CRISPR/Cas in Grapevine Genome Editing: The Best Is Yet to Come
Abstract
:1. Introduction
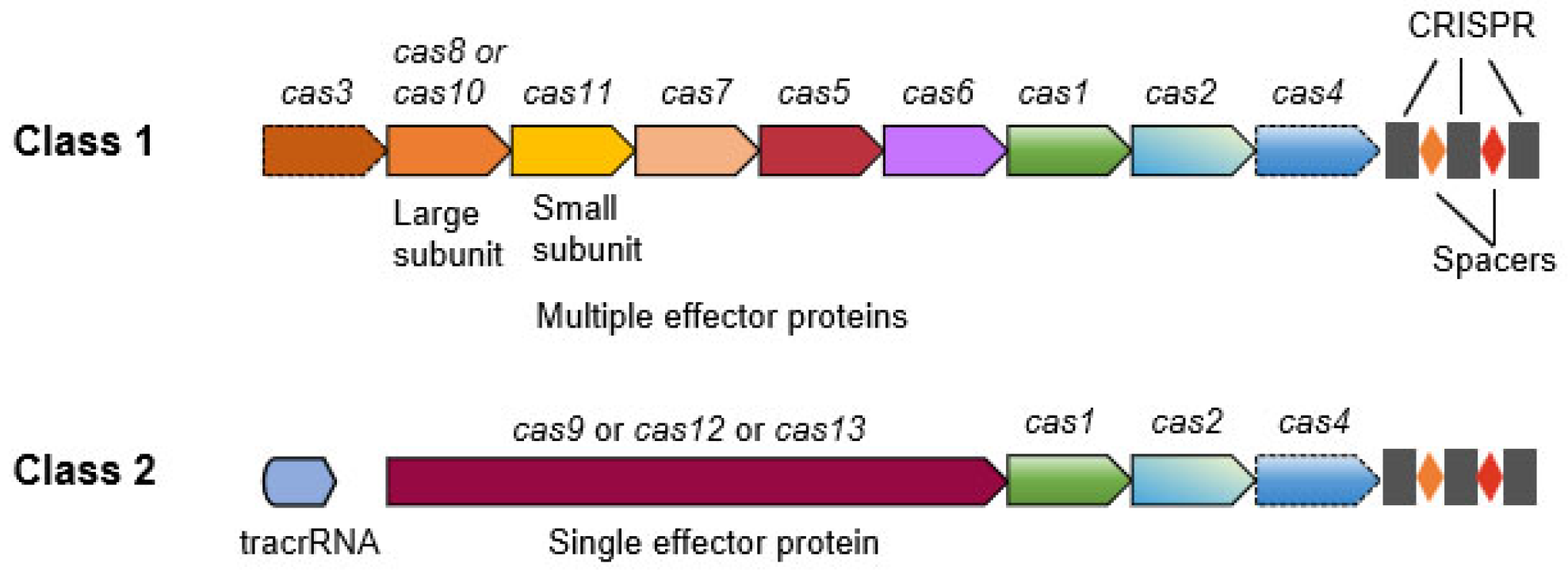
2. CRISPR/Cas Nucleases
2.1. DNA-Targeting Cas Proteins
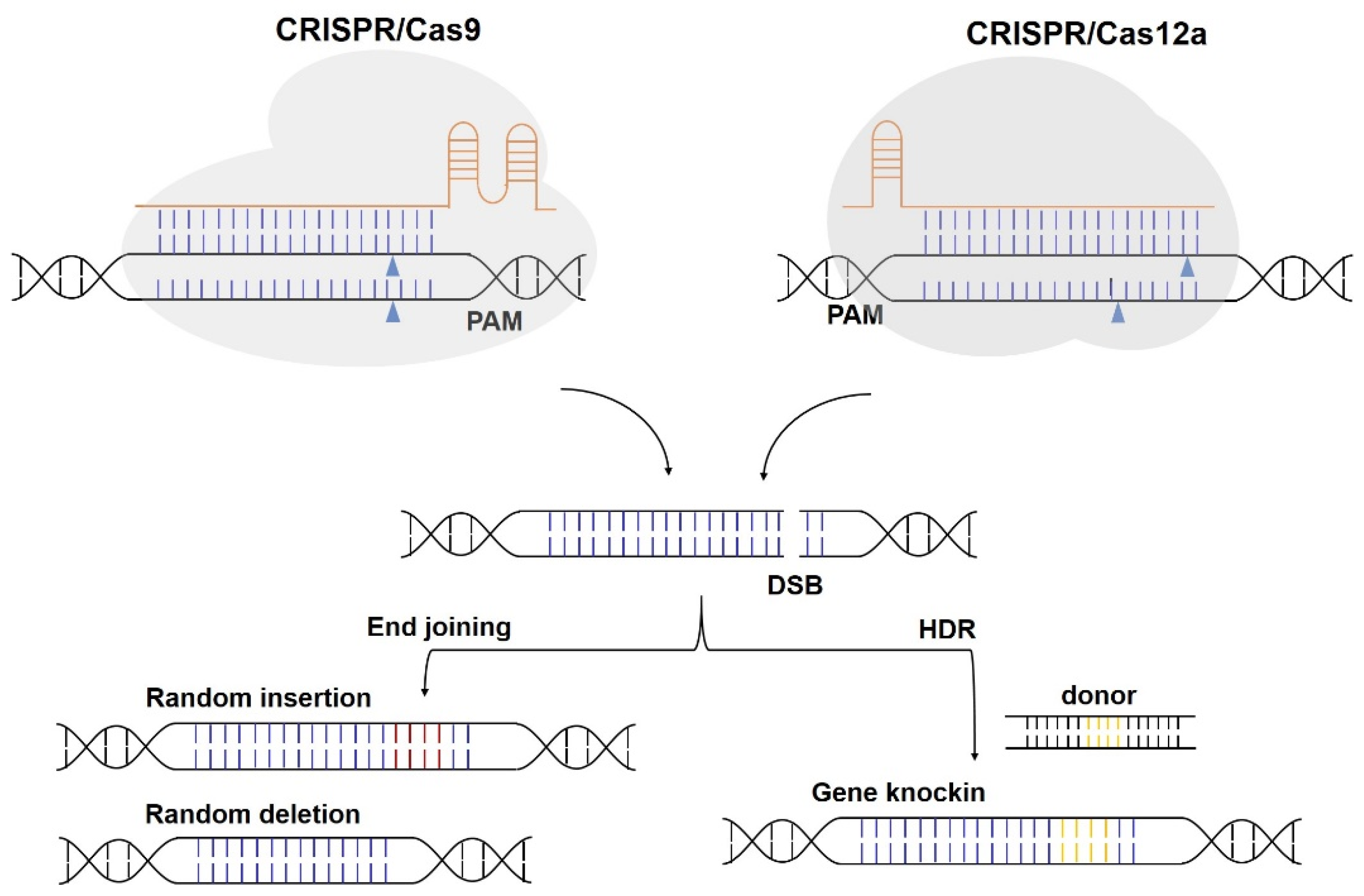
2.1.1. Cas9 Nucleases
2.1.2. Cas12 Nucleases
2.1.3. Base Editors
2.1.4. Prime Editors
2.2. RNA-Targeting Cas Proteins
2.2.1. Cas13 Nucleases
2.2.2. RNA-Targeting Cas9
3. Genome Editing in Grapevine
3.1. Proof-of-Concept Studies on CRISPR/Cas9
3.2. Applications of CRISPR/Cas9 in Grapevine
3.3. Optimization of CRISPR/Cas9 for Grapevine Genome Editing
3.4. Multiplex Genome Editing in Grapevine
3.5. CRISPR/dCas9-Mediated Gene Activation
3.6. Base Editing in Grapevine
3.7. Other CRISPR/Cas Systems Used in Grapevine
3.8. Online Tools for Guide RNA Design
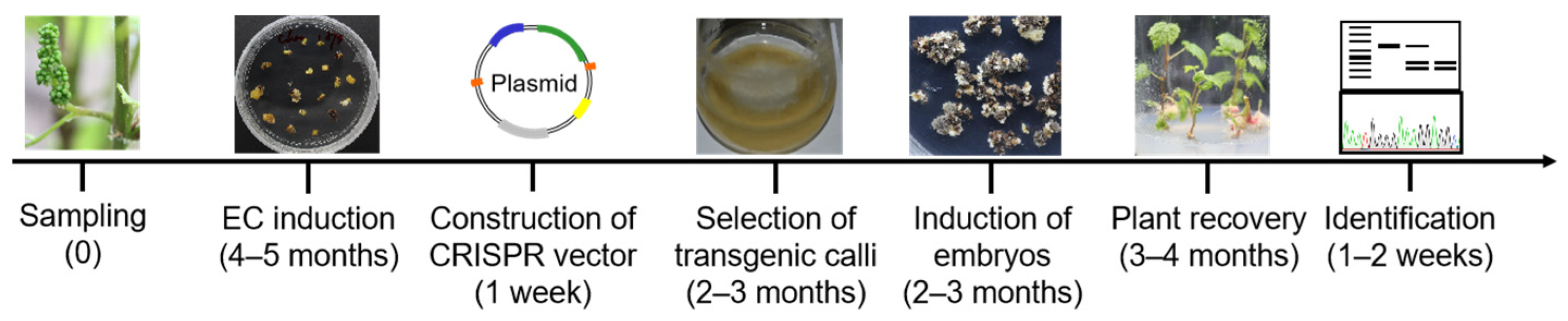
4. Challenges for Grapevine Genome Editing
4.1. Lack of Efficient System for Testing Cas Effectors
4.2. Limited Available Explants
4.3. Delivery of CRISPR/Cas Reagents and Plant Regeneration
5. Regulation of Gene-Edited Grapevine Plants
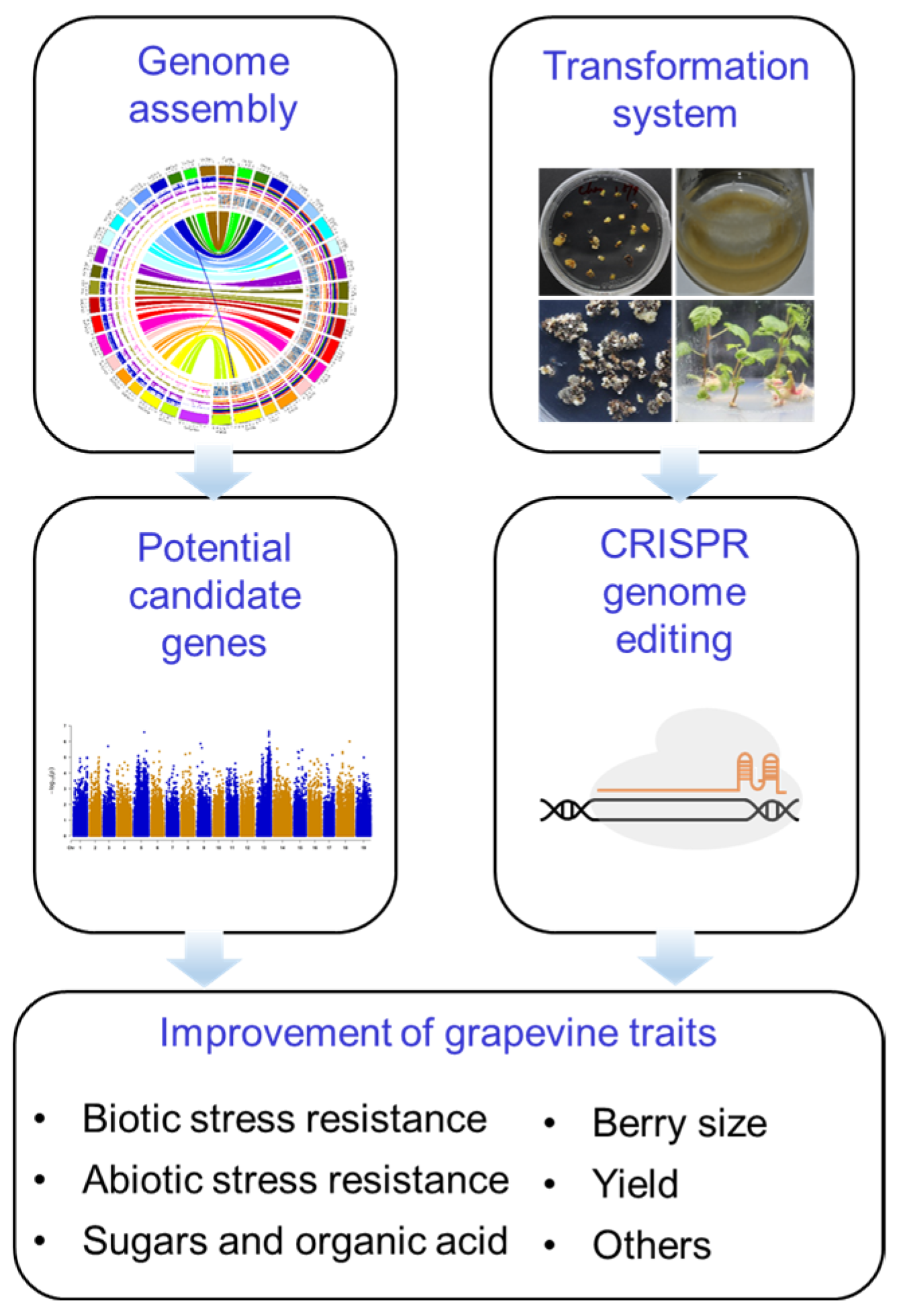
6. Future Perspectives
6.1. Taking Advantage of the Model System
6.2. Exploiting the Precise Genome Editing
6.3. Accelerating the Improvement of Grapevine Traits
6.4. Generating Transgene-Free, Edited Grapevine Plants
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the Iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc.Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Ren, C.; Li, H.; Liu, Y.; Li, S.; Liang, Z. Highly efficient activation of endogenous gene in grape using CRISPR/dCas9-based transcriptional activators. Hortic. Res. 2022, 9, uhab037. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wheatley, M.; Meakem, V.; Galarneau, E.; Gutierrez, B.; Zhong, G.Y. Editing VvDXS1 for the creation of muscat flavour in Vitis vinifera cv. Scarlet Royal. Plant Biotechnol. J. 2024, 22, 1610–1621. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Shamshirgaran, Y.; Liu, J.; Sumer, H.; Verma, P.J.; Taheri-Ghahfarokhi, A. Tools for Efficient Genome Editing; ZFN, TALEN, and CRISPR. Methods Mol. Biol. 2022, 2495, 29–46. [Google Scholar] [PubMed]
- Taheri-Ghahfarokhi, A.; Taylor, B.J.M.; Nitsch, R.; Lundin, A.; Cavallo, A.L.; Madeyski-Bengtson, K.; Karlsson, F.; Clausen, M.; Hicks, R.; Mayr, L.M.; et al. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 2018, 46, 8417–8434. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Schiml, S.; Fauser, F.; Puchta, H. The CRISPR/Cas9 system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014, 80, 1139–1150. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Rusk, N. CRISPRs and epigenome editing. Nat. Methods 2014, 11, 28. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR/Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Lee, S.; Kim, Y.; Park, J.; Min, S.; Choi, J.W.; Huang, T.P.; Yoon, S.; Liu, D.R.; Kim, H.H. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 2020, 4, 111–124. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR/Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR/Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR/Cas9 systems enable specific editing of the human genome. Mol. Ter. 2016, 24, 636–644. [Google Scholar] [CrossRef]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Chatterjee, P.; Jakimo, N.; Jacobson, J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018, 4, eaau0766. [Google Scholar] [CrossRef]
- Edraki, A.; Mir, A.; Ibraheim, R.; Gainetdinov, I.; Yoon, Y.; Song, C.Q.; Cao, Y.; Gallant, J.; Xue, W.; Rivera-Pérez, J.A.; et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 2019, 73, 714–726.e4. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR/Cas9 nucleases with no detectable genome-wide of-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Kulcsár, P.I.; Tálas, A.; Huszár, K.; Ligeti, Z.; Tóth, E.; Weinhardt, N.; Fodor, E.; Welker, E. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol. 2017, 18, 190. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specifc SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR/Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 2016, 6, 38169. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17103. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, C.; Liu, Q.; Fu, Y.; Wang, K. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genom. 2017, 44, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Zhang, Y.; You, Q.; Tang, X.; Ren, Q.; Liu, S.; Yang, L.; Wang, Y.; Liu, X.; Liu, B.; et al. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol. Plant 2018, 11, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Pietralla, J.; Capdeville, N.; Schindele, P.; Puchta, H. Optimizing ErCas12a for efficient gene editing in Arabidopsis thaliana. Plant Biotechnol. J. 2024, 22, 401–412. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, Q.; Tang, X.; Liu, S.; Malzahn, A.A.; Zhou, J.; Wang, J.; Yin, D.; Pan, C.; Yuan, M.; et al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 2021, 12, 1944. [Google Scholar] [CrossRef]
- Zhang, L.; Li, G.; Zhang, Y.; Cheng, Y.; Roberts, N.; Glenn, S.E.; DeZwaan-McCabe, D.; Rube, H.T.; Manthey, J.; Coleman, G. Boosting genome editing efficiency in human cells and plants with novel LbCas12a variants. Genome Biol. 2023, 24, 102. [Google Scholar] [CrossRef]
- Huang, H.; Huang, G.; Tan, Z.; Hu, Y.; Shan, L.; Zhou, J.; Zhang, X.; Ma, S.; Lv, W.; Huang, T.; et al. Engineered Cas12a-Plus nuclease enables gene editing with enhanced activity and specificity. BMC Biol. 2022, 20, 91. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.K.; Armstrong, B.; Schindele, P.; Puchta, H. Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. Plant Biotechnol. J. 2021, 19, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef]
- Tak, Y.E.; Kleinstiver, B.P.; Nuñez, J.K.; Hsu, J.Y.; Horng, J.E.; Gong, J.; Weissman, J.S.; Joung, J.K. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 2017, 14, 1163–1166. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Liu, Y.; Yang, B.; Wang, X.; Wei, J.; Lu, Z.; Zhang, Y.; Wu, J.; Huang, X.; et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Xu, P.; Faure, G.; Maguire, S.; Kannan, S.; Altae-Tran, H.; Vo, S.; Desimone, A.; Macrae, R.K.; Zhang, F. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 2023, 620, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Druteika, G.; Bigelyte, G.; Budre, K.; Zedaveinyte, R.; Silanskas, A.; Kazlauskas, D.; Venclovas, Č.; Siksnys, V. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 2021, 599, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grunewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef]
- Ren, B.; Yan, F.; Kuang, Y.; Li, N.; Zhang, D.; Zhou, X.; Lin, H.; Zhou, H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant 2018, 11, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.L.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950–954. [Google Scholar] [CrossRef]
- Neugebauer, M.E.; Hsu, A.; Arbab, M.; Krasnow, N.A.; McElroy, A.N.; Pandey, S.; Doman, J.L.; Huang, T.P.; Raguram, A.; Banskota, S.; et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 2023, 41, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, B.; Ru, G.; Meng, H.; Yan, Y.; Hong, M.; Zhang, D.; Luan, C.; Zhang, S.; Wu, H.; et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat. Biotechnol. 2023, 41, 663–672. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Sretenovic, S.; Ren, Q.; Yang, L.; Bao, Y.; Qi, C.; Yuan, M.; He, Y.; Liu, S.; Liu, X.; et al. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 2019, 12, 1027–1036. [Google Scholar] [CrossRef]
- Wang, M.; Xu, Z.; Gosavi, G.; Ren, B.; Cao, Y.; Kuang, Y.; Zhou, C.; Spetz, C.; Yan, F.; Zhou, X.; et al. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J. 2020, 18, 1645–1647. [Google Scholar] [CrossRef]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.K. Precise A·T to G·C base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Lam, D.K.; Rees, H.A.; Sola-Esteves, N.M.; Barrera, L.A.; Born, D.A.; Edwards, A.; Gehrke, J.M.; Lee, S.J.; Liquori, A.J.; et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020, 38, 892–900. [Google Scholar] [CrossRef]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E.; et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020, 38, 883–891. [Google Scholar] [CrossRef]
- Wei, C.; Wang, C.; Jia, M.; Guo, H.X.; Luo, P.Y.; Wang, M.G.; Zhu, J.K.; Zhang, H. Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J. Integr. Plant Biol. 2021, 63, 1595–1599. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zeng, D.; Zhao, Y.; Wang, Y.; Liu, T.; Li, S.; Xue, Y.; Luo, Y.; Xie, X.; Chen, L.; et al. PhieABEs: A PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 2022, 20, 934–943. [Google Scholar] [CrossRef]
- Yan, D.; Ren, B.; Liu, L.; Yan, F.; Li, S.; Wang, G.; Sun, W.; Zhou, X.; Zhou, H. High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant 2021, 14, 722–731. [Google Scholar] [CrossRef]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Shen, R.; Li, Z.; Yao, Q.; Zhang, X.; Zhong, D.; Tan, X.; Song, M.; Han, H.; Zhu, J.K.; et al. Efficient C-to-G editing in rice using an optimized base editor. Plant Biotechnol. J. 2022, 20, 1238–1240. [Google Scholar] [CrossRef]
- Tong, H.; Wang, X.; Liu, Y.; Liu, N.; Li, Y.; Luo, J.; Ma, Q.; Wu, D.; Li, J.; Xu, C.; et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat Biotechnol. 2023, 41, 1080–1084. [Google Scholar] [CrossRef]
- Tong, H.; Liu, N.; Wei, Y.; Zhou, Y.; Li, Y.; Wu, D.; Jin, M.; Cui, S.; Li, H.; Li, G.; et al. Programmable deaminase-free base editors for G-to-Y conversion by engineered glycosylase. Natl. Sci. Rev. 2023, 10, nwad143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, B.; Chen, L.; Xie, L.; Yu, W.; Wang, Y.; Li, L.; Yin, S.; Yang, L.; Hu, H.; et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020, 38, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zong, Y.; Jin, S.; Zhu, H.; Lin, D.; Li, S.; Qiu, J.L.; Wang, Y.; Gao, C. SWISS: Multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol. 2020, 21, 141. [Google Scholar] [CrossRef]
- Liang, Y.; Xie, J.; Zhang, Q.; Wang, X.; Gou, S.; Lin, L.; Chen, T.; Ge, W.; Zhuang, Z.; Lian, M.; et al. AGBE: A dual deaminase-mediated base editor by fusing CGBE with ABE for creating a saturated mutant population with multiple editing patterns. Nucleic Acids Res. 2022, 50, 5384–5399. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, C.; He, Y.; Li, S.; Yan, L.; Li, Y.; Zhu, Z.; Xia, L. Plant base editing and prime editing: The current status and future perspectives. J. Integr. Plant Biol. 2023, 65, 444–467. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, G.; Huang, S.; Li, X.; Wang, X.; Li, G.; Chi, T.; Chen, Y.; Huang, X.; Wang, X. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021, 31, 1134–1136. [Google Scholar] [CrossRef]
- Li, X.; Zhou, L.; Gao, B.Q.; Li, G.; Wang, X.; Wang, Y.; Wei, J.; Han, W.; Wang, Z.; Li, J.; et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat. Commun. 2022, 13, 1669. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chen, W.; Suiter, C.C.; Lee, C.; Chardon, F.M.; Yang, W.; Leith, A.; Daza, R.M.; Martin, B.; Shendure, J. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 2022, 40, 218–226. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, X.O.; Weng, Z.; Xue, W. Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 2022, 40, 227–234. [Google Scholar] [CrossRef]
- Wang, J.; He, Z.; Wang, G.; Zhang, R.; Duan, J.; Gao, P.; Lei, X.; Qiu, H.; Zhang, C.; Zhang, Y.; et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 2022, 19, 331–340. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J. Mol. Biol. 2019, 431, 66–87. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Terns, M.P. CRISPR-based Technologies: Impact of RNA-targeting Systems. Mol. Cell 2018, 72, 404–412. [Google Scholar] [CrossRef]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar]
- Liu, Y.; Chen, Z.; He, A.; Zhan, Y.; Li, J.; Liu, L.; Wu, H.; Zhuang, C.; Lin, J.; Zhang, Q.; et al. Targeting cellular mRNAs translation by CRISPR/Cas9. Sci. Rep. 2016, 6, 29652. [Google Scholar] [CrossRef] [PubMed]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169. [Google Scholar] [CrossRef]
- Strutt, S.C.; Torrez, R.M.; Kaya, E.; Negrete, O.A.; Doudna, J.A. RNA-dependent RNA targeting by CRISPR/Cas9. eLife 2018, 7, e32724. [Google Scholar] [CrossRef] [PubMed]
- Dugar, G.; Leenay, R.T.; Eisenbart, S.K.; Bischler, T.; Aul, B.U.; Beisel, C.L.; Sharma, C.M. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell 2018, 69, 893–905.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Ren, C.; Zhong, G.Y.; Yang, L.; Li, S.; Liang, Z. Identification of genomic sites for CRISPR/Cas9-based genome editing in the Vitis vinifera genome. BMC Plant Biol. 2016, 16, 96. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, e0177966. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef]
- Wan, D.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef]
- Olivares, F.; Loyola, R.; Olmedo, B.; Miccono, M.L.Á.; Aguirre, C.; Vergara, R.; Riquelme, D.; Madrid, G.; Plantat, P.; Mora, R.; et al. CRISPR/Cas9 Targeted Editing of Genes Associated with Fungal Susceptibility in Vitis vinifera L. cv. Thompson Seedless Using Geminivirus-Derived Replicons. Front. Plant Sci. 2021, 12, 791030. [Google Scholar] [CrossRef]
- Li, M.Y.; Jiao, Y.T.; Wang, Y.T.; Zhang, N.; Wang, B.B.; Liu, R.Q.; Yin, X.; Xu, Y.; Liu, G.T. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Djennane, S.; Gersch, S.; Le-Bohec, F.; Piron, M.C.; Baltenweck, R.; Lemaire, O.; Merdinoglu, D.; Hugueney, P.; Nogué, F.; Mestre, P. CRISPR/Cas9 editing of Downy mildew resistant 6 (DMR6-1) in grapevine leads to reduced susceptibility to Plasmopara viticola. J. Exp. Bot. 2024, 75, 2100–2112. [Google Scholar] [CrossRef]
- Giacomelli, L.; Zeilmaker, T.; Giovannini, O.; Salvagnin, U.; Masuero, D.; Franceschi, P.; Vrhovsek, U.; Scintilla, S.; Rouppe van der Voort, J.; Moser, C. Simultaneous editing of two DMR6 genes in grapevine results in reduced susceptibility to downy mildew. Front. Plant Sci. 2023, 14, 1242240. [Google Scholar] [CrossRef]
- Ren, F.; Ren, C.; Zhang, Z.; Duan, W.; Lecourieux, D.; Li, S.; Liang, Z. Efficiency Optimization of CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape. Front. Plant Sci. 2019, 10, 612. [Google Scholar] [CrossRef]
- Medvedieva, M.O.; Blume, Y.B. Legal regulation of plant genome editing with the CRISPR/Cas9 technology as an example. Cytol. Genet. 2018, 52, 204–212. [Google Scholar] [CrossRef]
- Su, W.; Xu, M.; Radani, Y.; Yang, L. Technological development and application of plant genetic transformation. Int. J. Mol. Sci. 2023, 24, 10646. [Google Scholar] [CrossRef]
- Ren, C.; Guo, Y.; Kong, J.; Lecourieux, F.; Dai, Z.; Li, S.; Liang, Z. Knockout of VvCCD8 gene in grapevine affects shoot branching. BMC Plant Biol. 2020, 20, 47. [Google Scholar] [CrossRef] [PubMed]
- Sunitha, S.; Rock, C.D. CRISPR/Cas9-mediated targeted mutagenesis of TAS4 and MYBA7 loci in grapevine rootstock 101-14. Transgenic Res. 2020, 29, 355–367. [Google Scholar] [CrossRef]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 52. [Google Scholar] [CrossRef]
- Iocco-Corena, P.; Chaïb, J.; Torregrosa, L.; Mackenzie, D.; Thomas, M.R.; Smith, H.M. VviPLATZ1 is a major factor that controls female flower morphology determination in grapevine. Nat. Commun. 2021, 12, 6995. [Google Scholar] [CrossRef]
- Tu, M.; Fang, J.; Zhao, R.; Liu, X.; Yin, W.; Wang, Y.; Wang, X.; Wang, X.; Fang, Y. CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera). Hortic. Res. 2022, 9, uhac022. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Lu, M.; Li, X.; Sun, H.; Cheng, Z.; Miao, Y.; Fu, Y.; Zhang, X. An Efficient Agrobacterium rhizogenes-Mediated Hairy Root Transformation Method in a Soybean Root Biology Study. Int. J. Mol. Sci. 2022, 23, 12261. [Google Scholar] [CrossRef]
- Yang, Y.; Ke, J.; Han, X.; Wuddineh, W.A.; Song, G.Q.; Zhong, G.Y. Removal of a 10-kb Gret1 transposon from VvMybA1 of Vitis vinifera cv. Chardonnay. Hortic. Res. 2022, 9, uhac201. [Google Scholar] [CrossRef]
- Nakajima, I.; Kawahigashi, H.; Nishitani, C.; Azuma, A.; Haji, T.; Toki, S.; Endo, M. Targeted deletion of grape retrotransposon associated with fruit skin color via CRISPR/Cas9 in Vitis labrascana ‘Shine Muscat’. PLoS ONE 2023, 18, e0286698. [Google Scholar] [CrossRef]
- Clemens, M.; Faralli, M.; Lagreze, J.; Bontempo, L.; Piazza, S.; Varotto, C.; Malnoy, M.; Oechel, W.; Rizzoli, A.; Dalla Costa, L. VvEPFL9-1 Knock-Out via CRISPR/Cas9 Reduces Stomatal Density in Grapevine. Front. Plant Sci. 2022, 13, 878001. [Google Scholar] [CrossRef]
- Scintilla, S.; Salvagnin, U.; Giacomelli, L.; Zeilmaker, T.; Malnoy, M.A.; Rouppe van der Voort, J.; Moser, C. Regeneration of non-chimeric plants from DNA-free edited grapevine protoplasts. Front. Plant Sci. 2022, 13, 1078931. [Google Scholar] [CrossRef] [PubMed]
- Villette, J.; Lecourieux, F.; Bastiancig, E.; Héloir, M.C.; Poinssot, B. New improvements in grapevine genome editing: High efficiency biallelic homozygous knock-out from regenerated plantlets by using an optimized zCas9i. Plant Methods 2024, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mansour, H.; Wang, T.; Poojari, S.; Li, F. Naked-Eye Detection of Grapevine Red-Blotch Viral Infection Using a Plasmonic CRISPR Cas12a Assay. Anal. Chem. 2019, 91, 11510–11513. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.; Dean, N. Dramatic improvement of CRISPR/Cas9 editing in Candida albicans by increased single guide RNA expression. mSphere 2017, 2, e00385-16. [Google Scholar] [CrossRef]
- Long, L.; Guo, D.D.; Gao, W.; Yang, W.W.; Hou, L.P.; Ma, X.N.; Miao, Y.C.; Botella, J.R.; Song, C.P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef]
- Li, B.; Fu, C.; Zhou, J.; Hui, F.; Wang, Q.; Wang, F.; Wang, G.; Xu, Z.; Che, L.; Yuan, D.; et al. Highly Efficient Genome Editing Using Geminivirus-Based CRISPR/Cas9 System in Cotton Plant. Cells 2022, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Mao, Y.; Ha, S.; Liu, W.; Botella, J.R.; Zhu, J.K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016, 35, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, T.; Yang, N.; Xu, M.; Yan, L.; Wang, L.; Wang, R.; Zhao, Y. Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J. Genet. Genom. 2017, 44, 469–472. [Google Scholar]
- Tang, X.; Ren, Q.; Yang, L.; Bao, Y.; Zhong, Z.; He, Y.; Liu, S.; Qi, C.; Liu, B.; Wang, Y.; et al. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 2019, 17, 1431–1445. [Google Scholar] [CrossRef]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef]
- Shi, L.; Su, J.; Cho, M.J.; Song, H.; Dong, X.; Liang, Y.; Zhang, Z. Promoter editing for the genetic improvement of crops. J. Exp. Bot. 2023, 74, 4349–4366. [Google Scholar] [CrossRef]
- Xing, S.; Chen, K.; Zhu, H.; Zhang, R.; Zhang, H.; Li, B.; Gao, C. Fine-tuning sugar content in strawberry. Genome Biol. 2020, 21, 230. [Google Scholar] [CrossRef]
- Zinelabidine, L.H.; Torres-Pérez, R.; Grimplet, J.; Baroja, E.; Ibáñez, S.; Carbonell-Bejerano, P.; Martínez-Zapater, J.M.; Ibáñez, J.; Tello, J. Genetic variation and association analyses identify genes linked to fruit set-related traits in grapevine. Plant Sci. 2021, 306, 110875. [Google Scholar]
- Tello, J.; Torres-Pérez, R.; Flutre, T.; Grimplet, J.; Ibáñez, J. VviUCC1 Nucleotide Diversity, Linkage Disequilibrium and Association with Rachis Architecture Traits in Grapevine. Genes 2020, 11, 598. [Google Scholar] [CrossRef]
- Ren, C.; Lin, Y.; Li, H.; Li, S.; Liang, Z. Targeted genome editing in grape using multiple CRISPR-guided editing systems. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ren, C.; Gathunga, E.K.; Li, X.; Li, H.; Kong, J.; Dai, Z.; Liang, Z. Efficient genome editing in grapevine using CRISPR/LbCas12a system. Mol. Hortic. 2023, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.; Hao, X.; Liu, Z.; Liu, M.; Wang, J.; Liu, L.; Liu, N.; Song, R.; Zhang, J.; Fang, Y.; et al. Engineering CRISPR immune systems conferring GLRaV-3 resistance in grapevine. Hortic. Res. 2022, 9, uhab023. [Google Scholar] [CrossRef]
- Wang, Y.; Lecourieux, F.; Zhang, R.; Dai, Z.; Lecourieux, D.; Li, S.; Liang, Z. Data Comparison and Software Design for Easy Selection and Application of CRISPR-based Genome Editing Systems in Plants. Genom. Proteom. Bioinf. 2021, 19, 937–948. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A Convenient Software Toolkit for CRISPR-Based Genome Editing. Mol. Plant. 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and mTALE-Act Systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Trinidad, J.L.; Longkumer, T.; Kohli, A. Rice Protoplast Isolation and Transfection for Transient Gene Expression Analysis. Methods Mol. Biol. 2021, 2238, 313–324. [Google Scholar]
- Najafi, S.; Bertini, E.; D’Incà, E.; Fasoli, M.; Zenoni, S. DNA-free genome editing in grapevine using CRISPR/Cas9 ribonucleoprotein complexes followed by protoplast regeneration. Hortic. Res. 2022, 10, uhac240. [Google Scholar] [CrossRef]
- Kong, Q.; Li, J.; Wang, S.; Feng, X.; Shou, H. Combination of Hairy Root and Whole-Plant Transformation Protocols to Achieve Efficient CRISPR/Cas9 Genome Editing in Soybean. Plants 2023, 12, 1017. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Bahramnejad, B.; Douleti Baneh, H.; Emamifar, A.; Goodwin, P.H. Hairy root culture optimization and resveratrol production from Vitis vinifera subsp. sylvesteris. World J. Microbiol. Biotechnol. 2017, 33, 67. [Google Scholar] [CrossRef]
- Tisserant, L.P.; Aziz, A.; Jullian, N.; Jeandet, P.; Clément, C.; Courot, E.; Boitel-Conti, M. Enhanced Stilbene Production and Excretion in Vitis vinifera cv Pinot Noir Hairy Root Cultures. Molecules 2016, 21, 1703. [Google Scholar] [CrossRef] [PubMed]
- Franks, T.K.; Powell, K.S.; Choimes, S.; Marsh, E.; Iocco, P.; Sinclair, B.J.; Ford, C.M.; van Heeswijck, R. Consequences of transferring three sorghum genes for secondary metabolite (cyanogenic glucoside) biosynthesis to grapevine hairy roots. Transgenic Res. 2006, 15, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Cutanda-Perez, M.C.; Ageorges, A.; Gomez, C.; Vialet, S.; Terrier, N.; Romieu, C.; Torregrosa, L. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 2009, 69, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Ruffa, P.; Vallania, R.; Gribaudo, I. Somatic embryogenesis from whole flowers, anthers and ovaries of grapevine (Vitis spp.). Plant Cell Tiss. Org. Cult. 2007, 90, 79–83. [Google Scholar] [CrossRef]
- Dai, L.; Zhou, Q.; Li, R.; Du, Y.; He, J.; Wang, D.; Cheng, S.; Zhang, J.; Wang, Y. Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell Tiss. Org. Cult. 2015, 121, 397–412. [Google Scholar] [CrossRef]
- Iocco, P.; Franks, T.; Thomas, M. Genetic transformation of major wine grape cultivars of Vitis vinifera L. Transgenic Res. 2001, 10, 105–112. [Google Scholar] [CrossRef]
- Gambino, G.; Nuzzo, F.; Moine, A.; Chitarra, W.; Pagliarani, C.; Petrelli, A.; Boccacci, P.; Delliri, A.; Velasco, R.; Nerva, L.; et al. Genome editing of a recalcitrant wine grape genotype by lipofectamine-mediated delivery of CRISPR/Cas9 ribonucleoproteins to protoplasts. Plant J. 2024, 119, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Hou, Q.; Wei, X.; Qi, Y.; Pu, A.; Wu, S.; An, X.; Wan, X. Promoting genotype-independent plant transformation by manipulating developmental regulatory genes and/or using nanoparticles. Plant Cell Rep. 2023, 42, 1395–1417. [Google Scholar] [CrossRef]
- Vidal, J.R.; Kikkert, J.R.; Wallace, P.G.; Reisch, B.I. High-efficiency biolistic co-transformation and regeneration of ‘Chardonnay’ (Vitis vinifera L.) containing npt-II and antimicrobial peptide genes. Plant Cell Rep. 2003, 22, 252–260. [Google Scholar] [CrossRef]
- Vidal, J.R.; Kikkert, J.R.; Donzelli, B.D.; Wallace, P.G.; Reisch, B.I. Biolistic transformation of grapevine using minimal gene cassette technology. Plant Cell Rep. 2006, 25, 807–814. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 2018, 54, 240–252. [Google Scholar] [CrossRef]
- Chen, J.; Tomes, S.; Gleave, A.P.; Hall, W.; Luo, Z.; Xu, J.; Yao, J.L. Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic. Res. 2022, 9, uhab014. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef]
- Wang, B.; Huang, J.; Zhang, M.; Wang, Y.; Wang, H.; Ma, Y.; Zhao, X.; Wang, X.; Liu, C.; Huang, H.; et al. Carbon dots enable efficient delivery of functional DNA in plants. ACS Appl. Bio Mater. 2020, 3, 8857–8864. [Google Scholar] [CrossRef]
- Caradus, J.R. Processes for regulating genetically modified and gene edited plants. GM Crops Food 2023, 14, 1–41. [Google Scholar] [CrossRef]
- Eckerstorfer, M.F.; Engelhard, M.; Heissenberger, A.; Simon, S.; Teichmann, H. Plants developed by new genetic modification techniques-comparison of existing regulatory frameworks in the EU and NonEU countries. Front. Bioeng. Biotechnol. 2019, 7, 26. [Google Scholar] [CrossRef]
- McHughen, A. A critical assessment of regulatory triggers for products of biotechnology: Product vs. process. GM Crops Food 2016, 7, 125–158. [Google Scholar] [CrossRef]
- Broll, H.; Braeuning, A.; Lampen, A. European Court of Justice decision for genome editing: Consequences on food/feed risk assessment and detection. Food Control 2019, 104, 288–291. [Google Scholar] [CrossRef]
- Puchta, H. Regulation of gene-edited plants in Europe: From the valley of tears into the shining sun? Abiotech 2023, 5, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A.K. Global regulation of genetically modified crops aimed the gene edited crop boom-A review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Chakravorty, N.; Bansal, K.C. Genome editing for food, nutrition, and health. Nucleus 2024, 67, 1–4. [Google Scholar] [CrossRef]
- Tsuda, M.; Watanabe, K.N.; Ohsawa, R. Regulatory status of genome-edited organisms under the Japanese cartagena act. Front. Bioeng. Biotechnol. 2019, 7, 387. [Google Scholar] [CrossRef]
- Lemke, S.L. Gene editing in plants. a nutrition professional’s guide to the science, regulatory, and social considerations. Nutr. Today 2022, 57, 57–63. [Google Scholar] [CrossRef]
- Bohle, F.; Schneider, R.; Mundorf, J.; Zühl, L.; Simon, S.; Engelhard, M. Where does the EU-path on new genomic techniques lead us? Front. Genome Ed. 2024, 6, 1377117. [Google Scholar] [CrossRef]
- Chaïb, J.; Torregrosa, L.; Mackenzie, D.; Corena, P.; Bouquet, A.; Thomas, M.R. The grape microvine -a model system for rapid forward and reverse genetics of grapevines. Plant J. 2010, 62, 1083–1092. [Google Scholar] [CrossRef]
- Dai, X.; Chen, X.; Fang, Q.; Li, J.; Bai, Z. Inducible CRISPR genome-editing tool: Classifications and future trends. Crit. Rev. Biotechnol. 2018, 38, 573–586. [Google Scholar] [CrossRef]
- Wang, X.; Ye, L.; Lyu, M.; Ursache, R.; Löytynoja, A.; Mähönen, A.P. An inducible genome editing system for plants. Nat. Plants 2020, 6, 766–772. [Google Scholar] [CrossRef]
- Dong, Y.; Duan, S.; Xia, Q.; Liang, Z.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunić, G.; Bert, P.F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, K.; Li, H.; Kuang, Y.; Liang, Z. Biography of Vitis genomics: Recent advances and prospective. Hortic. Res. 2024, 11, uhae128. [Google Scholar] [CrossRef] [PubMed]
- Uddin, A.; Gallardo, R.K.; Rickard, B.; Alston, J.; Sambucci, O. Consumer acceptance of new plant-breeding technologies: An application to the use of gene editing in fresh table grapes. PLoS ONE 2022, 17, e0270792. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 20155. [Google Scholar] [CrossRef]
- Tricoli, D.M.; Debernardi, J.M. An efficient protoplast-based genome editing protocol for Vitis species. Hortic. Res. 2023, 11, uhad266. [Google Scholar] [CrossRef]

| Effector | Target | Explant | Delivery Method | Editing Type | Trait | Phenotype | Reference |
|---|---|---|---|---|---|---|---|
| SpCas9 | IdnDH | EC of Chardonnay | Agrobacterium tumefaciens | KO | Tartaric acid synthesis | Decreased TA content | [16] |
| MLO7 | Protoplasts from Chardonnay EC | PEG | KO | Powdery mildew resistance | ND | [17] | |
| VvPDS | EC of Neo Muscat | Agrobacterium tumefaciens | KO | Carotenoid biosynthesis | Albino phenotype | [117] | |
| VvPDS | EC of Chardonnay and 41B | Agrobacterium tumefaciens | KO | Carotenoid biosynthesis | ND | [124] | |
| VvPDS | Protoplasts from EC of 101-14, Cabernet Sauvignon, Chardonnay, Merlot, Thompson Seedless, Colombard, GRN1, V. arizonica, and Pixie | PEG | KO | Carotenoid biosynthesis | ND | [125] | |
| VvPDS | EC of Nebbiolo | PEG and lipofectamines | KO | Carotenoid biosynthesis | Albino phenotype | [126] | |
| VvWRKY52 | EC of Thompson Seedless | Agrobacterium tumefaciens | KO | Botrytis cinerea resistance | Enhanced resistance | [118] | |
| CCD8 | EC of 41B | Agrobacterium tumefaciens | KO | Shoot branching | Increased branches | [127] | |
| VvPR4b | EC of Thompson Seedless | Agrobacterium tumefaciens | KO | Downy mildew resistance | Decreased resistance | [121] | |
| VvMLO3, VvMLO4 | EC of Thompson Seedless | Agrobacterium tumefaciens | KO | Powdery mildew resistance | Enhanced resistance | [119] | |
| TAS4, MYBA7 | EC of rootstock 101-14 | Agrobacterium tumefaciens | KO | Anthocyanin accumulation | No accumulation | [128] | |
| PDS TMT1, TMT2 | EC of 41B | Agrobacterium tumefaciens | KO | Carotenoid biosynthesis sugar accumulation | Albino phenotype Decreased sugar content | [129] | |
| VvAIR12, VvSWEET4, VvLIN2, VvDEL1 | EC of Thompson Seedless | Agrobacterium tumefaciens | KO | Botrytis cinerea resistance | Enhanced resistance | [120] | |
| VvPLATZ1 | EC of microvine, genotype 04C023V0006 (H/H) | Agrobacterium tumefaciens | KO | Female flower morphology | Reflex stamens | [130] | |
| VvbZIP36 | EC of Thompson Seedless | Agrobacterium tumefaciens | KO | Anthocyanin accumulation | Increased anthocyanin content | [131] | |
| VvDMR6 | EC of Crimson seedless and Sugraone | PEG | KO | Downy mildew resistance | ND | [132] | |
| VvMLO6 | Powdery mildew resistance | ND | |||||
| VvMYBA1 | EC of Chardonnay | Agrobacterium tumefaciens | KO | Anthocyanin accumulation | ND | [133] | |
| EC of Shine Muscat | ND | [134] | |||||
| VvEPFL9-1 | EC of Sugraone | Agrobacterium tumefaciens | KO | Stomata formation | Reduced stomatal density | [135] | |
| VvDMR6-1 | EC of 41B | Agrobacterium tumefaciens | KO | Downy mildew resistance | Enhanced resistance | [122] | |
| GFP | Protoplasts from Thompson Seedless EC | PEG | KO | GFP fluorescence | Loss of GFP fluorescence | [136] | |
| VvDMR6-1, VvDMR6-2 | EC of Crimson seedless and Sugraone | Agrobacterium tumefaciens | KO | Downy mildew resistance | Enhanced resistance | [123] | |
| zCas9i | LysM receptor-like kinase gene (Vitvi05g00623) | EC of Chardonnay | Agrobacterium tumefaciens | KO | Immune response | ND | [137] |
| LbCas12a | TMT1, TMT2, DFR1 | EC of 41B | Agrobacterium tumefaciens | KO | Sugar accumulation flavonoid accumulation | Altered sugar and flavonoid contents | [138] |
| PE | VvDXS1 | EC of Scarlet Royal | Agrobacterium tumefaciens | Base editing | Monoterpenes biosynthesis | Increased monoterpenes content | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, C.; Mohamed, M.S.M.; Aini, N.; Kuang, Y.; Liang, Z. CRISPR/Cas in Grapevine Genome Editing: The Best Is Yet to Come. Horticulturae 2024, 10, 965. https://doi.org/10.3390/horticulturae10090965
Ren C, Mohamed MSM, Aini N, Kuang Y, Liang Z. CRISPR/Cas in Grapevine Genome Editing: The Best Is Yet to Come. Horticulturae. 2024; 10(9):965. https://doi.org/10.3390/horticulturae10090965
Chicago/Turabian StyleRen, Chong, Mohamed Salaheldin Mokhtar Mohamed, Nuremanguli Aini, Yangfu Kuang, and Zhenchang Liang. 2024. "CRISPR/Cas in Grapevine Genome Editing: The Best Is Yet to Come" Horticulturae 10, no. 9: 965. https://doi.org/10.3390/horticulturae10090965








