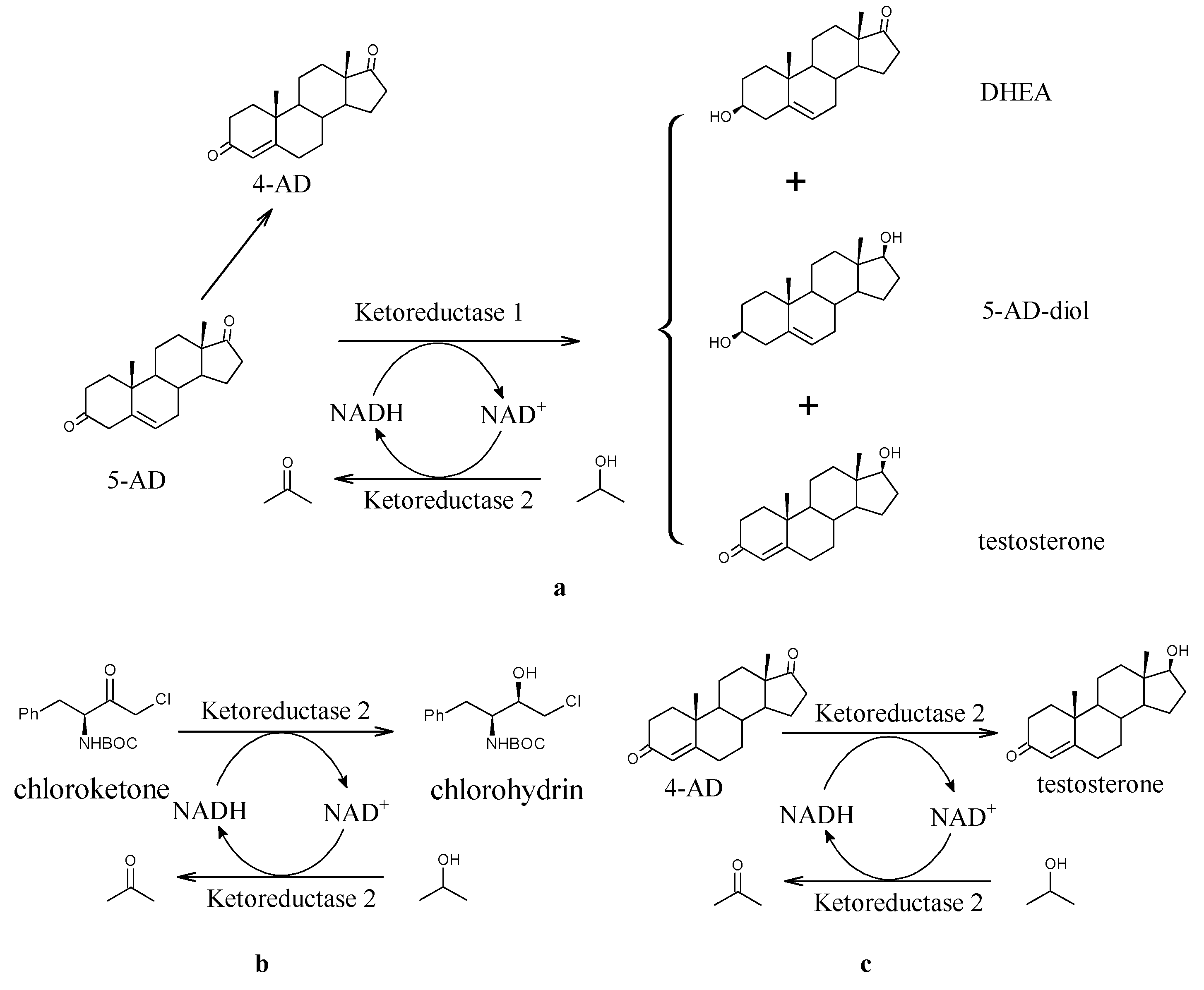
Testosterone Biosynthesis from 4-Androstene-3,17-Dione Catalyzed via Bifunctional Ketoreductase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Media
2.2. Cultivation
2.3. Enzyme Activity Assay
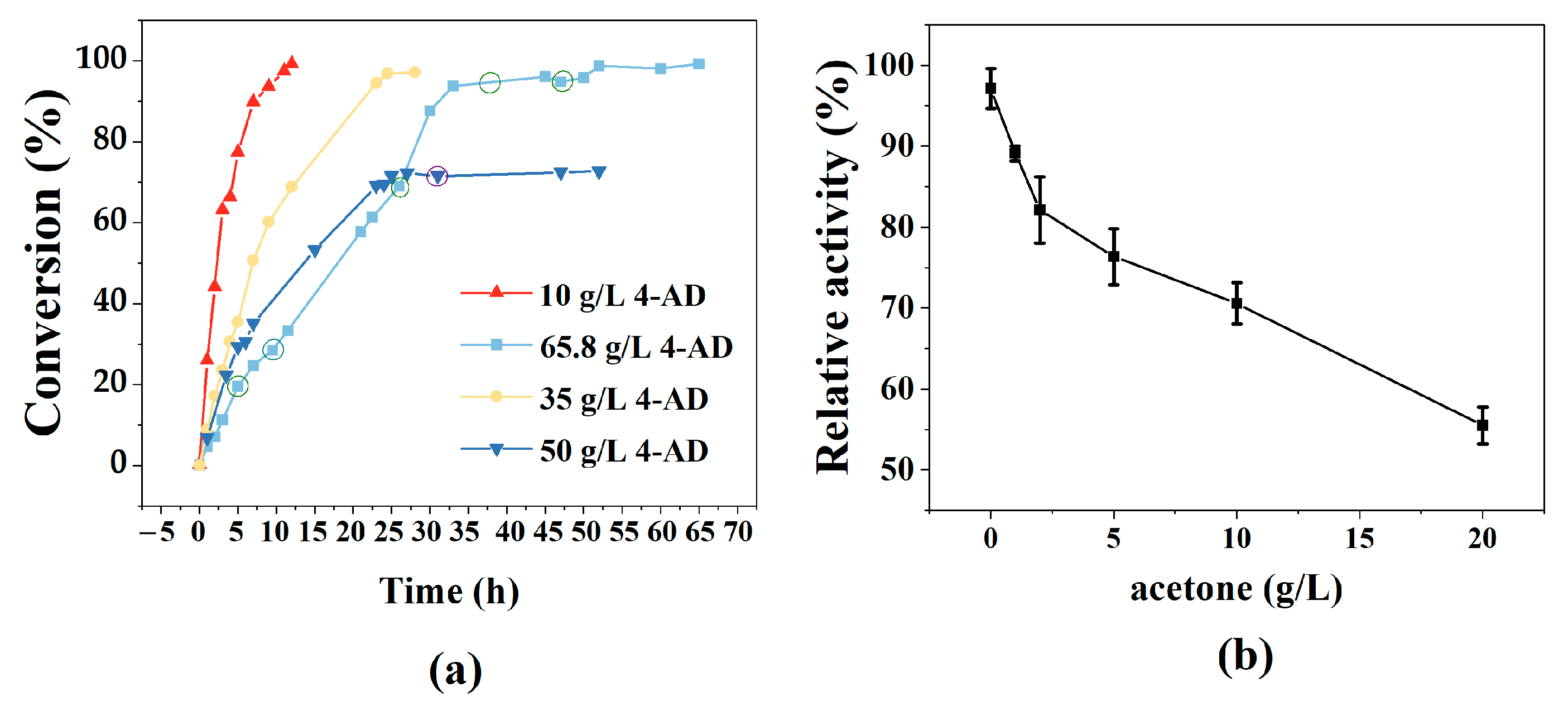
2.4. Biosynthesis and Isolation of Testosterone
2.5. Analysis
3. Results and Discussion
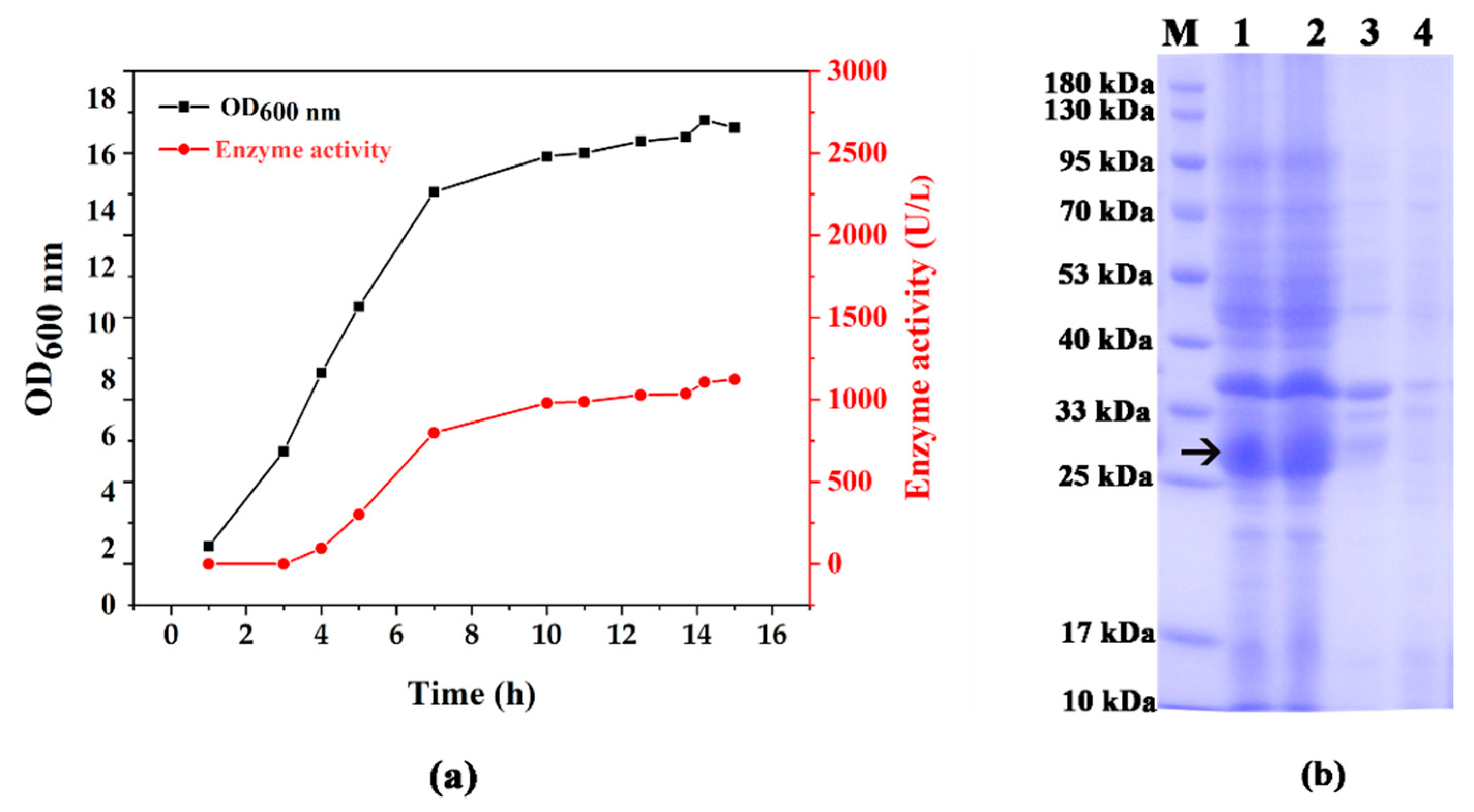
3.1. Fermentation and Protein Expression
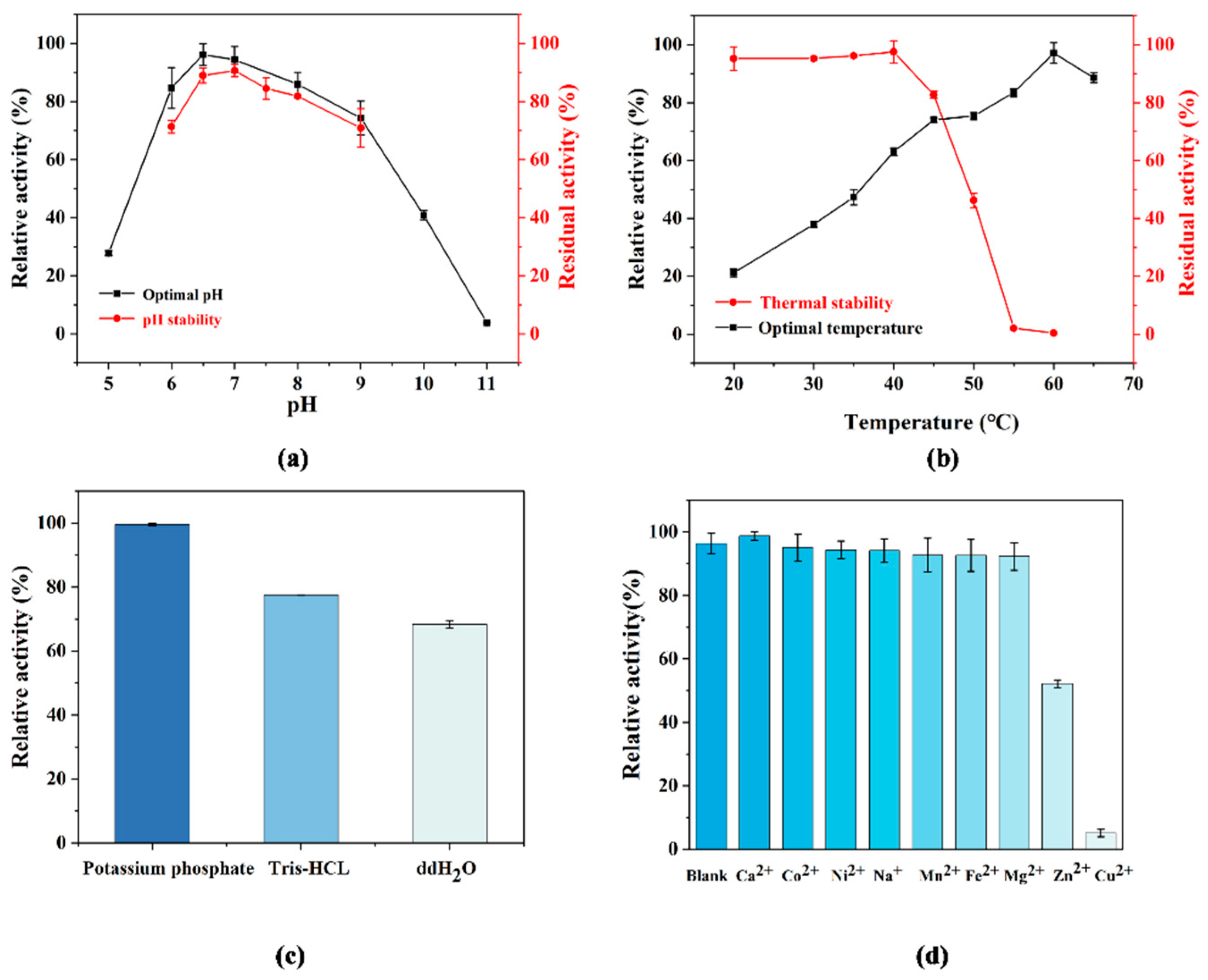
3.2. Enzymatic Properties
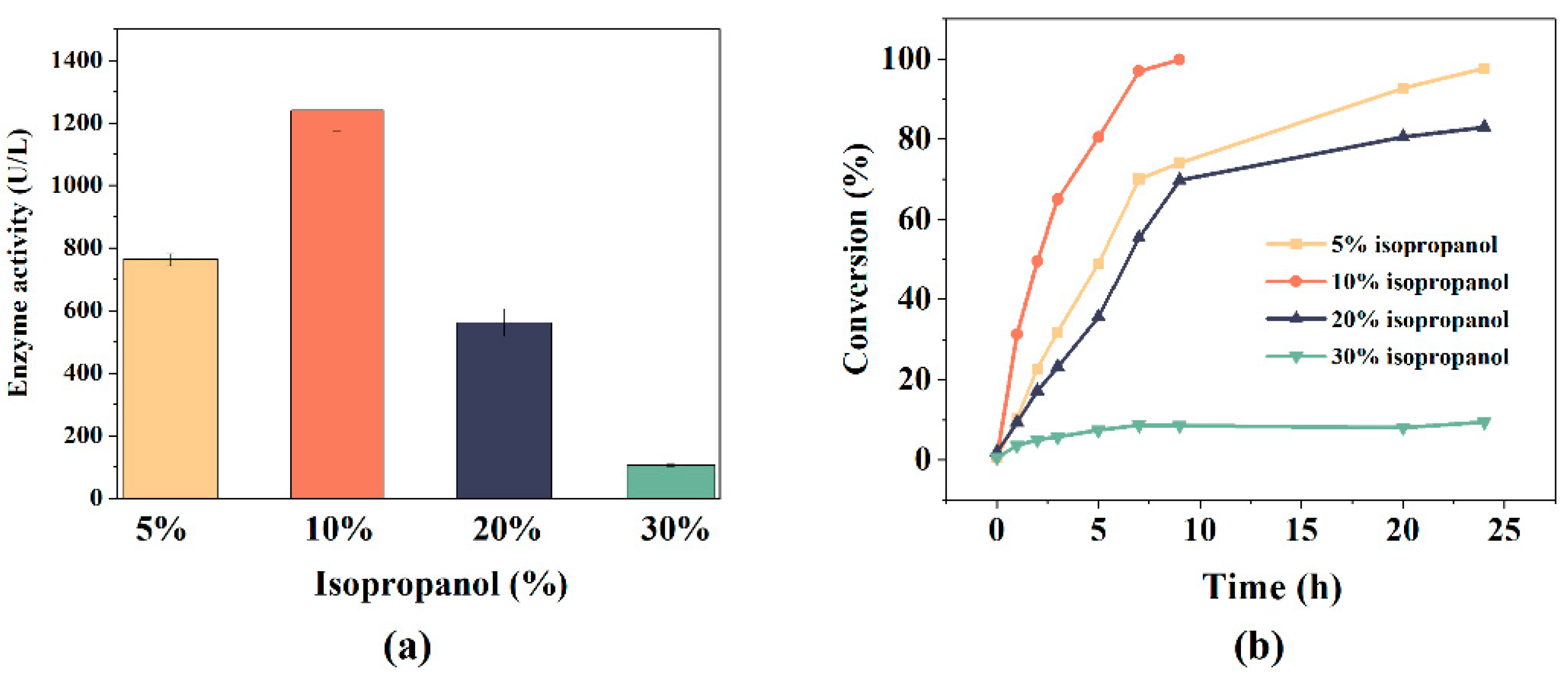
3.3. Optimization of NADH Regeneration Conditions
3.4. Production of TS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tauchen, J.; Jurasek, M.; Huml, L.; Rimpelova, S. Medicinal Use of Testosterone and Related Steroids Revisited. Molecules 2021, 26, 1032. [Google Scholar] [CrossRef] [PubMed]
- Nieschlag, E.; Nieschlag, S. Testosterone: The Medical and Cultural History of Testosterone and the Testes; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Fessner, N.D.; Srdič, M.; Weber, H.; Schmid, C.; Schönauer, D.; Schwaneberg, U.; Glieder, A. Preparative-Scale Production of Testosterone Metabolites by Human Liver Cytochrome P450 Enzyme 3A4. Adv. Synth. Catal. 2020, 362, 2725–2738. [Google Scholar] [CrossRef]
- Ihara, M.; Sudow, I.; Fukumoto, K.; Kametani, T. ChemInform Abstract: Stereoselective Total Synthesis of Testosterone and Androsterone via A/B-Ring Construction of the Steroidal Ring System by Intramolecular Diels-Alder Reaction. Chem. Informationsdienst 1986, 17, 117–123. [Google Scholar] [CrossRef]
- Ihara, M.; Tokunaga, Y.; Taniguchi, N.; Fukumoto, K. Stereoselective synthesis of (+)-testosterone via intramolecular 13-dipolar cycloadditxon of nitrile oxide. Tetrahedron 1991, 47, 6635–6648. [Google Scholar] [CrossRef]
- Andryushina, V.A.; Morozova, L.S.; Grinenko, G.S. Synthesis of testosterone from androst-4-ene-3,17-dione. Pharm. Chem. J. 1987, 21, 883–884. [Google Scholar] [CrossRef]
- Bhatti, H.N.; Khera, R.A. Biological transformations of steroidal compounds: A review. Steroids 2012, 77, 1267–1290. [Google Scholar] [CrossRef] [PubMed]
- Egorova, O.V.; Nikolayeva, V.M.; Sukhodolskaya, G.V.; Donova, M.V. Transformation of C19-steroids and testosterone production by sterol-transforming strains of Mycobacterium spp. J. Mol. Catal. B Enzym. 2009, 57, 198–203. [Google Scholar] [CrossRef]
- Tekucheva, D.N.; Nikolayeva, V.M.; Karpov, M.V.; Timakova, T.A.; Shutov, A.V.; Donova, M.V. Bioproduction of testosterone from phytosterol by Mycolicibacterium neoaurum strains: “one-pot”, two modes. Bioresour. Bioprocess. 2022, 9, 116. [Google Scholar] [CrossRef]
- Fogal, S.; Motterle, R.; Castellin, A.; Arvotti, G.; Bergantino, E. Biocatalyzed synthesis of testosterone. Chem. Eng. Trans. 2010, 20, 61–66. [Google Scholar]
- Meier, M.; Moeller, G.; Adamski, J. Perspectives in Understanding the Role of Human 17β-Hydroxysteroid Dehydrogenases in Health and Disease. Ann. N. Y. Acad. Sci. 2010, 1155, 15–24. [Google Scholar] [CrossRef]
- Shao, M.; Zhang, X.; Rao, Z.; Xu, M.; Yang, T.; Li, H.; Xu, Z.; Yang, S. Efficient testosterone production by engineered Pichia pastoris co-expressing human 17β-hydroxysteroid dehydrogenase type 3 and Saccharomyces cerevisiae glucose 6-phosphate dehydrogenase with NADPH regeneration. Green Chem. 2016, 18, 1774–1784. [Google Scholar] [CrossRef]
- Ding, J.; You, S.; Zhang, J.; Zhang, H.; Wang, H.; Zhang, W.; Qi, W.; Su, R.; He, Z. Rational design of 17β-hydroxysteroid dehydrogenase type3 for improving testosterone production with an engineered Pichia pastoris. Bioresour. Technol. 2021, 341, 125833. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cabezon, L.; Galan, B.; Garcia, J.L. Engineering Mycobacterium smegmatis for testosterone production. Microb. Biotechnol. 2017, 10, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Sood, U.; Singh, Y.; Shakarad, M.; Lal, R. Highlight on Engineering Mycobacterium smegmatisfor testosterone production. Microb. Biotechnol. 2017, 10, 73–75. [Google Scholar] [CrossRef]
- Guevara, G.; Olortegui Flores, Y.; Fernandez de Las Heras, L.; Perera, J.; Navarro Llorens, J.M. Metabolic engineering of Rhodococcus ruber Chol-4: A cell factory for testosterone production. PLoS ONE 2019, 14, e0220492. [Google Scholar] [CrossRef] [PubMed]
- Tekucheva, D.N.; Fokina, V.V.; Nikolaeva, V.; Shutov, A.A.; Karpov, M.V.; Donova, M.V. Cascade Biotransformation of Phytosterol to Testosterone by Mycolicibacterium neoaurum VKM Ac-1815D and Nocardioides simplex VKM Ac-2033D Strains. Microbiology 2022, 91, 303–312. [Google Scholar] [CrossRef]
- Peltoketo, H.; Luu-The, V.; Simard, J.; Adamski, J. 17 beta-Hydroxysteroid dehydrogenase (HSD)/17-ketosteroid reductase (KSR) family; nomenclature and main characteristics of the 17HSD/KSR enzymes. J. Mol. Endocrinol. 1999, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Breitling, R.; Laubner, D.; Adamski, J. Structure-based phylogenetic analysis of short-chain alcohol dehydrogenases and reclassification of the 17beta-hydroxysteroid dehydrogenase family. Mol. Biol. Evol. 2001, 18, 2154–2161. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Chen, X.; Feng, J.; Wang, M.; Wu, Q.; Zhu, D. Modulating the active site lid of an alcohol dehydrogenase from Ralstonia sp. enabled efficient stereospecific synthesis of 17β-hydroxysteroids. Enzym. Microb. Technol. 2021, 149, 109837. [Google Scholar] [CrossRef]
- Su, B.-M.; Zhao, H.-R.; Xu, L.; Xu, X.-Q.; Wang, L.-C.; Lin, J.; Lin, W. Construction of an Efficient Non-natural Enzyme System for Preparation of Testosterone in High Space-Time Yield. ACS Sustain. Chem. Eng. 2022, 10, 3373–3382. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Cai, G.; Peng, C.; Zhong, Y.; Luo, X. Coupled fermentation-bioconversion process for production of chiral α-chlorohydrin with recombinant ketoreductase. Process Biochem. 2019, 76, 34–39. [Google Scholar] [CrossRef]
- Krause, M.; Neubauer, A.; Neubauer, P. The fed-batch principle for the molecular biology lab: Controlled nutrient diets in ready-made media improve production of recombinant proteins in Escherichia coli. Microb. Cell Factories 2016, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, Z.; Wang, X.; Selvaraj, J.N.; Zhang, G. Genetic engineering modification and fermentation optimization for extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ma, Q.; Lu, J.; Xue, Y.; Xue, C. Optimisation for subcritical fluid extraction of 17-methyltestosterone with 1,1,1,2-tetrafluoroethane for HPLC analysis. Food Chem. 2012, 135, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Fryszkowska, A.; Peterson, J.; Davies, N.L.; Dewar, C.; Evans, G.; Bycroft, M.; Triggs, N.; Fleming, T.; Gorantla, S.S.C.; Hoge, G.; et al. Development of a Chemoenzymatic Process for Dehydroepiandrosterone Acetate Synthesis. Org. Process Res. Dev. 2016, 20, 1520–1528. [Google Scholar] [CrossRef]
- Bernard, A.; Payton, M. Fermentation and growth of Escherichia coli for optimal protein production. Curr. Protoc. Protein. Sci. 1995, 5, 5.3.1–5.3.18. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Payton, M. Selection of Escherichia coli expression systems. Curr. Protoc. Protein. Sci. 1995, 5, 5.2.1–5.2.18. [Google Scholar] [CrossRef]


| Substrate Concentration (g/L) | Time (h) | Conversion (%) | Product Purity (%) | Product Yield (%) |
|---|---|---|---|---|
| 10 | 9 | 99.29 | 99.79 | 97.94 |
| 35 | 24 | 98.12 | 99.81 | 91.59 |
| 65.8 | 52 | 98.73 | 99.82 | 92.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Mei, G.; Zhao, J.; Zhang, S.; Qin, W.; Sheng, Q.; Yang, Z. Testosterone Biosynthesis from 4-Androstene-3,17-Dione Catalyzed via Bifunctional Ketoreductase. Fermentation 2023, 9, 998. https://doi.org/10.3390/fermentation9120998
Wei Y, Mei G, Zhao J, Zhang S, Qin W, Sheng Q, Yang Z. Testosterone Biosynthesis from 4-Androstene-3,17-Dione Catalyzed via Bifunctional Ketoreductase. Fermentation. 2023; 9(12):998. https://doi.org/10.3390/fermentation9120998
Chicago/Turabian StyleWei, Yi, Guangyao Mei, Jinlin Zhao, Shaoyang Zhang, Wenping Qin, Qing Sheng, and Zhongyi Yang. 2023. "Testosterone Biosynthesis from 4-Androstene-3,17-Dione Catalyzed via Bifunctional Ketoreductase" Fermentation 9, no. 12: 998. https://doi.org/10.3390/fermentation9120998
APA StyleWei, Y., Mei, G., Zhao, J., Zhang, S., Qin, W., Sheng, Q., & Yang, Z. (2023). Testosterone Biosynthesis from 4-Androstene-3,17-Dione Catalyzed via Bifunctional Ketoreductase. Fermentation, 9(12), 998. https://doi.org/10.3390/fermentation9120998




