Exploring the Influence of Culture Environment on the Yield of Volvariella volvacea Based on Microbiomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Compost Substrate Formula, Cultivation Method, and Sampling
2.2. Metagenomic Sequencing
2.3. 16S rDNA Sequencing
2.4. Metagenomic Analysis
2.5. 16S rDNA Analysis
3. Results
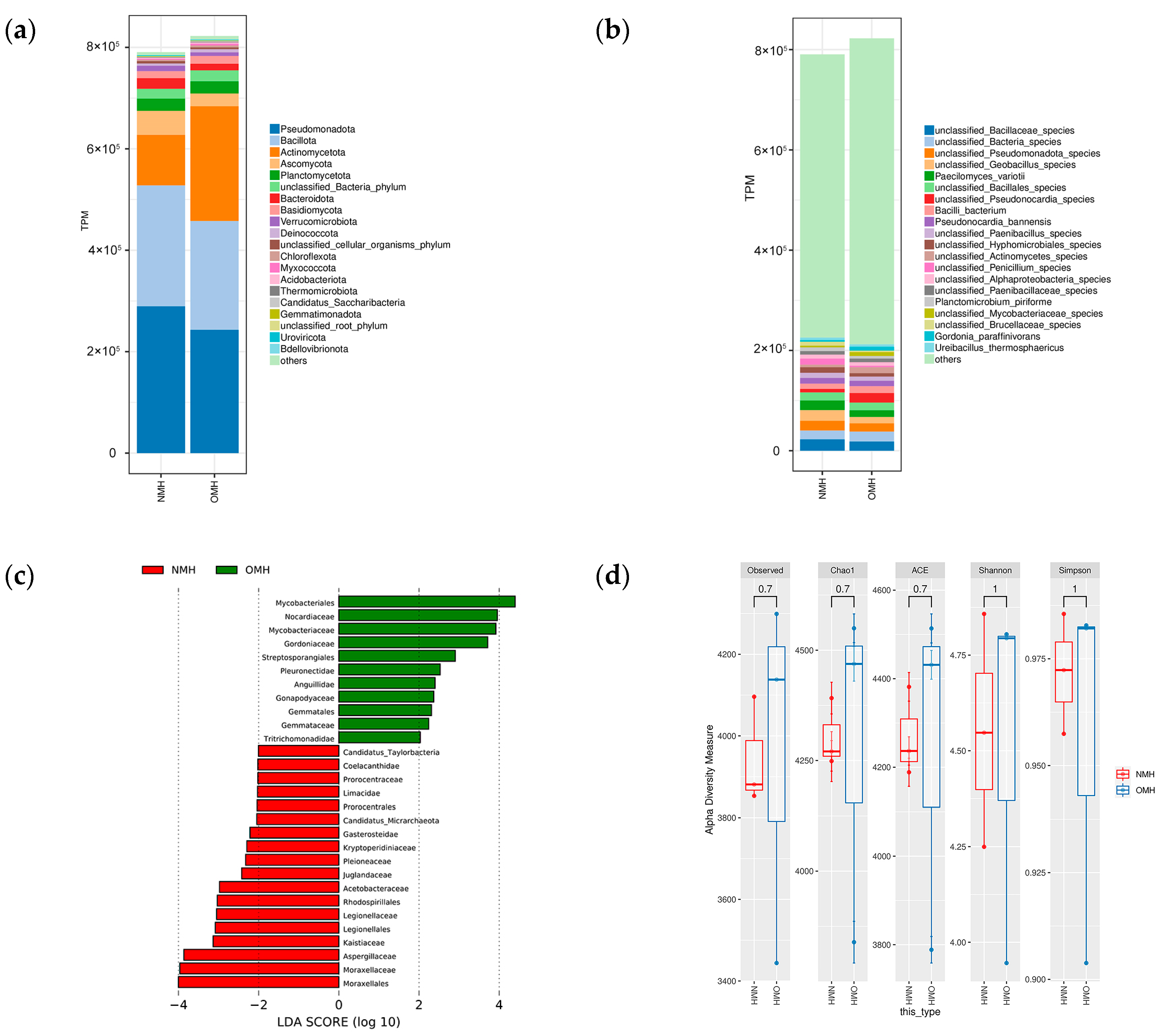
3.1. Illumina Sequencing of Culture Environment and Substrate Microbiota of Volvariella volvacea
3.2. Microbial Community Structure and Diversity in Different Culture Environments
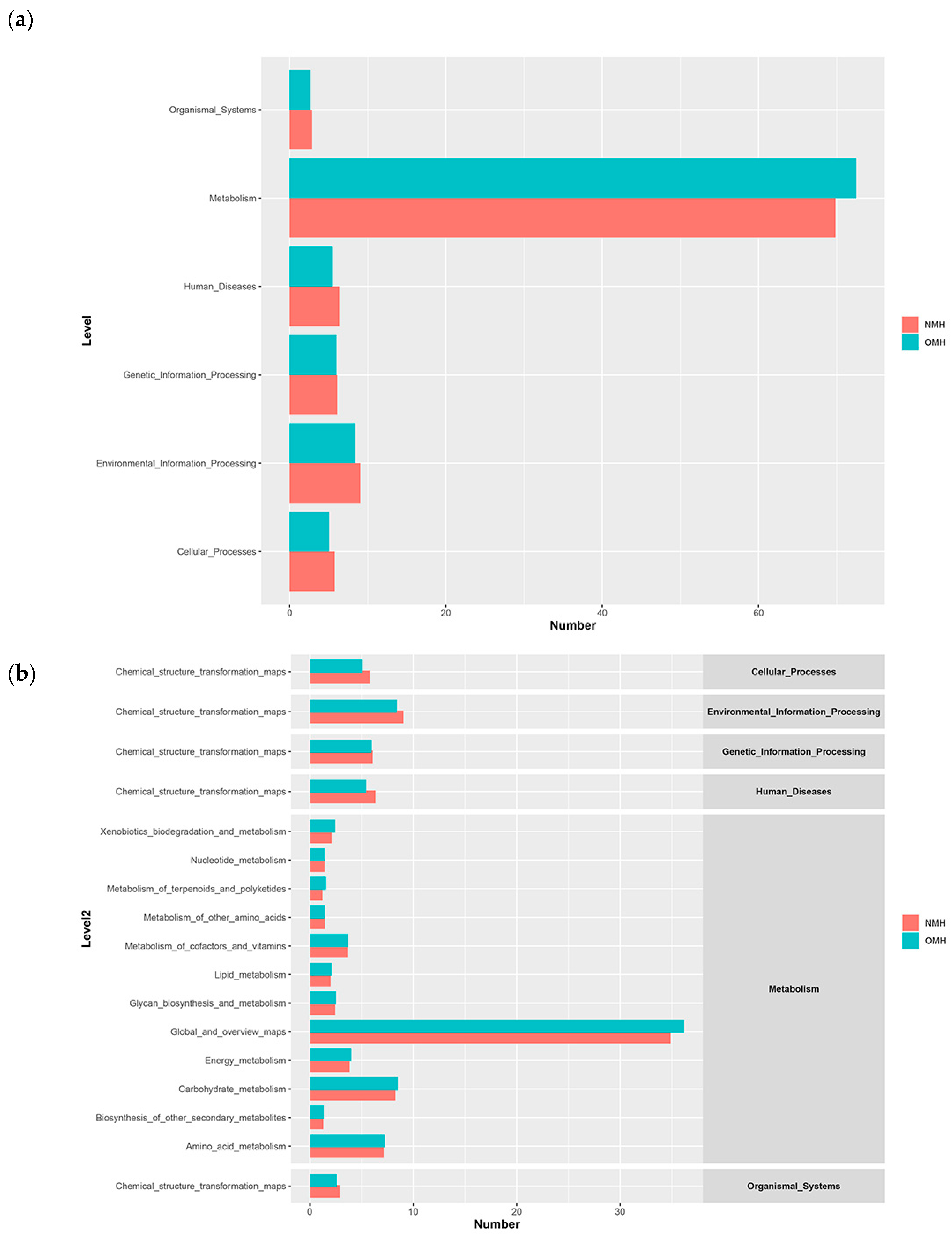
3.3. Microbial Metabolism in Different Culture Environments
3.4. Microbial Community Function in Different Culture Environments
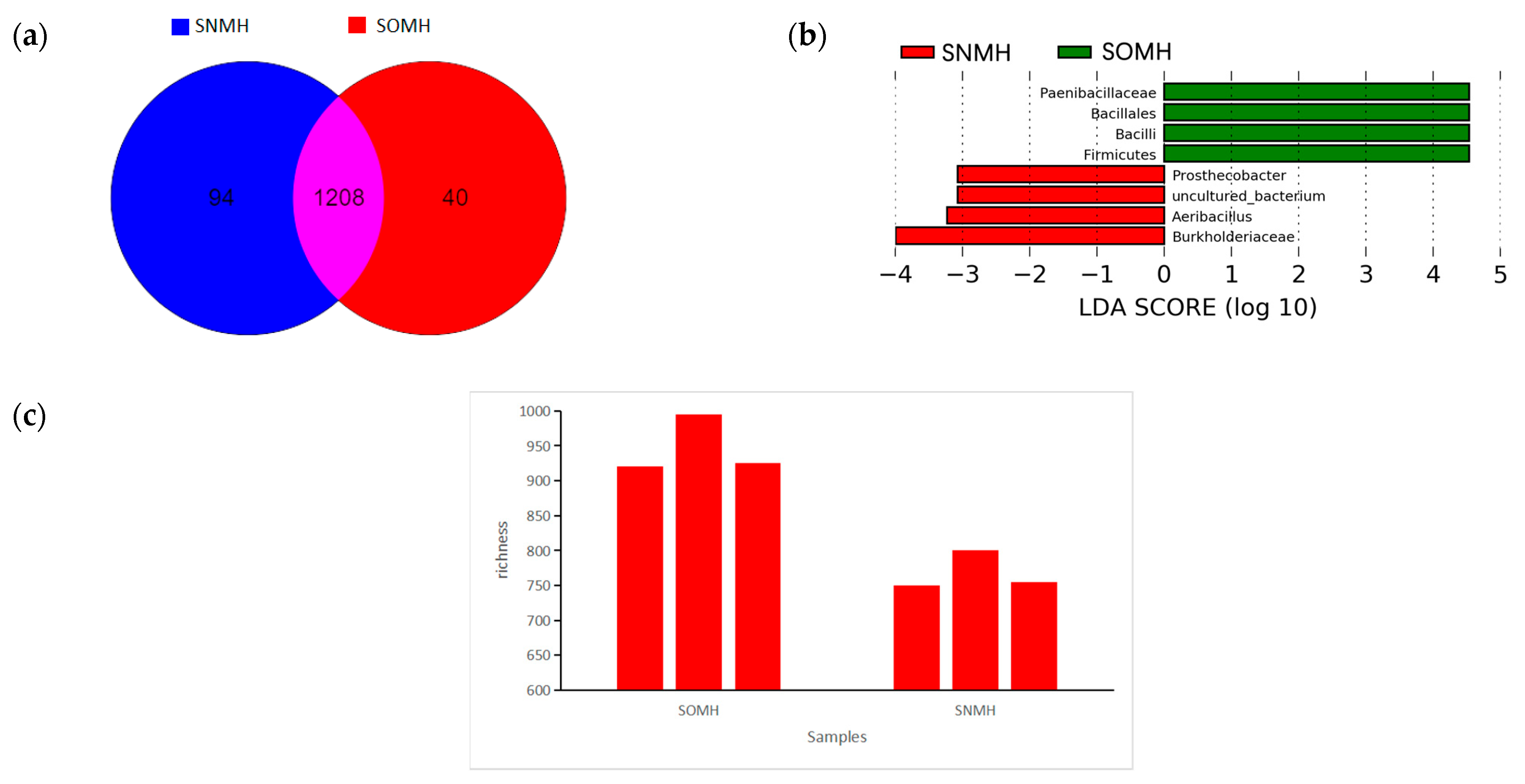
3.5. The Difference of Microbial Composition and Structure in the Substrate of Volvariella volvacea in Different Culture Environments
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.Q.; Geng, W.; Shen, Y.Q.; Wang, Y.L.; Dai, Y.C. Edible mushroom cultivation for food security and rural development in China: Bio-innovation, technological dissemination and marketing. Sustainability 2014, 6, 2961–2973. [Google Scholar] [CrossRef]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainsworth and Bisby’s Dictionary of the Fungi; CABI: Wallingford, UK, 2008. [Google Scholar]
- Sakinah, M.J.N.; Misran, A.; Mahmud, T.M.M.; Abdullah, S. A review: Production and postharvest management of Volvariella volvacea. Int. Food Res. J. 2019, 26, 367–376. [Google Scholar]
- Luo, Y.D. Study on Growth Characteristics of Mushroom and V1 Strain Cultivated on Cassava Skin. Master’s Thesis, Guangxi University China, Nanning, China, 2008. [Google Scholar]
- Wu, E.S.; Li, H.; Lv, J.; Li, Y.J.; Zhao, X.Y. Comparative analysis of nutritional components of 5 different varieties of straw mushroom. J. Edible Med. Fungi 2023, 31, 323–326. (In Chinese) [Google Scholar]
- Thapakorn, K.; Thanawan, P. Lead accumulation in the straw mushroom, Volvariella volvacea, from lead contaminated rice straw and stubble. Bull. Environ. Contam. Toxicol. 2013, 91, 231–234. [Google Scholar]
- Gong, M.; Zheng, T.T.; Wang, Y.; Wu, Y.Y.; Guo, Q.; Su, E.Z.; Zou, G.; Tan, Q.; Bao, D.P. The serine/threonine protein kinase VSKM1 promotes cold stress resistance of the postharvest Volvariella volvacea by enhancing mitoribosome activity. Postharvest Biol. Technol. 2023, 204, 112465. [Google Scholar] [CrossRef]
- Mathew, J.; Sudheesh, N.P.; Rony, K.A.; Smina, T.P.; Janardhanan, K.K. Antioxidant and antitumor activities of cultured mycelium of culinary-medicinal paddy straw mushroom Volvariella volvacea (Bull.: Fr.) singer (agaricomycetideae). Int. J. Med. Mushrooms 2008, 10, 139–147. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, S.J.; Zha, L.; Zhang, Y.R.; Li, Z.P.; Yu, C.X.; Chen, M.J. Influence of cellulase gene expression and cellulolytic activity on cellulose utilization in different Volvariella volvacea strains. Sci. Hortic. 2020, 261, 108956. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Data. Available online: http://www.fao.org/faostat/en/#home (accessed on 20 October 2023).
- Bao, D.P.; Gong, M.; Zheng, H.J.; Chen, M.J.; Zhang, L.; Wang, H.; Jiang, J.P.; Wu, L.; Zhu, Y.Q.; Zhu, G.; et al. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS ONE 2013, 8, e58294. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Chen, B.Z.; Mukhtar, I.; Xie, B.G.; Li, Z.; Meng, L. Characterization and expression pattern of homeobox transcription factors in fruiting body development of straw mushroom Volvariella volvacea. Fungal Biol. 2018, 123, 95–102. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, S.F.; Luo, D.D.; Mao, P.; Rosazlina, R.; Martin, F.; Xu, L.L. Decline in Morel Production upon Continuous Cropping Is Related to Changes in Soil Mycobiome. J. Fungi 2023, 9, 492. [Google Scholar] [CrossRef]
- Lan, Y.F.; Cong, Q.Q.; Wang, Q.W.; Tang, L.N.; Li, X.M.; Yu, Q.W.; Cui, X.; An, X.R.; Kong, F.H.; Li, X.D. First report of Cladobotryum protrusum causing cobweb disease on cultivated Morchella importuna. Plant Dis. 2020, 104, 977. [Google Scholar] [CrossRef]
- Wang, X.X.; Peng, J.Y.; Sun, L.; Bonito, G.; Guo, Y.X.; Li, Y.; Fu, Y.P. Genome sequencing of Paecilomyces penicillatus provides insights into its phylogenetic placement and mycoparasitism mechanisms on morel mushrooms. Pathogens 2021, 9, 834. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Liu, T.H.; Yu, Y.; Tang, J.; Jiang, L.; Martin, F.M.; Peng, W.H. Morel production related to soil microbial diversity and evenness. Microbiol. Spect 2021, 9, e0022921. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.M.; Jayawardena, R.S.; Thongklang, N.; Lv, M.L.; Zhu, X.T.; Zhao, Q. Morel production associated with soil nitrogen-fixing and nitrifying microorganisms. J. Fungi 2022, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Shao, G.G.; Zhou, T.; Fan, Q.H.; Yang, N.L.; Cui, M.; Zhang, J.W.; Wu, X.L.; Zhang, B.X.; Zhang, R.Y. Dazomet changes microbial communities and improves morel mushroom yield under continuous cropping. Front. Microbiol. 2023, 14, 1200226. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Y.; Gao, H.L.; Li, X.M.; Xiong, Y. Analysis of continuous cropping obstacles and prevention techniques of edible fungi. J. Yunnan Minzu Univ. 2023, 32, 558–562+588. (In Chinese) [Google Scholar]
- Chen, L.D.; Yan, M.; Qian, X.; Yang, Z.W.; Xu, Y.F.; Wang, T.J.; Cao, J.X.; Sun, S.J. Bacterial Community Composition in the Growth Process of Pleurotus eryngii and Growth-Promoting Abilities of Isolated Bacteria. Front. Microbiol. 2022, 13, 787628. [Google Scholar] [CrossRef] [PubMed]
- Halsey, J.A.; Silva, M.D.P.E.; Andreote, F.D. Bacterial selection by mycospheres of Atlantic rainforest mushrooms. Antonie Van Leeuwenhoek 2016, 109, 1353–1365. [Google Scholar] [CrossRef]
- Noble, R.; Dobrovin-Pennington, A.; Hobbs, P.J.; Pederby, J.; Rodger, A. Volatile C8 compounds and pseudomonads influence primordium formation of Agaricus bisporus. Mycologia 2009, 101, 583–591. [Google Scholar] [CrossRef]
- Noble, R.; Fermor, T.R.; Lincoln, S.; Dobrovin-Pennington, A.; Evered, C.; Mead, A.; Li, R. Primordia initiation of mushroom (Agaricus bisporus) strains on axenic casing materials. Mycologia 2003, 95, 620–629. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef]
- Qin, G.j.; Wang, X.; Chen, Q.J.; Zhang, G.Q. Changes in the main lignocellulosic degrading enzymes and material components during the production of Agaricus bisporus using different formula culture materials. Appl. J. Environ. Biol. 2017, 23, 1035–1041. (In Chinese) [Google Scholar]
- Bishop, E.L.; Pecchia, J.A.; Wilkinson, V.; Royse, D.J. Effects of Spent Mushroom Compost (SMC) as an Ingredient in Phase I Compost on Production of Agaricus bisporus. Compos. Sci. Util. 2016, 24, 246–258. [Google Scholar] [CrossRef]
- Zhang, H.L.; Wei, J.K.; Wang, Q.H.; Yang, R.; Gao, X.; Sang, Y.X.; Cai, P.P.; Zhang, G.Q.; Chen, Q.J. Lignocellulose utilization and bacterial communities of millet straw based mushroom (Agaricus bisporus) production. Sci. Rep. 2019, 9, 1151. [Google Scholar] [CrossRef]
- Loong, S.K.; Khor, C.S.; Jafar, F.L.; AbuBakar, S. Utility of 16S rDNA Sequencing for Identification of Rare Pathogenic Bacteria. J. Clin. Lab. Anal. 2016, 30, 1056–1060. [Google Scholar] [CrossRef]
- Luu, T.-T.-H.; Bui, D.-K.; Huynh, N.; Le, T.-L.; Green, I.D. Effect of the Cultivation Technology on the Yield of Paddy Straw Mushroom (Volvariella volvacea). Korean J. Mycol. 2022, 50, 161–171. [Google Scholar]
- Chiu, S.W.; Law, S.C.; Ching, M.L.; Cheung, K.W.; Chen, M.J. Themes for mushroom exploitation in the 21st century: Sustainability, waste management, and conservation. J. Gen. Appl. Microbiol. 2000, 46, 269–282. [Google Scholar] [CrossRef]
- Vieira, F.R.; Pecchia, J.A. Bacterial Community Patterns in the Agaricus bisporus Cultivation System, from Compost Raw Materials to Mushroom Caps. Microb. Ecol. 2022, 84, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Li, F.; Xu, X.; Liu, Y.C.; Kong, X.Q.; Chen, J.Q.; Liu, T.; Chen, L.D. Study on the community structure and function of symbiotic bacteria from different growth and developmental stages of Hypsizygus marmoreus. BMC Microbiol. 2020, 20, 311. [Google Scholar] [CrossRef]
- Palangi, V.; Kaya, A.; Kaya, A.; Giannenas, I. Ecofriendly Usability of Mushroom Cultivation Substrate as a Ruminant Feed: Anaerobic Digestion Using Gas Production Techniques. Animals 2022, 12, 1583. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, S.J.; Zhang, Y.R.; Sun, Y.H.; Huang, J.C.; Yu, C.X.; Chen, M.J. Progress of the application of microbial diversity analysis technology to the analysis of edible fungi fermentation media. Chin. J. Edible Fungi 2019, 26, 148–156. (In Chinese) [Google Scholar]
- Leao, I.; de Carvalho, T.B.; Henriques, V.; Ferreira, C.; Sampaio-Maia, B.; Manaia, C.M. Pseudomonadota in the oral cavity: A glimpse into the environment-human nexus. Appl. Microbiol. Biotechnol. 2022, 107, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.C.; Ahn, Y.N.; Khan, S.M.; Chung, I.M.; Won, H.Y.; Jhune, C.S.; Park, Y.J. The microbial population in the air of cultivation facility of oyster mushrooms. J. Microbiol. 2012, 50, 1053–1057. [Google Scholar] [CrossRef]
- Wee, W.Y.; Chew, X.Y.; Taheri, S.; Li Tan, X.; Teo, C.H. Whole genome sequencing and phylogenomic analyses of a novel glufosinate-tolerant Pseudomonas species. 3 Biotech 2022, 12, 123. [Google Scholar] [CrossRef]
- Nagesh, M.; Javeed, S.; Ramanujam, B.; Rangeswaran, R. Suitability of soil types for Paecilomyces lilacinus and Pochonia chlamydosporia and their performance against root-knot nematode, Meloidogyne incognita on Lycopersicon esculentum in glasshouse. Indian J. Agric. Sci. 2013, 83, 826–830. [Google Scholar]
- Taechowisan, T.; Peberdy, J.F.; Lumyong, S. Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J. Microbiol. Biotechnol. 2003, 19, 381–385. [Google Scholar] [CrossRef]
- Ye, J.L.; Bu, Y.N.; He, M.T.; Wu, Y.F.; Yang, X.T.; Zhang, L.L.; Song, X.Y. Genome-wide analysis of invertase gene family in wheat (Triticum aestivum L.) indicates involvement of TaCWINVs in pollen development. Plant Growth Regul. 2022, 98, 189. [Google Scholar] [CrossRef]
- Zhang, C.H.; Zhang, G.; Wen, Y.M.; Yang, Y.Q.; Liu, Y.F.; Zhang, H.; Wang, Z.H.; Qiu, L.Y. The role of 1-aminocyclopropane-1-carboxylic acid in the chemoattractant hyphae of Agaricus bisporus of Pseudomonas putida. J. North China Agric. Sci. 2019, 34, 209–218. (In Chinese) [Google Scholar]
- Shi, Q.C.; Abdel-Hamid, A.M.; Sun, Z.Y.; Cheng, Y.F.; Tu, T.; Cann, I.; Yao, B.; Zhu, W.Y. Carbohydrate-binding modules facilitate the enzymatic hydrolysis of lignocellulosic biomass: Releasing reducing sugars and dissociative lignin available for producing biofuels and chemicals. Biotechnol. Adv. 2023, 65, 108126. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.R. Influence of Different Treatment of Culture Material on Sterilization Effect in Edible Fungi Production. Master’s Thesis, Shanxi Agricultural University, Jinzhong, China, 2016. [Google Scholar]
- Luthfi, A.A.I.; Manaf, S.F.A.; Illias, R.M.; Harun, S.; Mohammad, A.W.; Jahim, J.M. Biotechnological route for sustainable succinate production utilizing oil palm frond and kenaf as potential carbon sources. Appl. Microbiol. Biotechnol. 2017, 101, 3055–3075. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Si, H.Y.; Fan, Y.B.; Wang, B.; Hua, D.L.; Wang, Z.X.; Dong, C.W. Microbiological Community Analysis of the Composting of Poplar Processing Residues. IOP Conf. Ser. Earth Environ. Sci. 2021, 781, 052025. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Thai, M. Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl. Microbiol. Biotechnol. 2018, 102, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Naraian, R. Enhanced growth and yield of oyster mushroom by growth-promoting bacteria Glutamicibacter arilaitensis MRC119. J. Basic Microbiol. 2021, 61, 45–54. [Google Scholar] [CrossRef]
- Barrett, C.F.; Parker, M.A. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 2006, 72, 1198–1206. [Google Scholar] [CrossRef] [PubMed]




| Sample | Contig | ||||
|---|---|---|---|---|---|
| Contig Number | N50 (bp) | Min (bp) | Max (bp) | Average Length (bp) | |
| NMH1 | 632,326 | 1172 | 200 | 547,145 | 896.3 |
| NMH2 | 507,465 | 756 | 200 | 190,352 | 709.5 |
| NMH3 | 710,665 | 876 | 200 | 308,874 | 764.6 |
| OMH1 | 541,423 | 1004 | 200 | 185,247 | 847.8 |
| OMH2 | 692,502 | 758 | 200 | 281,280 | 718.6 |
| OMH3 | 814,113 | 799 | 200 | 307,081 | 742.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, J.; Kang, L.; Peng, Y.; Ye, L.; Zhou, H.; Liu, M. Exploring the Influence of Culture Environment on the Yield of Volvariella volvacea Based on Microbiomics. Horticulturae 2024, 10, 204. https://doi.org/10.3390/horticulturae10030204
Liu Z, Wang J, Kang L, Peng Y, Ye L, Zhou H, Liu M. Exploring the Influence of Culture Environment on the Yield of Volvariella volvacea Based on Microbiomics. Horticulturae. 2024; 10(3):204. https://doi.org/10.3390/horticulturae10030204
Chicago/Turabian StyleLiu, Zhu, Jianhao Wang, Linzhi Kang, Yangyang Peng, Luyao Ye, Hui Zhou, and Ming Liu. 2024. "Exploring the Influence of Culture Environment on the Yield of Volvariella volvacea Based on Microbiomics" Horticulturae 10, no. 3: 204. https://doi.org/10.3390/horticulturae10030204
APA StyleLiu, Z., Wang, J., Kang, L., Peng, Y., Ye, L., Zhou, H., & Liu, M. (2024). Exploring the Influence of Culture Environment on the Yield of Volvariella volvacea Based on Microbiomics. Horticulturae, 10(3), 204. https://doi.org/10.3390/horticulturae10030204




