Synthesis and Application of Natural Deep Eutectic Solvents (NADESs) for Upcycling Horticulture Residues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Chemicals
2.1.2. Horticulture Residue
2.2. Methodology
2.2.1. Preparation of Natural Deep Eutectic Solvents (NADESs)
2.2.2. Characterization of NADESs
Density
Water Activity
Viscosity
pH
Spectroscopic Technique
2.3. Application of NADESs for Bioactive Molecule Extraction
2.3.1. Experimental Design
2.3.2. Optimization
2.4. Extraction Procedure
2.5. Antioxidant Power
2.5.1. Total Phenolic Content (TPC)
2.5.2. Total Flavonoid Content (TFC)
2.6. Antiradical Scavenging Activities
2.6.1. Ferric Reducing Antioxidant Power (FRAP)
2.6.2. Cupric Ion Reducing Antioxidant Capacity (CUPRAC) Assay
2.6.3. 2,2-Diphenyl-2-Picrylhydrazyl (DPPH) Assay
2.7. Statistical Analysis
3. Results
3.1. Synthesis of NADESs
3.2. Characterization of NADESs
3.2.1. Physical Properties
3.2.2. Molecular Properties
3.3. Application of NADESs for Extraction
3.3.1. Extraction Yields
3.3.2. Effects of Extraction Conditions on Antioxidant Yield
Effects of Extraction Conditions on TPC
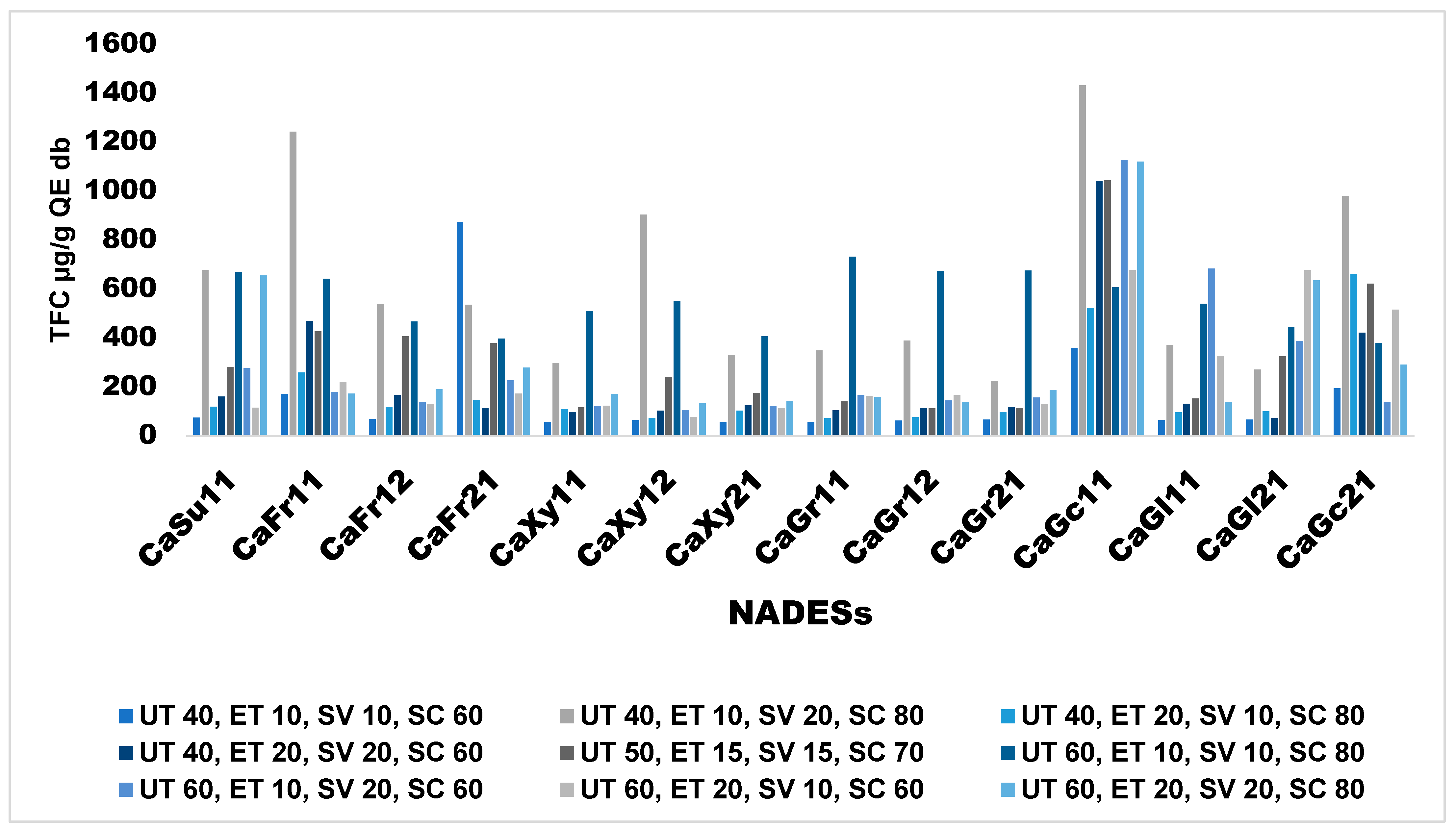
Effects of Extraction Conditions on TFC
3.3.3. Effects of Extraction Conditions on Antiradical Scavenging Activity
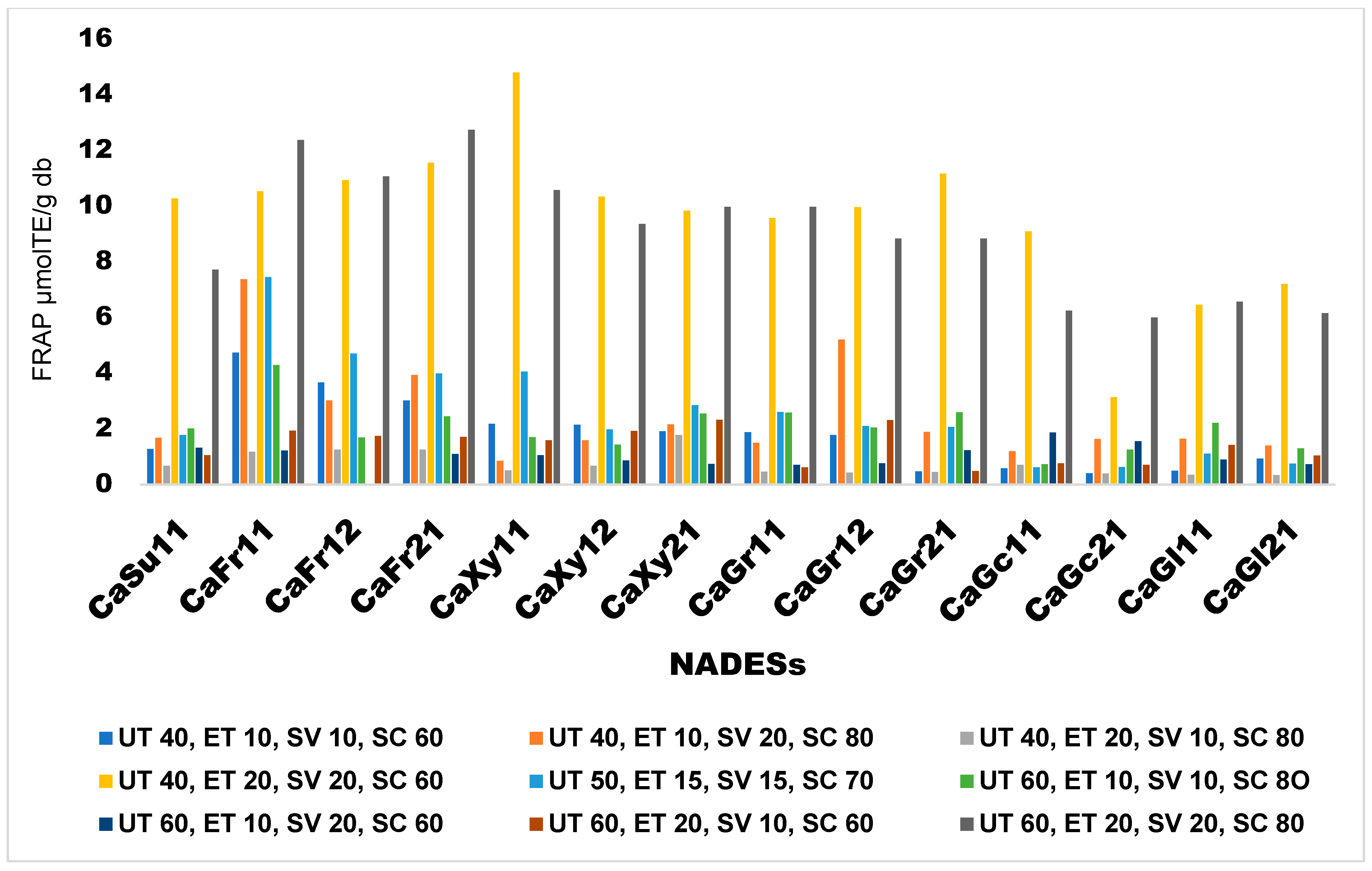
Effects of Extraction Conditions on FRAP
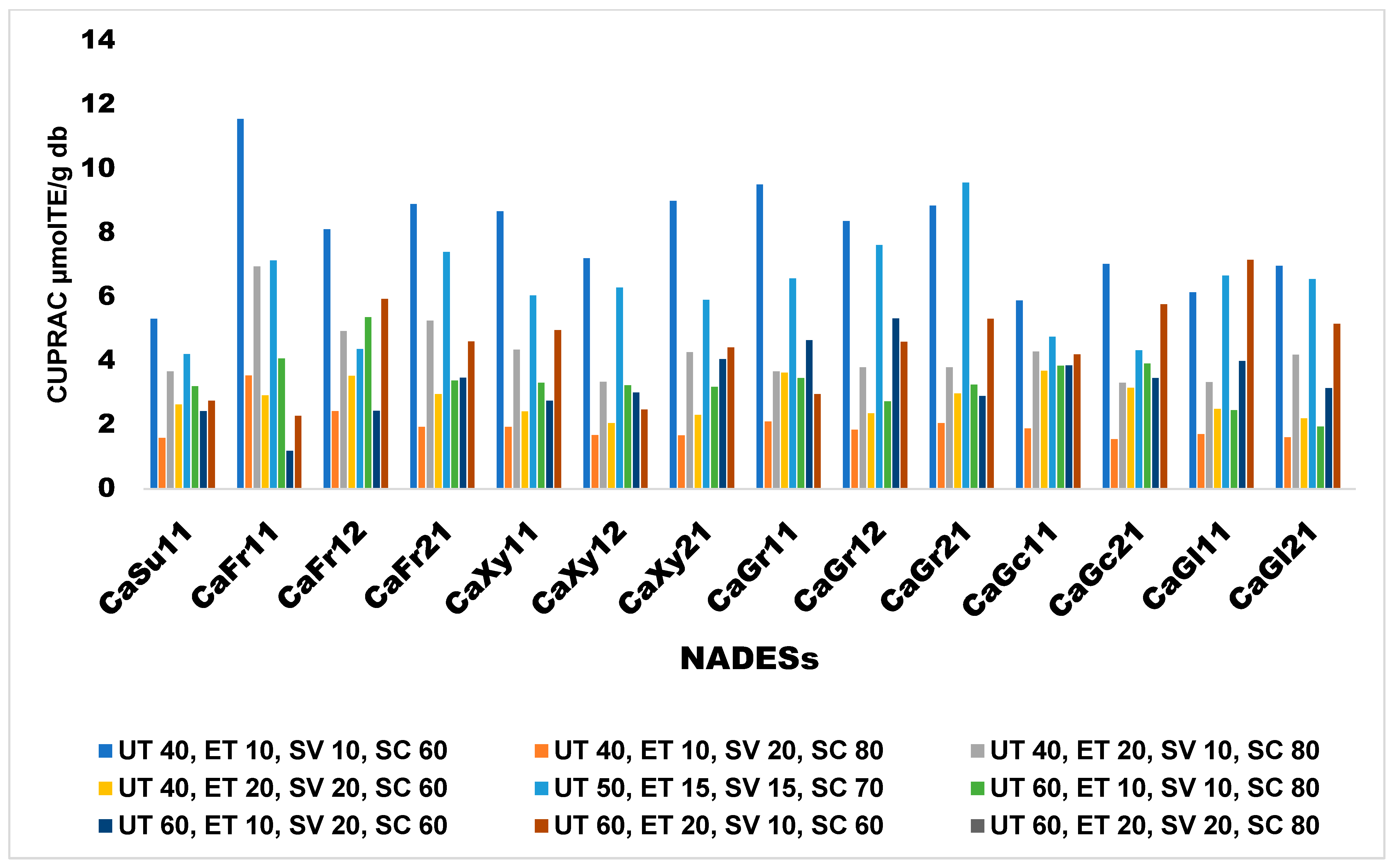
Effects of Extraction Conditions on CUPRAC
3.4. Rotatable Central Composite Response Surface Methodology (RCCRSM)
3.5. Optimization of the Extraction Conditions and Validation of the Models
4. Discussions
4.1. Synthesis of NADESs
4.2. Characterization of NADES
4.2.1. Physical Properties of NADESs
Density
Water Activity
Viscosity
pH
4.3. Molecular Properties
4.4. Application of NADESs for Extraction
4.4.1. Extraction Yields
4.4.2. Effects of Extraction Conditions on Antioxidants
4.4.3. Efficiency of NADESs as Solvent for Polyphenol Extraction
4.5. Effects of Extraction Conditions on Antiradical Scavenging Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NADESs | Natural deep eutectic solvents |
| ANP | African nutmeg peel |
| UAE | Ultrasound-assisted extraction |
| UAS | Ultrasound-assisted synthesis |
| UT | Ultrasound (extraction) temperature |
| ET | Extraction time |
| SV | Solvent (NADES) volume |
| SC | Solvent (NADES) concentration |
| CUPRAC | Cupric ion reducing antioxidant capacity |
| FRAP | Ferric reducing antioxidant power |
| DPPH | 2,2-Diphenyl-2-picrylhydrazyl |
| ARA | Antiradical activity |
| TPC | Total phenolic content |
| TFC | Total flavonoid content |
| ChCl-LA | Choline chloride–lactic acid |
| HBA | Hydrogen bond acceptor |
| HBD | Hydrogen bond donor |
| MFAT | Many factors at a time |
| RCCRSM | Rotatable central composite response surface methodology |
| CHCL | Choline chloride |
| FTIR | Fourier transform infrared spectroscopy |
| LSD | Least significant difference |
| SP | Supplementary plot |
| SF | Supplementary figure |
References
- Sharma, A.; Soni, R.; Soni, S.K. From Waste to Wealth: Exploring Modern Composting Innovations and Compost Valorization. J. Mater. Cycles Waste Manag. 2024, 26, 20–48. [Google Scholar] [CrossRef]
- Waqas, M.; Hashim, S.; Humphries, U.W.; Ahmad, S.; Noor, R.; Shoaib, M.; Naseem, A.; Hlaing, P.T.; Lin, H.A. Composting Processes for Agricultural Waste Management: A Comprehensive Review. Processes 2023, 11, 731. [Google Scholar] [CrossRef]
- Okeke, U.J.; Micucci, M.; Mihaylova, D.; Cappiello, A. The Effects of Experimental Conditions on Extraction of Polyphenols from African Nutmeg Peels Using NADESs-UAE: A Multifactorial Modelling Technique. Sci. Rep. 2025, 15, 4890. [Google Scholar] [CrossRef]
- Zhuang, B.; Dou, L.-L.; Li, P.; Liu, E.-H. Deep Eutectic Solvents as Green Media for Extraction of Flavonoid Glycosides and Aglycones from Platycladi cacumen. J. Pharm. Biomed. Anal. 2017, 134, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zeng, J.; Tang, H.; Cheng, Y.; Tan, J.; Li, T.; Li, X.; He, J.; Zhang, Y. Effect of Deep Eutectic Solvent Extraction on Auricularia Auricula Polysaccharide Solubilization and Antioxidant Potential. Sustain. Chem. Pharm. 2023, 34, 101166. [Google Scholar] [CrossRef]
- Yoo, D.E.; Jeong, K.M.; Han, S.Y.; Kim, E.M.; Jin, Y.; Lee, J. Deep Eutectic Solvent-Based Valorization of Spent Coffee Grounds. Food Chem. 2018, 255, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Zannou, O.; Pashazadeh, H.; Ghellam, M.; Ibrahim, S.A.; Koca, I. Extraction of Anthocyanins from Borage (Echium amoenum) Flowers Using Choline Chloride and a Glycerol-Based, Deep Eutectic Solvent: Optimization, Antioxidant Activity, and In Vitro Bioavailability. Molecules 2021, 27, 134. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Wang, F.; Jian, Y.; Chernyshev, V.M.; Zhang, Y.; Wang, Z.; Yuan, Z.; Chen, X. Extraction of Flavonoids from Glycyrrhiza Residues Using Deep Eutectic Solvents and Its Molecular Mechanism. J. Mol. Liq. 2022, 363, 119848. [Google Scholar] [CrossRef]
- Yang, G.-Y.; Song, J.-N.; Chang, Y.-Q.; Wang, L.; Zheng, Y.-G.; Zhang, D.; Guo, L. Natural Deep Eutectic Solvents for the Extraction of Bioactive Steroidal Saponins from Dioscoreae Nipponicae Rhizoma. Molecules 2021, 26, 2079. [Google Scholar] [CrossRef]
- Wei, Z.; Qi, X.; Li, T.; Luo, M.; Wang, W.; Zu, Y.; Fu, Y. Application of Natural Deep Eutectic Solvents for Extraction and Determination of Phenolics in Cajanus Cajan Leaves by Ultra Performance Liquid Chromatography. Sep. Purif. Technol. 2015, 149, 237–244. [Google Scholar] [CrossRef]
- Rathee, P.; Sehrawat, R.; Rathee, P.; Khatkar, A.; Akkol, E.K.; Khatkar, S.; Redhu, N.; Türkcanoğlu, G.; Sobarzo-Sánchez, E. Polyphenols: Natural Preservatives with Promising Applications in Food, Cosmetics and Pharma Industries; Problems and Toxicity Associated with Synthetic Preservatives; Impact of Misleading Advertisements; Recent Trends in Preservation and Legislation. Materials 2023, 16, 4793. [Google Scholar] [CrossRef] [PubMed]
- Aires, A. Polyphenols Applications in Pharmaceutic and Cosmetic Industries. In Technologies to Recover Polyphenols from AgroFood By-Products and Wastes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 337–357. ISBN 978-0-323-85273-9. [Google Scholar]
- Clijnk, D.; Codera, V.; Pou, J.O.; Fernandez-Garcia, J.; Gonzalez-Olmos, R. Enhancing Circular Economy of Waste Refrigerants Management Using Deep Eutectic Solvents. Sustain. Mater. Technol. 2024, 41, e01062. [Google Scholar] [CrossRef]
- Gao, H.; Xiao, J.; Wang, Y.; Yu, Y.; Wu, J.; Zhang, J.; Wang, Z.; Wang, S.; Luo, Q. Advancing Circular Economy: Biorefinery of Agri-Waste via Green Multicomponent Deep Eutectic Solvent Pretreatment. Ind. Crops Prod. 2025, 226, 120673. [Google Scholar] [CrossRef]
- Gomez-Urios, C.; Puchades-Colera, P.; Frigola, A.; Esteve, M.J.; Blesa, J.; Lopez-Malo, D. Natural Deep Eutectic Solvents: A Paradigm of Stability and Permeability in the Design of New Ingredients. J. Mol. Liq. 2024, 412, 125864. [Google Scholar] [CrossRef]
- Hikmawanti, N.P.E.; Ramadon, D.; Jantan, I.; Mun’im, A. Natural Deep Eutectic Solvents (NADES): Phytochemical Extraction Performance Enhancer for Pharmaceutical and Nutraceutical Product Development. Plants 2021, 10, 2091. [Google Scholar] [CrossRef] [PubMed]
- Olfat, A.; Mostaghim, T.; Shahriari, S.; Salehifar, M. Extraction of Bioactive Compounds of Hypnea Flagelliformis by Ultrasound-Assisted Extraction Coupled with Natural Deep Eutectic Solvent and Enzyme Inhibitory Activity. Algal Res. 2024, 78, 103388. [Google Scholar] [CrossRef]
- Agiriga, A.; Siwela, M. Monodora myristica (Gaertn.) Dunal: A Plant with Multiple Food, Health and Medicinal Applications: A Review. Am. J. Food Technol. 2017, 12, 271–284. [Google Scholar] [CrossRef]
- Florindo, C.; Oliveira, F.S.; Rebelo, L.P.N.; Fernandes, A.M.; Marrucho, I.M. Insights into the Synthesis and Properties of Deep Eutectic Solvents Based on Cholinium Chloride and Carboxylic Acids. ACS Sustain. Chem. Eng. 2014, 2, 2416–2425. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, B.; Ferreira-Santos, P.; Alfonso, I.M.; Martínez, S.; Genisheva, Z.; Gullón, B. Deep Eutectic Solvents as a Green Tool for the Extraction of Bioactive Phenolic Compounds from Avocado Peels. Molecules 2022, 27, 6646. [Google Scholar] [CrossRef]
- Zhang, M.; Tian, R.; Han, H.; Wu, K.; Wang, B.; Liu, Y.; Zhu, Y.; Lu, H.; Liang, B. Preparation Strategy and Stability of Deep Eutectic Solvents: A Case Study Based on Choline Chloride-Carboxylic Acid. J. Clean. Prod. 2022, 345, 131028. [Google Scholar] [CrossRef]
- Kang, K.; Jia, X.; Zheng, K.; Wang, X.; Liu, B.; Hou, Y. Physical Properties of Natural Deep Eutectic Solvent and Its Application in Remediation of Heavy Metal Lead in Soil. J. Contam. Hydrol. 2023, 258, 104222. [Google Scholar] [CrossRef] [PubMed]
- Gomez, F.J.V.; Espino, M.; Fernández, M.A.; Silva, M.F. A Greener Approach to Prepare Natural Deep Eutectic Solvents. ChemistrySelect 2018, 3, 6122–6125. [Google Scholar] [CrossRef]
- Pelosi, C.; Gonzalez-Rivera, J.; Bernazzani, L.; Tiné, M.R.; Duce, C. Optimized Preparation, Thermal Characterization and Microwave Absorption Properties of Deep Eutectic Solvents Made by Choline Chloride and Hydrated Salts of Alkali Earth Metals. J. Mol. Liq. 2023, 371, 121104. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments, 9th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; ISBN 978-1-119-11347-8. [Google Scholar]
- Cabrera, L.; Xavier, L.; Zecchi, B. Extraction of Phenolic Compounds with Antioxidant Activity from Olive Pomace Using Natural Deep Eutectic Solvents: Modelling and Optimization by Response Surface Methodology. Discov. Food 2024, 4, 29. [Google Scholar] [CrossRef]
- Alburquerque, J. Agrochemical Characterisation of “Alperujo”, a Solid by-Product of the Two-Phase Centrifugation Method for Olive Oil Extraction. Bioresour. Technol. 2004, 91, 195–200. [Google Scholar] [CrossRef]
- Ianni, F.; Scandar, S.; Mangiapelo, L.; Blasi, F.; Marcotullio, M.C.; Cossignani, L. NADES-Assisted Extraction of Polyphenols from Coriander Seeds: A Systematic Optimization Study. Antioxidants 2023, 12, 2048. [Google Scholar] [CrossRef]
- Kivrak, İ.; Duru, M.E.; Öztürk, M.; Mercan, N.; Harmandar, M.; Topçu, G. Antioxidant, Anticholinesterase and Antimicrobial Constituents from the Essential Oil and Ethanol Extract of Salvia potentillifolia. Food Chem. 2009, 116, 470–479. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. [2] Ferric Reducing/Antioxidant Power Assay: Direct Measure of Total Antioxidant Activity of Biological Fluids and Modified Version for Simultaneous Measurement of Total Antioxidant Power and Ascorbic Acid Concentration. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 299, pp. 15–27. ISBN 978-0-12-182200-2. [Google Scholar]
- Lim, C.S.H.; Lim, S.L. Ferric Reducing Capacity Versus Ferric Reducing Antioxidant Power for Measuring Total Antioxidant Capacity. Lab. Med. 2013, 44, 51–55. [Google Scholar] [CrossRef]
- Apak, R.; Güçlü, K.; Özyürek, M.; Bektaşoğlu, B.; Bener, M. Cupric Ion Reducing Antioxidant Capacity Assay for Antioxidants in Human Serum and for Hydroxyl Radical Scavengers. In Advanced Protocols in Oxidative Stress II; Armstrong, D., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 594, pp. 215–239. ISBN 978-1-60761-410-4. [Google Scholar]
- Akyüz, E.; Türkoğlu, S.; Sözgen Başkan, K.; Tütem, E.; Apak, M.R. Comparison of Antioxidant Capacities and Antioxidant Components of Commercial Bitter Melon (Momordicacharantia L.) Products. Turk. J. Chem. 2020, 44, 1663–1673. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Wang, H.; Tao, Y.; Masuku, M.V.; Cao, J.; Yang, J.; Huang, K.; Ge, Y.; Yu, Y.; Xiao, Z.; Kuang, Y.; et al. Effects of Deep Eutectic Solvents on the Biotransformation Efficiency of ω-Transaminase. J. Mol. Liq. 2023, 377, 121379. [Google Scholar] [CrossRef]
- Hayyan, M.; Mbous, Y.P.; Looi, C.Y.; Wong, W.F.; Hayyan, A.; Salleh, Z.; Mohd-Ali, O. Natural Deep Eutectic Solvents: Cytotoxic Profile. SpringerPlus 2016, 5, 913. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Tailoring Properties of Natural Deep Eutectic Solvents with Water to Facilitate Their Applications. Food Chem. 2015, 187, 14–19. [Google Scholar] [CrossRef]
- Zaman, S.U.; Mehdi, M.S.; Umar, M.; Rafiq, S.; Saif-ur-Rehman; Zaman, M.K.U.; Javed, M.D.; Waseem, M.A.; Tahir, N. Preparation of Ammonium Persulfate/Glycerol Based Novel Deep Eutectic Solvent under Controlled Conditions; Characterizations, Physical Properties. J. Mol. Struct. 2023, 1283, 135265. [Google Scholar] [CrossRef]
- Rodriguez Rodriguez, N.; van den Bruinhorst, A.; Kollau, L.J.B.M.; Kroon, M.C.; Binnemans, K. Degradation of Deep-Eutectic Solvents Based on Choline Chloride and Carboxylic Acids. ACS Sustain. Chem. Eng. 2019, 7, 11521–11528. [Google Scholar] [CrossRef]
- Delgado-Mellado, N.; Larriba, M.; Navarro, P.; Rigual, V.; Ayuso, M.; García, J.; Rodríguez, F. Thermal Stability of Choline Chloride Deep Eutectic Solvents by TGA/FTIR-ATR Analysis. J. Mol. Liq. 2018, 260, 37–43. [Google Scholar] [CrossRef]
- Xu, F.-X.; Zhang, J.-Y.; Jin, J.; Li, Z.-G.; She, Y.-B.; Lee, M.-R. Microwave-Assisted Natural Deep Eutectic Solvents Pretreatment Followed by Hydrodistillation Coupled with GC-MS for Analysis of Essential Oil from Turmeric (Curcuma longa L.). J. Oleo Sci. 2021, 70, 1481–1494. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.M.; Jin, Y.; Yoo, D.E.; Han, S.Y.; Kim, E.M.; Lee, J. One-Step Sample Preparation for Convenient Examination of Volatile Monoterpenes and Phenolic Compounds in Peppermint Leaves Using Deep Eutectic Solvents. Food Chem. 2018, 251, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.C.; Ferrer, M.L.; Mateo, C.R.; Del Monte, F. Freeze-Drying of Aqueous Solutions of Deep Eutectic Solvents: A Suitable Approach to Deep Eutectic Suspensions of Self-Assembled Structures. Langmuir 2009, 25, 5509–5515. [Google Scholar] [CrossRef]
- Santana, A.P.R.; Mora-Vargas, J.A.; Guimarães, T.G.S.; Amaral, C.D.B.; Oliveira, A.; Gonzalez, M.H. Sustainable Synthesis of Natural Deep Eutectic Solvents (NADES) by Different Methods. J. Mol. Liq. 2019, 293, 111452. [Google Scholar] [CrossRef]
- Hsieh, Y.-H.; Li, Y.; Pan, Z.; Chen, Z.; Lu, J.; Yuan, J.; Zhu, Z.; Zhang, J. Ultrasonication-Assisted Synthesis of Alcohol-Based Deep Eutectic Solvents for Extraction of Active Compounds from Ginger. Ultrason. Sonochem. 2020, 63, 104915. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, J.; Zhang, H.; Dzah, C.S.; Zandile, M.; Duan, Y.; Ma, H.; Luo, X. Advances in Ultrasound Assisted Extraction of Bioactive Compounds from Cash Crops—A Review. Ultrason. Sonochem. 2018, 48, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Sardroudi, H.; Javadi, A.; Jafarizadeh-Malmiri, H.; Anarjan, N.; Mirzaei, H. Different Deep Eutectic Solvents in Lutein Extraction from Lyophilized Egg Yolk: Preparation, Screening and Characterization. Waste Biomass Valor 2023, 15, 1379–1389. [Google Scholar] [CrossRef]
- Yadav, A.; Kar, J.R.; Verma, M.; Naqvi, S.; Pandey, S. Densities of Aqueous Mixtures of (Choline Chloride+ethylene Glycol) and (Choline Chloride+malonic Acid) Deep Eutectic Solvents in Temperature Range 283.15–363.15K. Thermochim. Acta 2015, 600, 95–101. [Google Scholar] [CrossRef]
- Kalhor, P.; Ghandi, K. Deep Eutectic Solvents for Pretreatment, Extraction, and Catalysis of Biomass and Food Waste. Molecules 2019, 24, 4012. [Google Scholar] [CrossRef]
- Moradi, H.; Farzi, N. Experimental and Computational Assessment of the Physicochemical Properties of Choline Chloride/Ethylene Glycol Deep Eutectic Solvent in 1:2 and 1:3 Mole Fractions and 298.15–398.15 K. J. Mol. Liq. 2021, 339, 116669. [Google Scholar] [CrossRef]
- Dai, Y.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Natural Deep Eutectic Solvents as a New Extraction Media for Phenolic Metabolites in Carthamus tinctorius L. Anal. Chem. 2013, 85, 6272–6278. [Google Scholar] [CrossRef]
- Che Zain, M.S.; Yeoh, J.X.; Lee, S.Y.; Shaari, K. Physicochemical Properties of Choline Chloride-Based Natural Deep Eutectic Solvents (NaDES) and Their Applicability for Extracting Oil Palm Flavonoids. Sustainability 2021, 13, 12981. [Google Scholar] [CrossRef]
- Panić, M.; Radošević, K.; Radojčić Redovniković, I.; Zagajski Kučan, K.; Sander, A.; Halambek, J.; Prlić Kardum, J.; Mitar, A. Physicochemical Properties, Cytotoxicity, and Antioxidative Activity of Natural Deep Eutectic Solvents Containing Organic Acid. Chem. Biochem. Eng. Q. 2019, 33, 1–18. [Google Scholar] [CrossRef]
- Abbott, A.P.; Harris, R.C.; Ryder, K.S. Application of Hole Theory to Define Ionic Liquids by Their Transport Properties. J. Phys. Chem. B 2007, 111, 4910–4913. [Google Scholar] [CrossRef]
- Biernacki, K.; Souza, H.K.S.; Almeida, C.M.R.; Magalhães, A.L.; Gonçalves, M.P. Physicochemical Properties of Choline Chloride-Based Deep Eutectic Solvents with Polyols: An Experimental and Theoretical Investigation. ACS Sustain. Chem. Eng. 2020, 8, 18712–18728. [Google Scholar] [CrossRef]
- Neri, L.; Di Mattia, C.D.; Sacchetti, G.; Pittia, P.; Mastrocola, D. The Influence of Water Activity and Molecular Mobility on Pectinmethylesterase Activity in Salt and Glucose–Maltodextrin Model Systems. Food Bioprod. Process. 2018, 107, 1–9. [Google Scholar] [CrossRef]
- Sato, Y.; Miyawaki, O. Analysis of Hydration Parameter for Sugars Determined from Viscosity and Its Relationship with Solution Parameters. Food Chem. 2016, 190, 594–598. [Google Scholar] [CrossRef]
- Behboudi, E.; Shekaari, H.; Zafarani-Moattar, M.T. Water Activity in Aqueous Solution of Sucrose in the Presence of Some Deep Eutectic Solvents. J. Chem. Eng. Data 2021, 66, 1043–1054. [Google Scholar] [CrossRef]
- Sazali, A.L.; AlMasoud, N.; Amran, S.K.; Alomar, T.S.; Pa’ee, K.F.; El-Bahy, Z.M.; Yong, T.-L.K.; Dailin, D.J.; Chuah, L.F. Physicochemical and Thermal Characteristics of Choline Chloride-Based Deep Eutectic Solvents. Chemosphere 2023, 338, 139485. [Google Scholar] [CrossRef]
- Liu, C.; Fang, H.; Qiao, Y.; Zhao, J.; Rao, Z. Properties and Heat Transfer Mechanistic Study of Glycerol/Choline Chloride Deep Eutectic Solvents Based Nanofluids. Int. J. Heat Mass Transf. 2019, 138, 690–698. [Google Scholar] [CrossRef]
- Francisco, M.; van den Bruinhorst, A.; Kroon, M.C. New Natural and Renewable Low Transition Temperature Mixtures (LTTMs): Screening as Solvents for Lignocellulosic Biomass Processing. Green Chem. 2012, 14, 2153. [Google Scholar] [CrossRef]
- AlOmar, M.K.; Hayyan, M.; Alsaadi, M.A.; Akib, S.; Hayyan, A.; Hashim, M.A. Glycerol-Based Deep Eutectic Solvents: Physical Properties. J. Mol. Liq. 2016, 215, 98–103. [Google Scholar] [CrossRef]
- Florindo, C.; Branco, L.C.; Marrucho, I.M. Development of Hydrophobic Deep Eutectic Solvents for Extraction of Pesticides from Aqueous Environments. Fluid Phase Equilibria 2017, 448, 135–142. [Google Scholar] [CrossRef]
- Alcalde, R.; Gutiérrez, A.; Atilhan, M.; Aparicio, S. An Experimental and Theoretical Investigation of the Physicochemical Properties on Choline Chloride—Lactic Acid Based Natural Deep Eutectic Solvent (NADES). J. Mol. Liq. 2019, 290, 110916. [Google Scholar] [CrossRef]
- Shishov, A.; Pochivalov, A.; Dubrovsky, I.; Bulatov, A. Deep Eutectic Solvents with Low Viscosity for Automation of Liquid-Phase Microextraction Based on Lab-in-Syringe System: Separation of Sudan Dyes. Talanta 2023, 255, 124243. [Google Scholar] [CrossRef]
- Skulcova, A.; Russ, A.; Jablonsky, M.; Sima, J. The pH Behavior of Seventeen Deep Eutectic Solvents. BioRes 2018, 13, 5042–5051. [Google Scholar] [CrossRef]
- Lemaoui, T.; Abu Hatab, F.; Darwish, A.S.; Attoui, A.; Hammoudi, N.E.H.; Almustafa, G.; Benaicha, M.; Benguerba, Y.; Alnashef, I.M. Molecular-Based Guide to Predict the pH of Eutectic Solvents: Promoting an Efficient Design Approach for New Green Solvents. ACS Sustain. Chem. Eng. 2021, 9, 5783–5808. [Google Scholar] [CrossRef]
- Ghaedi, H.; Ayoub, M.; Sufian, S.; Lal, B.; Uemura, Y. Thermal Stability and FT-IR Analysis of Phosphonium-Based Deep Eutectic Solvents with Different Hydrogen Bond Donors. J. Mol. Liq. 2017, 242, 395–403. [Google Scholar] [CrossRef]
- Silva, D.T.D.; Pauletto, R.; Cavalheiro, S.D.S.; Bochi, V.C.; Rodrigues, E.; Weber, J.; Silva, C.D.B.D.; Morisso, F.D.P.; Barcia, M.T.; Emanuelli, T. Natural Deep Eutectic Solvents as a Biocompatible Tool for the Extraction of Blueberry Anthocyanins. J. Food Compos. Anal. 2020, 89, 103470. [Google Scholar] [CrossRef]
- Rashid, R.; Mohd Wani, S.; Manzoor, S.; Masoodi, F.A.; Masarat Dar, M. Green Extraction of Bioactive Compounds from Apple Pomace by Ultrasound Assisted Natural Deep Eutectic Solvent Extraction: Optimisation, Comparison and Bioactivity. Food Chem. 2023, 398, 133871. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, B.; Parkinson, C.; Gonzalez-Miquel, M. Extraction of Polyphenolic Antioxidants from Orange Peel Waste Using Deep Eutectic Solvents. Sep. Purif. Technol. 2018, 206, 1–13. [Google Scholar] [CrossRef]
- Vanda, H.; Dai, Y.; Wilson, E.G.; Verpoorte, R.; Choi, Y.H. Green Solvents from Ionic Liquids and Deep Eutectic Solvents to Natural Deep Eutectic Solvents. Comptes Rendus Chim. 2018, 21, 628–638. [Google Scholar] [CrossRef]
- Zannou, O.; Pashazadeh, H.; Ghellam, M.; Ali Redha, A.; Koca, I. Enhanced Ultrasonically Assisted Extraction of Bitter Melon (Momordica charantia) Leaf Phenolic Compounds Using Choline Chloride-Acetic Acid–Based Natural Deep Eutectic Solvent: An Optimization Approach and in Vitro Digestion. Biomass Conv. Bioref. 2022, 14, 11491–11503. [Google Scholar] [CrossRef]
- Pontes, P.V.d.A.; Czaikoski, A.; Almeida, N.A.; Fraga, S.; Rocha, L.d.O.; Cunha, R.L.; Maximo, G.J.; Batista, E.A.C. Extraction Optimization, Biological Activities, and Application in O/W Emulsion of Deep Eutectic Solvents-Based Phenolic Extracts from Olive Pomace. Food Res. Int. 2022, 161, 111753. [Google Scholar] [CrossRef]
- Bertolo, M.R.V.; Martins, V.C.A.; Plepis, A.M.G.; Bogusz, S. Utilization of Pomegranate Peel Waste: Natural Deep Eutectic Solvents as a Green Strategy to Recover Valuable Phenolic Compounds. J. Clean. Prod. 2021, 327, 129471. [Google Scholar] [CrossRef]
- Obluchinskaya, E.; Pozharitskaya, O.; Shevyrin, V.; Kovaleva, E.; Flisyuk, E.; Shikov, A. Optimization of Extraction of Phlorotannins from the Arctic Fucus Vesiculosus Using Natural Deep Eutectic Solvents and Their HPLC Profiling with Tandem High-Resolution Mass Spectrometry. Mar. Drugs 2023, 21, 263. [Google Scholar] [CrossRef] [PubMed]
- Koraqi, H.; Aydar, A.Y.; Khalid, W.; Ercisli, S.; Rustagi, S.; Ramniwas, S.; Pandiselvam, R. Ultrasound-Assisted Extraction with Natural Deep Eutectic Solvent for Phenolic Compounds Recovery from Rosa Damascene Mill.: Experimental Design Optimization Using Central Composite Design. Microchem. J. 2024, 196, 109585. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Ponmurugan, K.; Maran Jeganathan, P. Development and Validation of Ultrasound-Assisted Solid-Liquid Extraction of Phenolic Compounds from Waste Spent Coffee Grounds. Ultrason. Sonochemistry 2017, 34, 206–213. [Google Scholar] [CrossRef]
- Luque-García, J.L.; Luque De Castro, M.D. Ultrasound: A Powerful Tool for Leaching. TrAC Trends Anal. Chem. 2003, 22, 41–47. [Google Scholar] [CrossRef]
- Şahin, S.; Şamlı, R. Optimization of Olive Leaf Extract Obtained by Ultrasound-Assisted Extraction with Response Surface Methodology. Ultrason. Sonochemistry 2013, 20, 595–602. [Google Scholar] [CrossRef]
- Zhang, Z.-S.; Wang, L.-J.; Li, D.; Jiao, S.-S.; Chen, X.D.; Mao, Z.-H. Ultrasound-Assisted Extraction of Oil from Flaxseed. Sep. Purif. Technol. 2008, 62, 192–198. [Google Scholar] [CrossRef]
- Mohammadpour, H.; Sadrameli, S.M.; Eslami, F.; Asoodeh, A. Optimization of Ultrasound-Assisted Extraction of Moringa Peregrina Oil with Response Surface Methodology and Comparison with Soxhlet Method. Ind. Crops Prod. 2019, 131, 106–116. [Google Scholar] [CrossRef]
- Bener, M.; Şen, F.B.; Önem, A.N.; Bekdeşer, B.; Çelik, S.E.; Lalikoglu, M.; Aşçı, Y.S.; Capanoglu, E.; Apak, R. Microwave-Assisted Extraction of Antioxidant Compounds from by-Products of Turkish Hazelnut (Corylus avellana L.) Using Natural Deep Eutectic Solvents: Modeling, Optimization and Phenolic Characterization. Food Chem. 2022, 385, 132633. [Google Scholar] [CrossRef]
- Mišan, A.; Nađpal, J.; Stupar, A.; Pojić, M.; Mandić, A.; Verpoorte, R.; Choi, Y.H. The Perspectives of Natural Deep Eutectic Solvents in Agri-Food Sector. Crit. Rev. Food Sci. Nutr. 2020, 60, 2564–2592. [Google Scholar] [CrossRef]
- Macchioni, V.; Carbone, K.; Cataldo, A.; Fraschini, R.; Bellucci, S. Lactic Acid-Based Deep Natural Eutectic Solvents for the Extraction of Bioactive Metabolites of Humulus lupulus L.: Supramolecular Organization, Phytochemical Profiling and Biological Activity. Sep. Purif. Technol. 2021, 264, 118039. [Google Scholar] [CrossRef]
- Bus, K.; Szterk, A. Relationship between Structure and Biological Activity of Various Vitamin K Forms. Foods 2021, 10, 3136. [Google Scholar] [CrossRef] [PubMed]
- Teplova, V.V.; Isakova, E.P.; Klein, O.I.; Dergachova, D.I.; Gessler, N.N.; Deryabina, Y.I. Natural Polyphenols: Biological Activity, Pharmacological Potential, Means of Metabolic Engineering (Review). Appl. Biochem. Microbiol. 2018, 54, 221–237. [Google Scholar] [CrossRef]
- Mansinhos, I.; Gonçalves, S.; Rodríguez-Solana, R.; Ordóñez-Díaz, J.L.; Moreno-Rojas, J.M.; Romano, A. Ultrasonic-Assisted Extraction and Natural Deep Eutectic Solvents Combination: A Green Strategy to Improve the Recovery of Phenolic Compounds from Lavandula pedunculata Subsp. Lusitanica (Chaytor) Franco. Antioxidants 2021, 10, 582. [Google Scholar] [CrossRef]
- Wang, W.; An, M.; Zhao, G.; Wang, Y.; Yang, D.; Zhang, D.; Zhao, L.; Han, J.; Wu, G.; Bo, Y. Ultrasonic-Assisted Customized Natural Deep Eutectic Solvents Extraction of Polyphenols from Chaenomeles speciosa. Microchem. J. 2023, 193, 108952. [Google Scholar] [CrossRef]
- Kutlu, N.; Kamiloğlu, A.; Abca, T.E.; Yilmaz, Ö. Ultrasound-assisted Deep Eutectic Solvent Extraction of Bioactive Compounds from Persimmon Calyx. J. Food Sci. 2024, 89, 294–305. [Google Scholar] [CrossRef]

| Run | Coded | Uncoded | ||||
|---|---|---|---|---|---|---|
| A | B | C | Temperature (°C) | Time (h) | HBA:HBD Ratio | |
| 1 | 2 | 3 | 1 | 70 | 2.0 | 1:1 |
| 2 | 2 | 1 | 2 | 70 | 1.0 | 1:2 |
| 3 | 2 | 2 | 3 | 70 | 1.5 | 2:1 |
| 4 | 2 | 3 | 2 | 70 | 2.0 | 1:2 |
| 5 | 1 | 1 | 1 | 50 | 1.0 | 1:1 |
| 6 | 3 | 2 | 2 | 90 | 1.5 | 1:2 |
| 7 | 1 | 1 | 2 | 50 | 1.0 | 1:2 |
| 8 | 3 | 3 | 3 | 90 | 2.0 | 2:1 |
| 9 | 1 | 2 | 3 | 50 | 1.5 | 2:1 |
| 10 | 1 | 3 | 2 | 50 | 2.0 | 1:2 |
| 11 | 2 | 2 | 2 | 70 | 1.5 | 1:2 |
| 12 | 3 | 1 | 2 | 90 | 1.0 | 1:2 |
| 13 | 1 | 1 | 3 | 50 | 1.0 | 2:1 |
| 14 | 2 | 3 | 3 | 70 | 2.0 | 2:1 |
| 15 | 3 | 3 | 1 | 90 | 2.0 | 1:1 |
| 16 | 1 | 3 | 3 | 50 | 2.0 | 2:1 |
| 17 | 1 | 3 | 1 | 50 | 2.0 | 1:1 |
| 18 | 3 | 1 | 1 | 90 | 1.0 | 1:1 |
| 19 | 3 | 1 | 3 | 90 | 1.0 | 2:1 |
| 20 | 3 | 3 | 2 | 90 | 2.0 | 1:2 |
| 21 | 2 | 2 | 1 | 70 | 1.5 | 1:1 |
| 22 | 1 | 2 | 1 | 50 | 1.5 | 1:1 |
| 23 | 2 | 1 | 1 | 70 | 1.0 | 1:1 |
| 24 | 3 | 2 | 1 | 90 | 1.5 | 1:1 |
| 25 | 2 | 1 | 3 | 70 | 1.0 | 2.1 |
| 26 | 1 | 2 | 2 | 50 | 1.5 | 1:2 |
| 27 | 3 | 2 | 3 | 90 | 1.5 | 2:1 |
| Model ID | (A) Temperature (°C) | (B) Time (min) | (C) Volume (mL) | (D) Concentration (%) |
| (1) | − (40) | − (10) | − (10) | − (60) |
| c | − (40) | − (10) | + (20) | − (60) |
| ab | + (60) | + (20) | − (10) | − (60) |
| ac | + (60) | − (10) | +(20) | − (60) |
| ad | + (60) | − (10) | − (10) | + (80) |
| bc | − (40) | + (20) | + (20) | − (60) |
| cd | − (40) | − (10) | + (20) | + (80) |
| abcd | + (60) | + (20) | + (20) | + (80) |
| Ç | 0 (50) | 0 (15) | 0 (15) | 0 (70) |
| Ç | 0 (50) | 0 (15) | 0 (150) | 0 (70) |
| Variable | Coded Levels | ||||
|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | |
| Experimental Actual Values | |||||
| Ultrasound temperature (UT) (°C) | 30 | 40 | 50 | 60 | 70 |
| Extraction time (ET) (min) | 5 | 10 | 15 | 20 | 25 |
| Solvent volume (SV) (mL) | 5 | 10 | 15 | 20 | 25 |
| Solvent concentration (SC) (%) | 50 | 60 | 70 | 80 | 90 |
| S/No | Code | HBA | HBD | HBA:HBD Ratio | Synthesis Temperature (°C) | Synthesis Time (min) | Physical Appearance |
|---|---|---|---|---|---|---|---|
| 1 | CaSu11 | Citric acid | Sucrose | 1:1 | 70 | 60 | Orange-yellow |
| 2 | CaFr11 | Citric acid | Fructose | 1:1 | 70 | 60 | Faintly yellow |
| 3 | CaFr12 | Citric acid | Fructose | 1:2 | 70 | 60 | Yellow |
| 4 | CaFr21 | Citric acid | Fructose | 2:1 | 70 | 60 | Yellow |
| 5 | CaXy11 | Citric acid | Xylitol | 1:1 | 70 | 60 | Colorless |
| 6 | CaXy12 | Citric acid | Xylitol | 1:2 | 70 | 60 | Colorless |
| 7 | CaXy21 | Citric acid | Xylitol | 2:1 | 70 | 60 | Colorless |
| 8 | CaGr11 | Citric acid | Glycerol | 1:1 | 70 | 60 | Colorless |
| 9 | CaGr12 | Citric acid | Glycerol | 1:2 | 70 | 60 | Colorless |
| 10 | CaG21 | Citric acid | Glycerol | 2:1 | 70 | 60 | Colorless |
| 11 | CaGc11 | Citric acid | Glycine | 1:1 | 70 | 60 | Colorless |
| 12 | CaGc21 | Citric acid | Glycine | 2:1 | 70 | 60 | Colorless |
| 13 | CaGl11 | Citric acid | Glucose | 1:1 | 70 | 60 | Yellow |
| 14 | CaGl21 | Citric acid | Glucose | 2:1 | 70 | 60 | yellow |
| NADESs | HBA | HBD | Ratio | pH | Water Activity | Viscosity (pa. s) | Density (g/cm3) |
|---|---|---|---|---|---|---|---|
| CaSu11 | Citric acid | sucrose | 1:1 | 1.92 ± 0.00C | 0.73 ± 0.010F | 0.3545 ± 0.0003A | 1.3768 ± 0.0002A |
| CaFr11 | Citric acid | fructose | 1:1 | 1.75 ± 0.01H | 0.77 ± 0.006DE | 0.0805 ± 0.0001F | 1.3293 ± 0.0003B |
| CaFr12 | Citric acid | fructose | 1:2 | 1.90 ± 0.01C | 0.76 ± 0.010E | 0.1797 ± 0.0002B | 1.3768 ± 0.0008A |
| CaFr21 | Citric acid | fructose | 2:1 | 1.84 ± 0.00EF | 0.72 ± 0.010FG | 0.1221 ± 0.0001D | 1.3770 ± 0.0000A |
| CaXy11 | Citric acid | xylitol | 1:1 | 1.85 ± 0.05E | 0.78 ± 0.010CD | 0.0269 ± 0.0001J | 1.2818 ± 0.0004C |
| CaXy12 | Citric acid | xylitol | 1:2 | 1.90 ± 0.03C | 0.76 ± 0.010E | 0.0806 ± 0.0002F | 1.2818 ± 0.0008C |
| CaXy21 | Citric acid | xylitol | 2:1 | 1.89 ± 0.03CD | 0.72 ± 0.006FG | 0.0871 ± 0.0001E | 1.3293 ± 0.0002B |
| CaGr11 | Citric acid | glycerol | 1:1 | 1.68 ± 0.02I | 0.79 ± 0.000C | 0.0131 ± 0.0001M | 1.2344 ± 0.0004D |
| CaGr12 | Citric acid | glycerol | 1:2 | 1.86 ± 0.02DE | 0.77 ± 0.010DE | 0.0191 ± 0.0020L | 1.2344 ± 0.0006D |
| CaGr21 | Citric acid | glycerol | 2:1 | 1.78 ± 0.01GH | 0.77 ± 0.010DE | 0.0525 ± 0.0005H | 1.3293 ± 0.0003B |
| CaGc11 | Citric acid | glycine | 1:1 | 2.61 ± 0.04A | 0.83 ± 0.010A | 0.0204 ± 0.0000K | 1.3293 ± 0.0004B |
| CaGc21 | Citric acid | glycine | 2:1 | 2.39 ± 0.01B | 0.81 ± 0.000B | 0.0760 ± 0.0020G | 1.3293 ± 0.0004B |
| CaGl11 | Citric acid | glucose | 1:1 | 1.81 ± 0.01FG | 0.77 ± 0.010DE | 0.0438 ± 0.0001I | 1.2818 ± 0.0004C |
| CaGl21 | Citric acid | glucose | 2:1 | 1.71 ± 0.01I | 0.71 ± 0.010G | 0.1313 ± 0.0002C | 1.3293 ± 0.0003B |
| NADESs | PEAKS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1(OH) | 2(CH) | 3(RCOOR) | 4(C=C) | 5(=CH2) | 6 | 7 | 8 | 9(C-O) | 10(C=C; =CH2) | |
| CaSu11 | 3306 | 2936 | 1713 | 1639 | 1400 | 1333 | 1209 | 1101 | 1028 | 897 |
| CaFr11 | 3327 | 2941 | 1715 | 1655 | 1638 | 1398 | 1339 | 1217 | 1140 | 1101 |
| CaFr12 | 3288 | 2941 | 1717 | 1655 | 1647 | 1639 | 1398 | 1341 | 1217 | 1144 |
| CaFr21 | 3375 | 2943 | 1711 | 1638 | 1398 | 1319 | 1206 | 1130 | 1103 | 1057 |
| CaXy11 | 3321 | 2949 | 1713 | 1638 | 1398 | 1211 | 1126 | 1057 | 1001 | 876 |
| CaXy12 | 3298 | 2943 | 1713 | 1639 | 1398 | 1317 | 1211 | 1125 | 1096 | 1001 |
| CaXy21 | 3385 | 3246 | 2949 | 2565 | 1709 | 1632 | 1396 | 1204 | 1119 | 1043 |
| CaGr11 | 3358 | 2951 | 1713 | 1639 | 1396 | 1319 | 1211 | 1113 | 1042 | 991 |
| CaGr12 | 3287 | 2949 | 1717 | 1639 | 1396 | 1319 | 1209 | 1111 | 1040 | 991 |
| CaGr21 | 3368 | 2955 | 2581 | 1709 | 1638 | 1396 | 1317 | 1119 | 1043 | 989 |
| CaGc11 | 3374 | 3231 | 1711 | 1624 | 1508 | 1406 | 1319 | 1219 | 1125 | 1038 |
| CaGc21 | 3393 | 3208 | 2953 | 2615 | 1709 | 1624 | 1508 | 1398 | 1319 | 1209 |
| CaGl11 | 3321 | 2936 | 1713 | 1638 | 1398 | 1317 | 1213 | 1105 | 1076 | 1028 |
| CaGl21 | 3372 | 3218 | 2941 | 2585 | 1713 | 1636 | 1396 | 1315 | 1206 | 1109 |
| NADESs | Runs | TPC (mgGAE/g) | TFC (µgQE/g) | FRAP (µMTE/g) | CUPRAC (µMTE/g) |
|---|---|---|---|---|---|
| CaSu11 | 30 | 270.6B | 331.3BCD | 2.954BC | 3.147A |
| CaFr11 | 30 | 389.2A | 421.4BC | 5.848A | 4.718A |
| CaFr12 | 30 | 458.0A | 262.7CD | 4.278ABC | 4.341A |
| CaFr21 | 30 | 163.7C | 271.3CD | 4.571AB | 4.359A |
| CaXy11 | 30 | 174.3BC | 172.1D | 4.132ABC | 3.910A |
| CaXy12 | 30 | 136.7C | 249.2CD | 3.225ABC | 3.274A |
| CaXy21 | 30 | 125.3C | 175.0D | 3.696ABC | 3.934A |
| CaGr11 | 30 | 147.8C | 208.7D | 3.246ABC | 4.241A |
| CaGr12 | 30 | 167.6C | 199.0D | 3.552ABC | 4.101A |
| CaGr21 | 30 | 143.4C | 188.7D | 3.123ABC | 4.393A |
| CaGc11 | 30 | 146.93C | 897.3A | 2.236BC | 3.852A |
| CaGc21 | 30 | 123.0C | 482.6B | 1.641C | 3.790A |
| CaGl11 | 30 | 143.3C | 265.3CD | 2.222BC | 3.798A |
| CaGl21 | 30 | 139.0C | 330.1BCD | 2.057BC | 3.626A |
| Run | Exp. ID | Conditions ª | Response ɓ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| X1 | X2 | X3 | X4 | TPC (mgGAE/g) | TFC (µgQE/g) | CUPRAC (µMTE/g) | FRAP (µMTE/g) | DPPH (%) | ||
| 1 | abd | 60 (1) | 20 (1) | 10 (−1) | 80 (1) | 179.09 ± 0.75 | 61.328 ± 0.78 | 3.1702 ± 0.01 | 0.7490 ± 0.01 | 26.7 ± 0.1 |
| 2 | b | 40 (−1) | 20 (1) | 10 (−1) | 60 (−1) | 113.4 ± 2.47 | 116.41 ± 1.56 | 6.2274 ± 0.03 | 0.7387 ± 0.01 | 89.6 ± 0.1 |
| 3 | 0 | 50 (0) | 15 (0) | 15 (0) | 70 (0) | 260.05 ± 0.42 | 339.26 ± 1.17 | 4.8886 ± 0.04 | 1.0656 ± 0.01 | 89.0 ± 0.1 |
| 4 | (1) | 40 (−1) | 10 (−1) | 10 (−1) | 60 (−1) | 164.05 ± 1.58 | 60.026 ± 1.19 | 3.1864 ± 0.03 | 0.8105 ± 0.016 | 53.6 ± 0.1 |
| 5 | ab | 60 (1) | 20 (1) | 10 (−1) | 60 (−1) | 229.25 ± 0.28 | 75.65 ± 1.97 | 3.1702 ± 0.03 | 1.7003 ± 0.009 | 17.2 ± 0.2 |
| 6 | d | 40 (−1) | 10 (−1) | 10 (−1) | 80 (1) | 168.63 ± 0.98 | 546.61 ± 1.8 | 6.3889 ± 0.03 | 1.539 ± 0.018 | 14.6 ± 0.1 |
| 7 | cd | 40 (−1) | 10 (−1) | 20 (1) | 80 (1) | 635.29 ± 1.96 | 457.03 ± 10.9 | 8.0685 ± 0.06 | 2.7236 ± 0.009 | 18.4 ± 0.1 |
| 8 | bc | 40 (−1) | 20 (1) | 20 (1) | 60 (−1) | 650.33 ± 0.57 | 120.57 ± 1.8 | 6.4212 ± 0.03 | 10.267 ± 0.018 | 10.5 ± 0.4 |
| 9 | abc | 60 (1) | 20 (1) | 20 (1) | 60 (−1) | 221.24 ± 1.13 | 414.32 ± 1.8 | 6.292 ± 0.11 | 1.9903 ± 0.018 | 13.7 ± 0.2 |
| 10 | ad | 60 (1) | 10 (−1) | 10 (−1) | 80 (1) | 322.71 ± 0.28 | 284.51 ± 0.9 | 4.196 ± 0.01 | 1.2618 ± 0.009 | 7.33 ± 0.6 |
| 11 | bd | 40 (−1) | 20 (10 | 10 (−1) | 80 (1) | 183.5 ± 1.02 | 61.59 ± 1.8 | 3.2187 ± 0.01 | 0.7413 ± 0.009 | 19.2 ± 0.1 |
| 12 | ac | 60 (1) | 10 (−1) | 20 (1) | 60 (−1) | 695.42 ± 2.83 | 123.7 ± 1.8 | 6.2435 ± 0.03 | 1.4774 ± 0.009 | 21.5 ± 0.2 |
| 13 | abcd | 60 (1) | 20 (1) | 20 (1) | 80 (1) | 276.47 ± 0.98 | 122.14 ± 4.51 | 6.4212 ± 0.06 | 1.4364 ± 0.018 | 31.7 ± 0.2 |
| 14 | a | 60 (1) | 10 (−1) | 10 (−1) | 60 (−1) | 237.09 ± 1.98 | 59.245 ± 0.45 | 3.0895 ± 0.01 | 0.7439 ± 0.013 | 63.9 ± 0.1 |
| 15 | c | 40 (−1) | 10 (−1) | 20 (1) | 60 (−1) | 379.09 ± 1.13 | 831.77 ± 3.61 | 12.6163 ± 0.1 | 1.5441 ± 0.009 | 1.51 ± 0.1 |
| 16 | acd | 60 (1) | 10 (−1) | 20 (1) | 80 (1) | 406.54 ± 3.01 | 1313.0 ± 3.61 | 12.33 ± 0.01 | 2.9344 ± 0.018 | 24.9 ± 0.1 |
| 17 | bcd | 40 (−1) | 20 (1) | 20 (1) | 80 (1) | 620.92 ± 1.13 | 755.73 ± 7.22 | 12.52 ± 0.01 | 3.0164 ± 0.018 | 74.7 ± 0.1 |
| 18 | cα | 50 (0) | 15 (0) | 25 (α) | 70 (0) | 231.21 ± 1.42 | 345.7 ± 6.77 | 15.4877 ± 0.14 | 3.7449 ± 0.044 | 73.6 ± 0.3 |
| 19 | aα | 70 (α) | 15 (0) | 15 (0) | 70 (0) | 370.74 ± 1.95 | 91.99 ± 4.06 | 4.6827 ± 0.02 | 1.0735 ± 0.007 | 20.6 ± 0.2 |
| 20 | dα | 50 (0) | 15 (0) | 15 (0) | 90 (α) | 148.04 ± 0.85 | 290.23 ± 1.35 | 9.3653 ± 0.04 | 2.3854 ± 0.027 | 22.4 ± 0.4 |
| 21 | bα | 50 (0) | 25 (α) | 15 (0) | 70 (0) | 409.31 ± 3.06 | 347.27 ± 5.41 | 9.3411 ± 0.04 | 2.17 ± 0.013 | 15.5 ± 0.1 |
| 22 | −bα | 50 (0) | 5 (−α) | 15 (0) | 70 (0) | 302.45 ± 2.25 | 311.33 ± 5.41 | 9.2926 ± 0.08 | 2.2623 ± 0.013 | 17.9 ± 0.2 |
| 23 | −aα | 30 (−α) | 15 (0) | 15 (0) | 70 (0) | 100 ± 1.47 | 862.89 ± 5.41 | 9.6318 ± 0.04 | 2.1931 ± 0.013 | 3.15 ± 0.4 |
| 24 | −dα | 50 (0) | 15 (0) | 15 (0) | 50 (−α) | 332.84 ± 1.12 | 130.27 ± 0.68 | 4.8401 ± 0.06 | 1.1273 ± 0.02 | 20.2 ± 0.3 |
| 25 | −cα | 50 (0) | 15 (0) | 5 (−α) | 70 (0) | 147.22 ± 1.5 | 60.286 ± 0.45 | 3.1379 ± 0.01 | 0.521 ± 0.346 | 62.1 ± 0.1 |
| Model Comp ª. | Response | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TPC | TFC | CUPRAC | FRAP | DPPH | ||||||
| F-Value | p-Value | F-Value | p-Value | F-Value | p-Value | F-Value | p-Value | F-Value | p-Value | |
| X1 | 0.25 | 0.618 | 10.83 | 0.002 | 18.64 | 0.000 | 16.18 | 0.000 | 0.21 | 0.650 |
| X2 | 1.09 | 0.301 | 9.18 | 0.003 | 3.17 | 0.079 | 6.95 | 0.010 | 10.18 | 0.003 |
| X3 | 63.61 | 0.000 | 30.93 | 0.000 | 148.55 | 0.000 | 65.15 | 0.000 | 3.80 | 0.058 |
| X4 | 0.75 | 0.390 | 11.72 | 0.001 | 13.75 | 0.000 | 2.68 | 0.106 | 0.46 | 0.500 |
| X12 | 0.39 | 0.536 | 3.15 | 0.080 | 3.02 | 0.086 | 1.62 | 0.208 | 62.54 | 0.000 |
| X22 | 11.16 | 0.001 | 0.06 | 0.811 | 21.31 | 0.000 | 6.74 | 0.011 | 54.39 | 0.000 |
| X32 | 0.02 | 0.896 | 1.14 | 0.289 | 21.26 | 0.000 | 5.32 | 0.024 | 3.21 | 0.081 |
| X42 | 0.88 | 0.353 | 0.99 | 0.324 | 2.78 | 0.100 | 8.02 | 0.006 | 47.25 | 0.000 |
| X1X2 | 15.10 | 0.000 | 0.28 | 0.601 | 1.59 | 0.212 | 14.28 | 0.000 | 80.73 | 0.000 |
| X1X3 | 16.61 | 0.000 | 0.05 | 0.825 | 1.46 | 0.231 | 20.22 | 0.000 | 1.50 | 0.227 |
| X1X4 | 3.59 | 0.051 | 0.68 | 0.413 | 2.01 | 0.161 | 6.39 | 0.014 | 1.00 | 0.323 |
| X2X3 | 0.41 | 0.526 | 1.79 | 0.184 | 2.36 | 0.129 | 13.53 | 0.000 | - | - |
| X2X4 | 0.00 | 0.963 | 6.14 | 0.015 | 18.64 | 0.000 | 30.21 | 0.000 | - | - |
| X3X4 | 0.22 | 0.643 | 1.04 | 0.312 | 3.17 | 0.079 | 5.65 | 0.020 | - | - |
| model | 10.24 | 0.000 | 73.67 | 0.000 | 38.4 | 0.000 | 19.934 | 0.000 | 35.512 | 0.000 |
| Lack of fit | 0.62 | 0.843 | 0.028 | 0.963 | 0.270 | 0.801 | 0.24 | 0.968 | 0.71 | 0.812 |
| Pure error | 0.84 | 0.49 | 0.65 | 0.18 | 0.92 | |||||
| Coefficients ª | TPC | TFC | CUPRAC | FRAP | DPPH |
|---|---|---|---|---|---|
| (I) | −2111 | −930 | 38.1 | 0.5 | −1825 |
| X1 | 52.6 | −66.6 | −0.479 | −0.085 | 28.17 |
| X2 | 17.3 | 129 | −0.144 | 1.051 | 45.73 |
| X3 | 102.6 | 33 | −0.526 | 0.681 | 7.48 |
| X4 | 1.2 | 43.2 | −0.572 | −0.298 | 23.25 |
| X12 | 0.075 | 0.428 | 0.00326 | 0.0014 | −0.181 |
| X22 | 1.603 | 0.231 | 0.03465 | 0.01142 | −0.6751 |
| X32 | −0.063 | −1.032 | 0.03461 | 0.01073 | −0.1639 |
| X42 | 0.112 | −0.24 | 0.00313 | 0.00312 | −0.1573 |
| X1X2 | −1.221 | −0.332 | −0.00645 | −0.01086 | −0.4851 |
| X1X3 | −1.28 | 0.14 | −0.00627 | −0.01292 | −0.0662 |
| X1X4 | −0.312 | 0.26 | 0.00363 | 0.00363 | −0.0270 |
| X2X3 | −0.4 | −1.69 | −0.0157 | 0.02114 | - |
| X2X4 | −0.015 | −1.566 | −0.00587 | −0.0158 | - |
| X3X4 | −0.146 | 0.644 | 0.00778 | −0.00683 | - |
| R2 | 0.984 | 0.9999 | 0.918 | 1.00 | 0.9992 |
| R2adj | 0.9779 | 0.9998 | 0.887 | 1.00 | 0.9988 |
| R2Pred | 0.9767 | 0.9997 | 0.882 | 1.00 | 0.9981 |
| Parameters | Predicted Optimum Factors to Achieve the Desirability a | Maximum Desirability | |||
|---|---|---|---|---|---|
| UT (°C) | ET (min) | SV (mL) | SC (%) | ||
| TPC | 45 | 5 | 25 | 90 | 0.993634 |
| TFC | 30 | 5 | 25 | 90 | 0.900334 |
| CUPRAC | 30 | 5 | 25 | 90 | 1.000000 |
| FRAP | 45 | 25 | 25 | 50 | 0.960543 |
| DPPH | 50 | 15 | 15 | 70 | 0.931252 |
| NADESs | TPC (mgGAE/g) | TFC (µgQE/g) | CUPRAC (µMTE/g) | FRAP (µMTE/g) | DPPH (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fitted | Experimental | Fitted | Experimental | Fitted | Experimental | Fitted | Experimental | Fitted | Experimental | |
| CaSu11 | 732 ± 128 | 860.02 ± 4.7 B | 1061 ± 373 | 1176.0 ± 13 F | 23.27 ± 3.1 | 24.76 ± 0.39 H | 26.28 ± 3.1 | 26.17 ± 0.17 A | 79.18 ± 7.3 | 72.24 ± 0.15 J |
| CaFr11 | 1269 ± 332 | 1290.9 ± 5.6 A | 2329 ± 427 | 2398.7 ± 23 A | 38.03 ± 4.4 | 38.46 ± 0.44 A | 26.05 ± 3.3 | 26.15 ± 0.11 B | 86.40 ± 11 | 73.55 ± 0.24 I |
| CaFr12 | 711 ± 142 | 748.64 ± 12 C | 1449 ± 338 | 1446.0 ± 12 C | 26.20 ± 2.8 | 26.24 ± 0.31 F | 16.75 ± 1.3 | 17.31 ± 0.12 G | 89.65 ± 7.4 | 84.19 ± 0.35 F |
| CaFr21 | 272.9 ± 56.8 | 281.05 ± 18 L | 437.5 ± 84.3 | 442.13 ± 8.7 L | 27.71 ± 3.1 | 25.15 ± 0.34 G | 18.07 ± 1.3 | 18.45 ± 0.08 E | 87.68 ± 6.0 | 84.26 ± 0.36 F |
| CaXy11 | 375.0 ± 39.7 | 365.20 ± 11 K | 607 ± 124 | 618.27 ± 10 J | 28.34 ± 2.8 | 24.71 ± 0.49 H | 18.12 ± 1.5 | 19.21 ± 0.13 D | 81.81 ± 4.2 | 64.78 ± 0.25 K |
| CaXy12 | 577.1 ± 41.5 | 588.78 ± 9.4 F | 1694 ± 237 | 1777.1 ± 10 B | 25.96 ± 3.0 | 28.92 ± 0.53 E | 17.11 ± 1.7 | 18.13 ± 0.11 F | 94.20 ± 13 | 97.42 ± 0.40 A |
| CaXy21 | 294.8 ± 38.9 | 289.76 ± 6.7 L | 551 ± 112 | 584.85 ± 19 K | 28.58 ± 3.1 | 29.53 ± 0.55 D | 15.82 ± 1.4 | 16.19 ± 0.11 I | 85.25 ± 6.9 | 64.19 ± 0.33 L |
| CaGr11 | 507.7 ± 67.7 | 539.76 ± 16 G | 621 ± 168 | 633.85 ± 20 I | 31.72 ± 2.7 | 32.29 ± 0.52 B | 16.41 ± 1.0 | 17.04 ± 0.09 H | 86.43 ± 9.2 | 83.55 ± 0.25 G |
| CaGr12 | 748.5 ± 90.5 | 754.63 ± 7.1 C | 828 ± 130 | 869.57 ± 12 H | 29.92 ± 2.9 | 29.89 ± 0.32 CD | 17.07 ± 1.5 | 18.52 ± 0.18 E | 90.52 ± 6.2 | 89.71 ± 0.31 C |
| CaGr21 | 393.6 ± 56.8 | 396.24 ± 9.7 J | 681 ± 220 | 632.60 ± 10 I | 31.45 ± 3.0 | 29.92 ± 0.37 C | 26.59 ± 3.3 | 26.21 ± 0.12 C | 86.42 ± 7.8 | 80.93 ± 0.20 H |
| CaGc11 | 498.8 ± 90.5 | 475.22 ± 8.6 I | 1330 ± 417 | 1354.4 ± 15 D | 22.16 ± 3.2 | 22.78 ± 0.43 J | 16.30 ± 1.5 | 17.21 ± 0.22 G | 89.70 ± 12 | 86.08 ± 0.15 E |
| CaGc21 | 499.3 ± 63.9 | 502.72 ± 7.5 H | 1224 ± 251 | 1341.4 ± 8.8 E | 24.02 ± 2.9 | 23.89 ± 0.50 I | 7.92 ± 1.0 | 7.59 ± 0.14 M | 95.98 ± 5.4 | 90.05 ± 0.23 B |
| CaGl11 | 626.8 ± 69.7 | 660.13 ± 7.6 E | 1224 ± 149 | 1448.6 ± 10 C | 25.29 ± 3.2 | 25.40 ± 0.15 G | 8.11 ± 1.6 | 8.59 ± 0.13 L | 34.98 ± 6.0 | 34.77 ± 0.24 M |
| CaGl21 | 649.6 ± 52.0 | 688.45 ± 7.8 D | 1003 ± 142 | 1025.4 ± 9.4 G | 25.99 ± 3.0 | 25.50 ± 0.52 G | 10.64 ± 2.7 | 11.59 ± 0.14 K | 89.29 ± 7.2 | 86.49 ± 0.27 D |
| Water | - | 144.34 ± 8.0 M | - | 589.63 ± 11 K | - | 22.81 ± 0.46 J | - | 12.07 ± 0.14 J | - | 11.69 ± 0.24 N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okeke, U.J.; Micucci, M.; Mihaylova, D.; Cappiello, A. Synthesis and Application of Natural Deep Eutectic Solvents (NADESs) for Upcycling Horticulture Residues. Horticulturae 2025, 11, 439. https://doi.org/10.3390/horticulturae11040439
Okeke UJ, Micucci M, Mihaylova D, Cappiello A. Synthesis and Application of Natural Deep Eutectic Solvents (NADESs) for Upcycling Horticulture Residues. Horticulturae. 2025; 11(4):439. https://doi.org/10.3390/horticulturae11040439
Chicago/Turabian StyleOkeke, Udodinma Jude, Matteo Micucci, Dasha Mihaylova, and Achille Cappiello. 2025. "Synthesis and Application of Natural Deep Eutectic Solvents (NADESs) for Upcycling Horticulture Residues" Horticulturae 11, no. 4: 439. https://doi.org/10.3390/horticulturae11040439
APA StyleOkeke, U. J., Micucci, M., Mihaylova, D., & Cappiello, A. (2025). Synthesis and Application of Natural Deep Eutectic Solvents (NADESs) for Upcycling Horticulture Residues. Horticulturae, 11(4), 439. https://doi.org/10.3390/horticulturae11040439










