Shaping an Open Microbiome for Butanol Production through Process Control
Abstract
:1. Introduction
2. Methods
2.1. Schott Bottle Fermentation
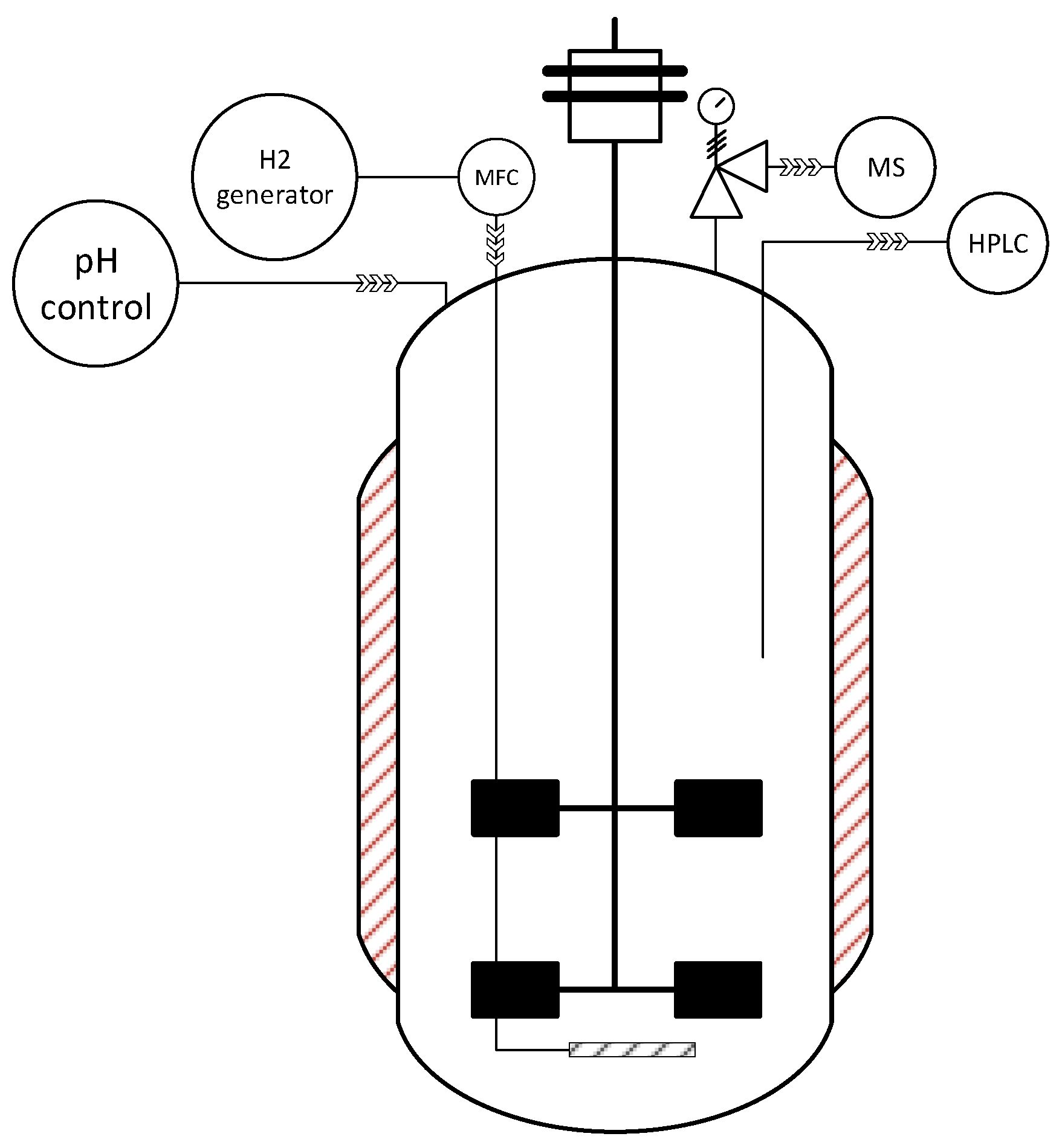
2.2. Bioreactor Fermentation
2.3. Analytical Methods
2.4. Thermodynamic Calculations
2.5. Carbon and Electron Balances
2.6. DNA Isolation and Amplicon Sequencing
2.7. Analysis of 16S rRNA Gene Amplicons
3. Results and Discussion
3.1. Butanol Production in 1 L Schott Bottles
3.2. Butanol Production under High Hydrogen Partial Pressure
3.3. Improved Butanol Formation Using a pH Controlled Bioreactor
3.4. Product Formation Controlled by Thermodynamics
3.5. Open Microbiome Analysis
4. Conclusions
- Schott bottle experiments showed butanol production from butyrate and hydrogen to a highest titer of 4.4 mM and volumetric productivity of 0.44 mmol L−1 d−1 of butanol. The use of a large inoculum size of anaerobic granular sludge (50% v/v) and lack of pH control contributed largely to by-product formation, with acetate as the most predominant measured by-product.
- A bioreactor operated at pH 5.5 and a of 2 bar showed an increase in butanol titer (10.9 mM) and volumetric productivity (0.68 mmol L−1 d−1); 2.98- and 4.65-fold increases from previously reported values, respectively. By-product formation from granular sludge was still prevalent, but directed towards ethanol production.
- Butyrate conversion is solely directed at butanol formation according to thermodynamics. Calculations of the actual Gibbs energy changes for the proposed catabolic reactions support the thermodynamic feasibility of by-product formation from bicarbonate in granular sludge, with the exception of lactate formation.
- Open microbiome analysis further supports exclusive butyrate conversion to butanol, probably by Mesotoga spp., and formation of by-products from residual carbon sources present in the inoculum. Reduced by-products such as ethanol and methane are most likely produced by Clostridium spp. and Methanobacterium spp., respectively.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | anaerobic digestion |
| VFA | volatile fatty acid |
| ABE | acetone, butanol, ethanol |
| HPLC | high-performance liquid chromatography |
| MS | mass spectrometer |
| MFC | mass flow controller |
| hydrogen partial pressure | |
| ∆G1 | actual Gibbs energy change |
| ∆G0 | standard Gibbs energy change |
| R | gas constant |
| T | temperature |
| stoichiometric coefficient of compound i | |
| concentration of compound i | |
| acid dissociation constant | |
| H2 | hydrogen |
| HCO3- | bicarbonate |
| CO2 | carbon dioxide |
| CH4 | methane |
| ASV | amplicon sequence variant |
| CCA | canonical correspondence analysis |
References
- Mansouri, S.S.; Udugama, I.A.; Cignitti, S.; Mitic, A.; Flores-Alsina, X.; Gernaey, K.V. Resource recovery from bio-based production processes: A future necessity? Curr. Opin. Chem. Eng. 2017, 18, 1–9. [Google Scholar] [CrossRef]
- International Energy Agency. Key World Energy Statistics; International Energy Agency: Paris, France, 2019.
- Dürre, P. Fermentative butanol production: Bulk chemical and biofuel. Ann. N. Y. Acad. Sci. 2008, 1125, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, A.; Mathimani, T.; Varjani, S.; Rene, E.R.; Kumar, G.; Kim, S.H.; Ponnusamy, V.K.; Yoon, J.J. Biobutanol as a promising liquid fuel for the future—Recent updates and perspectives. Fuel 2019, 253, 637–646. [Google Scholar] [CrossRef]
- Harvey, B.G.; Meylemans, H.A. The role of butanol in the development of sustainable fuel technologies. J. Chem. Technol. Biotechnol. 2011, 86, 2–9. [Google Scholar] [CrossRef]
- Pinto, T.; Flores-Alsina, X.; Gernaey, K.V.; Junicke, H. Alone or together? A review on pure and mixed microbial cultures for butanol production. Renew. Sustain. Energy Rev. 2021, 147, 111244. [Google Scholar] [CrossRef]
- Udugama, I.A.; Petersen, L.A.H.; Falco, F.C.; Junicke, H.; Mitic, A.; Alsina, X.F.; Mansouri, S.S.; Gernaey, K.V. Resource recovery from waste streams in a water-energy-food nexus perspective: Toward more sustainable food processing. Food Bioprod. Process. 2020, 119, 133–147. [Google Scholar] [CrossRef]
- Chen, H.; Hao, S.; Chen, Z.; O-Thong, S.; Fan, J.; Clark, J.; Luo, G.; Zhang, S. Mesophilic and thermophilic anaerobic digestion of aqueous phase generated from hydrothermal liquefaction of cornstalk: Molecular and metabolic insights. Water Res. 2020, 168, 115199. [Google Scholar] [CrossRef]
- Amha, Y.M.; Sinha, P.; Lagman, J.; Gregori, M.; Smith, A.L. Elucidating microbial community adaptation to anaerobic co-digestion of fats, oils, and grease and food waste. Water Res. 2017, 123, 277–289. [Google Scholar] [CrossRef]
- Feldman, H.; Flores-Alsina, X.; Ramin, P.; Kjellberg, K.; Jeppsson, U.; Batstone, D.J.; Gernaey, K.V. Modelling an industrial anaerobic granular reactor using a multi-scale approach. Water Res. 2017, 126, 488–500. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Woods, D.R. Acetone-butanol fermentation revisited. Microbiol. Rev. 1986, 50, 484–524. Available online: http://www.ncbi.nlm.nih.gov/pubmed/3540574 (accessed on 31 January 2018).
- Jankowska, E.; Duber, A.; Chwialkowska, J.; Stodolny, M.; Oleskowicz-Popiel, P. Conversion of organic waste into volatile fatty acids—The influence of process operating parameters. Chem. Eng. J. 2018, 345, 395–403. [Google Scholar] [CrossRef]
- Steinbusch, K.J.J.; Hamelers, H.V.M.; Buisman, C.J.N. Alcohol production through volatile fatty acids reduction with hydrogen as electron donor by mixed cultures. Water Res. 2008, 42, 4059–4066. [Google Scholar] [CrossRef] [PubMed]
- Junicke, H.; van Loosdrecht, M.C.M.; Kleerebezem, R. Kinetic and thermodynamic control of butyrate conversion in non-defined methanogenic communities. Appl. Microbiol. Biotechnol. 2016, 100, 915–925. [Google Scholar] [CrossRef] [Green Version]
- Kleerebezem, R.; van Loosdrecht, M.C.M. A Generalized Method for Thermodynamic State Analysis of Environmental Systems. Crit. Rev. Environ. Sci. Technol. 2010, 40, 1–54. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-thoughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 21 January 2021).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, G.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.5–6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 21 January 2021).
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoehler, T.M. Biological energy requirements as quantitative boundary conditions for life in the subsurface. Geobiology 2004, 2, 205–215. [Google Scholar] [CrossRef]
- Feldman, H.; Flores-Alsina, X.; Ramin, P.; Kjellberg, K.; Jeppsson, U.; Batstone, D.J.; Gernaey, K.V. Assessing the effects of intra-granule precipitation in a full-scale industrial anaerobic digester. Water Sci. Technol. 2019, 79, 1327–1337. [Google Scholar] [CrossRef]
- Mañas, A.; Pocquet, M.; Biscans, B.; Sperandio, M. Parameters influencing calcium phosphate precipitation in granular sludge sequencing batch reactor. Chem. Eng. Sci. 2012, 77, 165–175. [Google Scholar] [CrossRef]
- El-Zouheiri, F.A. Screening of Optimum Process Conditions for Butanol Production in Mixed Microbial Cultures; Danmarks Tekniske Universitet: Copenhagen, Denmark, 2019; Available online: https://findit.dtu.dk/en/catalog/2453508175 (accessed on 15 June 2021).
- Batstone, D.J.; Keller, J.; Angelidaki, I.; Kalyuzhnyi, S.V.; Pavlostathis, S.G.; Rozzi, A.; Sanders, W.T.M.; Siegrist, H.; Vavilin, V.A. The IWA Anaerobic Digestion Model No 1 (ADM1). Water Sci. Technol. 2002, 45, 65–73. [Google Scholar] [CrossRef]
- Cappelletti, M.; Zannoni, D.; Postec, A.; Ollivier, B. Members of the Order Thermotogales: From Microbiology to Hydrogen Production. In Microbial Bioenergy: Hydrogen Production; Springer: Dordrecht, The Netherlands, 2014; pp. 197–224. [Google Scholar] [CrossRef]
- Yamada, T.; Sekiguchi, Y. A naerolineaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: New York, NY, USA, 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Xu, P.-X.; Chai, L.-J.; Qiu, T.; Zhang, X.-J.; Lu, Z.-M.; Xiao, C.; Wang, S.-T.; Shen, C.-H.; Shi, J.-S.; Xu, Z.-H. Clostridium fermenticellae sp. nov., isolated from the mud in a fermentation cellar for the production of the Chinese liquor, baijiu. Int. J. Syst. Evol. Microbiol. 2019, 69, 859–865. [Google Scholar] [CrossRef]
- Oki, K.; Kudo, Y.; Watanabe, K. Lactobacillus saniviri sp. nov. and Lactobacillus senioris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 601–607. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.; Liu, X.; Dong, X. Methanobacterium beijingense sp. nov., a novel methanogen isolated from anaerobic digesters. Int. J. Syst. Evol. Microbiol. 2005, 55, 325–329. [Google Scholar] [CrossRef] [Green Version]
- Fournier, G.P.; Gogarten, J.P. Evolution of Acetoclastic Methanogenesis in Methanosarcina via Horizontal Gene Transfer from Cellulolytic Clostridia. J. Bacteriol. 2008, 190, 1124–1127. [Google Scholar] [CrossRef] [Green Version]
- Honda, T.; Fujita, T.; Tonouchi, A. Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int. J. Syst. Evol. Microbiol. 2013, 63, 3679–3686. [Google Scholar] [CrossRef] [PubMed]
- Props, R.; Kerckhof, F.M.; Rubbens, P.; de Vrieze, J.; Sanabria, E.H.; Waegeman, W.; Monsieurs, P.; Hammes, F.; Boon, N. Absolute quantification of microbial taxon abundances. ISME J. 2017, 11, 584–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Control | Experiment 50 | Experiment 15 | |
|---|---|---|---|
| Inoculum size (v/v) | 50% | 50% | 15% |
| Total suspended solids (g L−1) | 23.8 | 23.8 | 7.1 |
| Initial butyrate concentration (mM) | 0 | 50 | 50 |
| Medium | Yes | Yes | Yes |
| Hydrogen partial pressure (bar) | 1.5 | 1.5 | 1.5 |
| Control | Experiment 50 | Experiment 15 | |
|---|---|---|---|
| Butyrate (mM) | 0.52 | 41.70 | 47.36 |
| Products (mM) | |||
| Acetate | 14.33 | 19.58 | 5.19 |
| Butanol | 0.00 | 4.40 | 1.33 |
| i-Butyrate | 0.67 | 1.02 | 0.26 |
| Propionate | 0.82 | 1.81 | 0.34 |
| i-Valerate | 1.02 | 2.27 | 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, T.; Grimalt-Alemany, A.; Flores-Alsina, X.; Gavala, H.N.; Gernaey, K.V.; Junicke, H. Shaping an Open Microbiome for Butanol Production through Process Control. Fermentation 2022, 8, 333. https://doi.org/10.3390/fermentation8070333
Pinto T, Grimalt-Alemany A, Flores-Alsina X, Gavala HN, Gernaey KV, Junicke H. Shaping an Open Microbiome for Butanol Production through Process Control. Fermentation. 2022; 8(7):333. https://doi.org/10.3390/fermentation8070333
Chicago/Turabian StylePinto, Tiago, Antonio Grimalt-Alemany, Xavier Flores-Alsina, Hariklia N. Gavala, Krist V. Gernaey, and Helena Junicke. 2022. "Shaping an Open Microbiome for Butanol Production through Process Control" Fermentation 8, no. 7: 333. https://doi.org/10.3390/fermentation8070333
APA StylePinto, T., Grimalt-Alemany, A., Flores-Alsina, X., Gavala, H. N., Gernaey, K. V., & Junicke, H. (2022). Shaping an Open Microbiome for Butanol Production through Process Control. Fermentation, 8(7), 333. https://doi.org/10.3390/fermentation8070333








