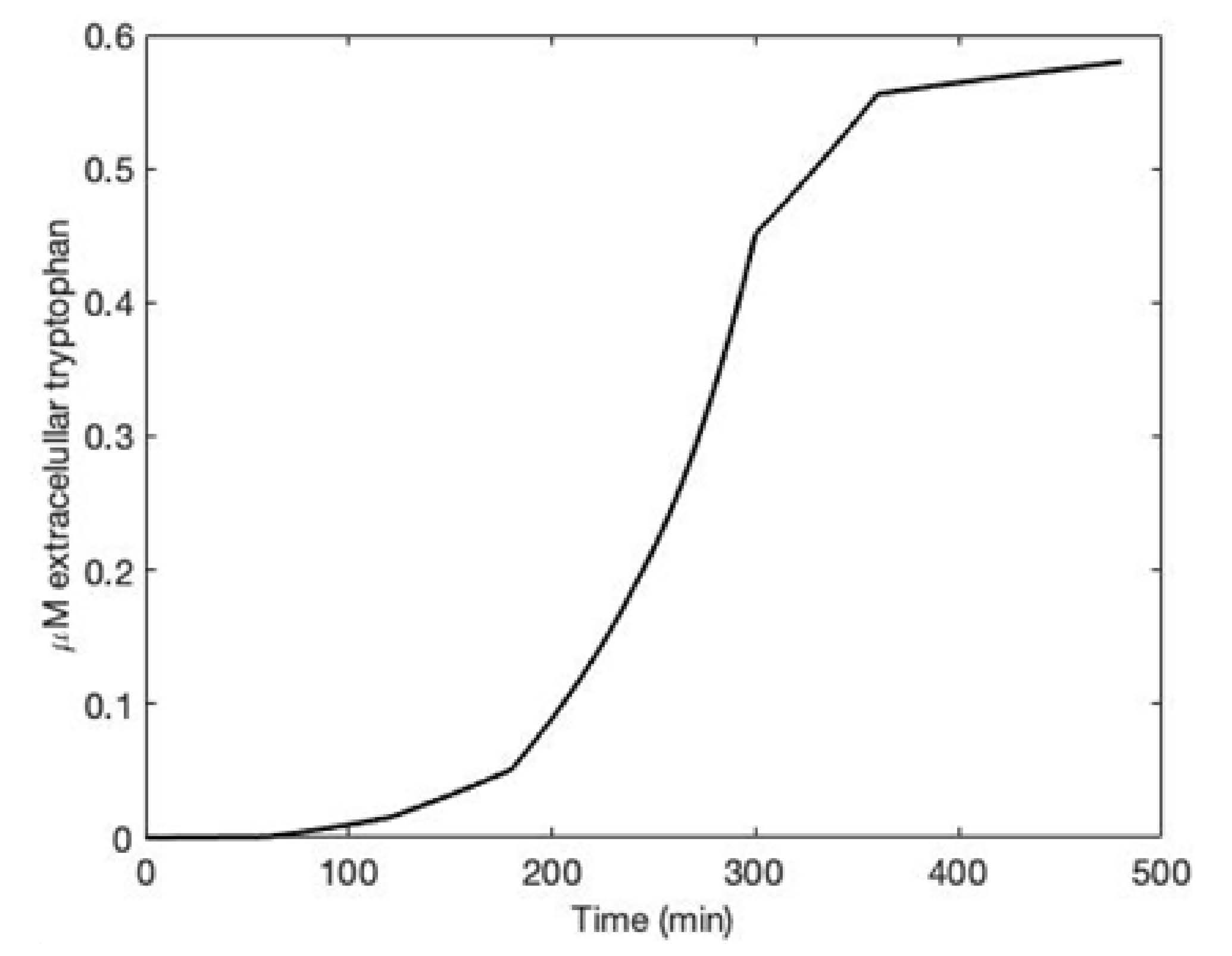
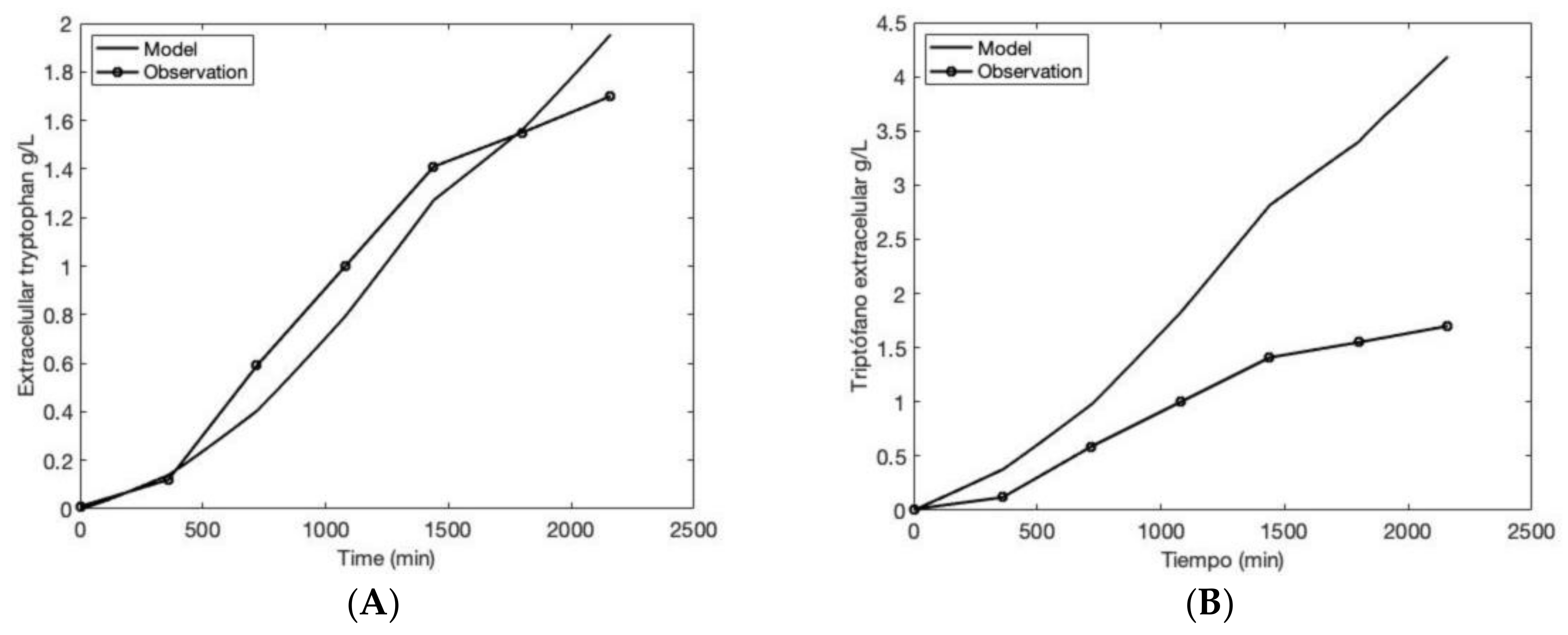
2.6. Model Validation with Experimental Reports
3. Results
3.1. Modifications to the Based Model
3.2. Comparing the Model to Experimental Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. L-tryptophan: Basic metabolic functions, behavioral research and therapeutic indications. Int. J. Tryptophan Res. 2009, 2, S2129. [Google Scholar] [CrossRef] [PubMed]
- Gosset, G. Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 2009, 20, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Crawford, I.P. Evolution of a biosynthetic pathway: The tryptophan paradigm. Annu. Rev. Microbiol. 1989, 43, 567–600. [Google Scholar] [CrossRef]
- Kaleta, C.; Schäuble, S.; Rinas, U.; Schuster, S. Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 2013, 8, 1105–1114. [Google Scholar] [CrossRef]
- Xiu, Z.L.; Zeng, A.P.; Deckwer, W.D. Model analysis concerning the effects of growth rate and intracellular tryptophan level on the stability and dynamics of tryptophan biosynthesis in bacteria. J. Biotechnol. 1997, 58, 125–140. [Google Scholar] [CrossRef]
- Bhartiya, S.; Rawool, S.; Venkatesh, K.V. Dynamic model of Escherichia coli tryptophan operon shows an optimal structural design. Eur. J. Biochem. 2003, 270, 2644–2651. [Google Scholar] [CrossRef]
- Sen, A.K.; Liu, W.M. Dynamic analysis of genetic control and regulation of amino acid synthesis: The tryptophan operon in Escherichia coli. Biotechnol. Bioeng. 1990, 35, 185–194. [Google Scholar] [CrossRef]
- Sinha, S. Theoretical study of tryptophan operon: Application in microbial technology. Biotechnol. Bioeng. 1988, 31, 117–124. [Google Scholar] [CrossRef]
- Xiu, Z.L.; Chang, Z.Y.; Zeng, A.P. Nonlinear dynamics of regulation of bacterial trp operon: Model analysis of integrated effects of repression, feedback inhibition, and attenuation. Biotechnol. Progr. 2002, 18, 686–693. [Google Scholar] [CrossRef]
- Simao, E.; Remy, E.; Thieffry, D.; Chaouiya, C. Qualitative modelling of regulated metabolic pathways: Application to the tryptophan biosynthesis in E. coli. Bioinformatics 2005, 21, ii190–ii196. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.K.; Kulasiri, D. On multiple regulatory mechanisms in the tryptophan operon system in Escherichia coli: In silico study of perturbation dynamics. In Silico Biol. 2008, 8, 485–510. [Google Scholar] [PubMed]
- Marín-Sanguino, A.; Torres, N.V. Optimization of tryptophan production in bacteria. Design of a strategy for genetic manipulation of the tryptophan operon for tryptophan flux maximization. Biotechnol. Progr. 2000, 16, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Deal, I.; Macauley, M.; Davies, R. Boolean models of the transport, synthesis, and metabolism of tryptophan in Escherichia Coli. arXiv 2022, arXiv:2022.11182. [Google Scholar]
- Schmid, J.W.; Mauch, K.; Reuss, M.; Gilles, E.D.; Kremling, A. Metabolic design based on a coupled gene expression—Metabolic network model of tryptophan production in Escherichia coli. Metab. Eng. 2004, 6, 364–377. [Google Scholar] [CrossRef]
- Sun, Y.; Mu, X.; Li, Z.; Teng, H.; Xiu, Z. Robustness and Nonlinear Dynamic Analysis for Trp Operon and Optimization of Tryptophan Production: An Integrated Model Considering Gene Regulation, Genes Interaction and Product Excretion. In Proceedings of the 2010 4th International Conference on Bioinformatics and Biomedical Engineering, Chengdu, China, 18–20 June 2010; pp. 1–7. [Google Scholar]
- Santillán, M.; Mackey, M.C. Dynamic regulation of the tryptophan operon: A modeling study and comparison with experimental data. Proc. Natl. Acad. Sci. USA 2001, 98, 1364–1369. [Google Scholar] [CrossRef]
- Santillan, M.; Zeron, E.S. Dynamic influence of feedback enzyme inhibition and transcription attenuation on the tryptophan operon response to nutritional shifts. J. Theor. Biol. 2004, 231, 287–298. [Google Scholar] [CrossRef]
- Drozdov-Tikhomirov, L.N.; Skurida, G.I. Mathematical model of tryptophan synthesis and excretion into the environment by E. coli cells. Mol. Biol. 1977, 11, 843–853. [Google Scholar]
- Boezi, J.A.; De Moss, R.D. Properties of a tryptophan transport system in Escherichia coli. Biochim. Biophys. Acta 1961, 49, 471–484. [Google Scholar] [CrossRef]
- Burrous, S.E.; DeMoss, R.D. Studies on tryptophan permease in Escherichia coli. Biochim. Biophys. Acta 1963, 73, 623–637. [Google Scholar] [CrossRef]
- Burkovski, A.; Krämer, R. Bacterial amino acid transport proteins: Occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 2002, 58, 265–274. [Google Scholar] [PubMed]
- Doroshenko, V.; Airich, L.; Vitushkina, M.; Kolokolova, A.; Livshits, V.; Mashko, S. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 2007, 275, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Forrest, L.R.; Krämer, R.; Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 2011, 1807, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Heinz, E.; Geck, P.; Wilbrandt, W. Coupling in secondary active transport: Activation of transport by co-transport and/or counter-transport with the fluxes of other solutes. Biochim. Biophys. Acta 1972, 255, 442–461. [Google Scholar] [CrossRef]
- Geck, P.; Heinz, E. Coupling in secondary transport Effect of electrical potentials on the kinetics of ion linked co-transport. Biochim. Biophys. Acta 1976, 443, 49–63. [Google Scholar] [CrossRef]
- Somerville, R. The Trp repressor, a ligand-activated regulatory protein. Prog. Nucleic Acid Res. Mol. Biol. 1992, 42, 1–38. [Google Scholar]
- Merino, E.; Jensen, R.A.; Yanofsky, C. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 2008, 11, 78–86. [Google Scholar] [CrossRef]
- Klig, L.S.; Crawford, I.P.; Yanofsky, C. Analysis of trp repressor-operator interaction by filter binding. Nucleic Acids Res. 1987, 15, 5339–5351. [Google Scholar] [CrossRef][Green Version]
- Gunsalus, R.P.; Miguel, A.G.; Gunsalus, G.L. Intracellular Trp repressor levels in Escherichia coli. J. Bacteriol. 1986, 167, 272–278. [Google Scholar] [CrossRef]
- Kasai, T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature 1974, 249, 523–527. [Google Scholar] [CrossRef]
- Yanofsky, C. Transcription attenuation: Once viewed as a novel regulatory strategy. J. Bacteriol. 2000, 182, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Henkin, T.M.; Yanofsky, C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays 2002, 24, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.I.; Crawford, I.P. Anthranilate synthetase: Partial purification and some kinetic studies on the enzyme from Escherichia coli. J. Biol. Chem. 1966, 241, 5577–5584. [Google Scholar] [CrossRef]
- Pittard, J.; Yang, J. Biosynthesis of the aromatic amino acids. EcoSal Plus 2008, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Bliss, R.D.; Painter, P.R.; Marr, A.G. Role of feedback inhibition in stabilizing the classical operon. J. Theor. Biol. 1982, 97, 177–193. [Google Scholar] [CrossRef]
- Orozco-Gómez, D.I.; Sosa-Hernández, J.E.; Gallardo-Navarro, Ó.A.; Santana-Solano, J.; Santillán, M. Bistable behaviour and medium-dependent post-translational regulation of the tryptophanase operon regulatory pathway in Escherichia coli. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Balcells, C.; Pastor, I.; Vilaseca, E.; Madurga, S.; Cascante, M.; Mas, F. Macromolecular crowding effect upon in vitro enzyme kinetics: Mixed activation–diffusion control of the oxidation of NADH by pyruvate catalyzed by lactate dehydrogenase. J. Phys. Chem. B. 2014, 118, 4062–4068. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Fundamentals of Enzyme Kinetics, 1st ed.; John Wiley & Sons: London, UK, 2013; pp. 76–89. [Google Scholar]
- Gu, P.; Yang, F.; Kang, J.; Wang, Q.; Qi, Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 2012, 11, 30. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; d’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Maier, R.M. Bacterial growth. In Environmental Microbiology, 2nd ed.; Maier, R.M., Pepper, I.L., Gerba, C.P., Eds.; Academic Press: London, UK, 2015; pp. 37–56. [Google Scholar]
- Ito, J.; Yanofsky, C. The nature of the anthranilic acid synthetase complex of Escherichia coli. J. Biol. Chem. 1966, 241, 4112–4114. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Effect of gene knockouts of L-tryptophan uptake system on the production of L-tryptophan in Escherichia coli. Process Biochem. 2012, 47, 340–344. [Google Scholar] [CrossRef]
- Santillán, M.; Mackey, M.C.; Zeron, E.S. Origin of bistability in the lac operon. Biophys. J. 2007, 92, 3830–3842. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Yanofsky, C. Anthranilate synthetase, an enzyme specified by the tryptophan operon of Escherichia coli: Comparative studies on the complex and the subunits. J. Bacteriol. 1969, 97, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Gosset, G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate: Sugar phosphotransferase system. Microb. Cell Fact. 2005, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lerner, C.G.; Inouye, M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990, 18, 4631. [Google Scholar] [CrossRef]
- Malan, T.P.; Kolb, A.; Buc, H.; McClure, W.R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J. Mol. Biol. 1984, 180, 881–909. [Google Scholar] [CrossRef]
- Baev, M.V.; Baev, D.; Radek, A.J.; Campbell, J.W. Growth of Escherichia coli MG1655 on LB medium: Determining metabolic strategy with transcriptional microarrays. Appl. Microbiol. Biotechnol. 2006, 71, 323–328. [Google Scholar] [CrossRef]
- Maria, G.; Gijiu, C.L.; Maria, C.; Tociu, C. Interference of the oscillating glycolysis with the oscillating tryptophan synthesis in the E. coli cells. Comput. Chem. Eng. 2018, 108, 395–407. [Google Scholar] [CrossRef]
- Maria, G.; Renea, L. Tryptophan Production Maximization in a Fed-Batch Bioreactor with Modified E. coli Cells, by Optimizing Its Operating Policy Based on an Extended Structured Cell Kinetic Model. Bioengineering 2021, 8, 210. [Google Scholar] [CrossRef]
- Bremer, H.; Dennis, P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 2008, 3. [Google Scholar] [CrossRef]
- Yoshida, H.; Shimada, T.; Ishihama, A. Coordinated hibernation of transcriptional and translational apparatus during growth transition of Escherichia coli to stationary phase. mSystems 2018, 3, e00057-18. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.N.; Lee, Y.; Hussein, R. Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Finkel, S.E. Long-term survival during stationary phase: Evolution and the GASP phenotype. Nat. Rev. Microbiol. 2006, 4, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]


| Estimated by | Kinetic Constants | Rate Constants | Dimensionless Constants |
|---|---|---|---|
| Santillán and Zeron [18] | Rtot ≈ 0.8 μM | ρ ≈ 2.4 × 102 μM min−1 | |
| KP ≈ 4.5 × 10−2 μM | KM ≈ 5.1 min−1 | ||
| KI≈ 4.1 μM | γM ≈ 0.69 min−1 | ||
| P ≈ 3 μM | kE ≈ 30 min−1 | ||
| Kρ ≈ 10 μM | γE ≈ 0 min−1 | ||
| Otot ≈ 4 × 10−3 μM | kT ≈ 7.4 × 104 min−1 | ||
| KR ≈ 2 × 10−4 μM | |||
| τE ≈ 1 min | |||
| KT ≈ 40 μM | |||
| Santillán and Mackey [17] | c ≈ 0.04 μM | b ≈ 0.85 | |
| Santillán et al. [45] | KG2 = 2.6 μM | # kmlac = 0.18 min−1 | Pp = 0.127 |
| kpc = 30 | |||
| nh = 1.3 | |||
| Orozco-Gómez et al. [37] | KG =11 μM | nG = 4 | |
| nW = 4 | |||
| K1 = 14 | |||
| λ = 7 | |||
| K2 = 9 | |||
| This work | C ≈ 8.5 μM | ktnaA ≈ 8.7066 min−1 | |
| K’x ≈ 168 μM | ktnaB ≈ 1750 min−1 | ||
| K2′x = 762.6 mM | Vmax ≈ 161.96 mM min−1 | ||
| K3′x ≈ 135.3 mM | Vmax2 ≈ 522.2 M min−1 | ||
| Kx ≈ 700 μM | Vmax3 ≈ 21,609 mM min−1 | ||
| K2x ≈ 41 mM | |||
| K3x ≈ 16.4 mM | |||
| km1′ ≈ 0.2 μM | |||
| km2′ ≈ 10 mM | |||
| km3′ ≈ 12.3 mM | |||
| OP = 0.0125 μM | |||
| OAB ≈ 4 × 10−3 μM | |||
| KW = 60 μM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro-López, D.A.; González de la Vara, L.E.; Santillán, M.; Martínez-Antonio, A. A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli. Fermentation 2022, 8, 560. https://doi.org/10.3390/fermentation8100560
Castro-López DA, González de la Vara LE, Santillán M, Martínez-Antonio A. A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli. Fermentation. 2022; 8(10):560. https://doi.org/10.3390/fermentation8100560
Chicago/Turabian StyleCastro-López, Diego Andrés, Luis E. González de la Vara, Moises Santillán, and Agustino Martínez-Antonio. 2022. "A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli" Fermentation 8, no. 10: 560. https://doi.org/10.3390/fermentation8100560
APA StyleCastro-López, D. A., González de la Vara, L. E., Santillán, M., & Martínez-Antonio, A. (2022). A Molecular Dynamic Model of Tryptophan Overproduction in Escherichia coli. Fermentation, 8(10), 560. https://doi.org/10.3390/fermentation8100560








