Abstract
Epigallocatechin gallate (EGCG) is an important contributor to bitterness and astringency in summer tea leaves; however, the transcriptional regulatory mechanisms of EGCG biosynthesis remain unclear. In this study, EGCG content was significantly decreased after foliar spraying with nano-Se fertilizers in tea leaves. A WRKY transcription factor (TF), CsWRKY70, was found to be positively related to EGCG content. The open reading frame of CsWRKY70 was 891 bp encoding 296 amino acids. CsWRKY70 is localized to the nucleus and has transcriptional activation activity. The electrophoretic mobility shift assay indicated that CsWRKY70 can directly bind to the promoters of CsLAR and CsUGT84A containing W-box (5′-C/TTGACT/C-3′) sequences. Dual-luciferase reporter experiment verified that CsWRKY70 activated CsLAR and CsUGT84A expressions in tobacco leaves. In summary, these results demonstrated that CsWRKY70 may reduce EGCG biosynthesis by inhibiting the CsLAR and CsUGT84A expressions under nano-Se treatment. Our findings provide new insight into the regulatory mechanism of WRKY TFs involved in catechin biosynthesis and offer a theoretical basis for breeding low or high EGCG content tea cultivars.
1. Introduction
Tea (Camellia sinensis) is the most popular beverage in the world due to its unique flavor and multiple health benefits. Tea quality is related to several chemical compounds in tea leaves, including tea polyphenols, amino acids, and caffeine. Catechins are key phenolic compounds in tea leaves, accounting for 50–60% of the total tea polyphenols, and are crucial in the formation of tea flavor [1]. Epigallocatechin gallate (EGCG), accounting for 50–80% of total catechins, is known to process numerous health benefits, including antioxidant [2], anti-inflammatory [3], and antiviral [4] properties. In general, catechin content in tea leaves increases with the temperature increasing, resulting in a high level of catechin in summer tea leaves [5]. However, an excessive level of catechin aggravates the bitterness and astringency of tea infusion, causing a decline in tea quality [6]. Our previous study has demonstrated that moderate and high concentrations of nano-Se can improve summer tea quality by decreasing tea polyphenols, catechins, and caffeine contents and increasing theanine content in summer tea leaves (Tables S1 and S2). However, the transcriptional regulatory mechanisms of EGCG biosynthesis remain unclear.
Catechins are the final products of the flavonoid biosynthesis pathway, which is derived from phenylpropanoid. EGCG biosynthetic pathway is controlled by several crucial genes. It has been demonstrated that non-galloylated catechins, such as catechin (C), gallocatechin (GC), epicatechin (EC), and epigallocatechin (EGC), are synthesized by several crucial enzymes, including anthocyanidin reductase (ANR), leucocyanidin reductase (LAR), and anthocyanidin synthase (ANS) [7]. Further, β-Glucogallin, a galloyl donor associated with the biosynthesis of EGCG, is synthesized by glucose and gallic acid in tea leaves through the UDP-glucose: galloyl-1-O-β-D-glucosyltransferase (CsUGT84A) [8]. Finally, type 1A serine carboxypeptidase-like acyltransferases (CsSCPL1A) convert non-galloylated catechins into galloylated catechins, such as ECG and EGCG [9]. Therefore, nano-Se may regulate EGCG accumulation in tea leaves by affecting the expressions of these crucial genes.
Transcription factors (TFs) are protein molecules with special structures that can specifically bind to the cis-acting elements in the promoter region of functional genes. TFs can affect target gene expression by activating or inhibiting the activity of the promoter of the structural gene [10]. According to the characteristics of conserved domains, TFs can be divided into multiple families, including MYB [11], bHLH [12], WRKY [13], NAC [14], and bZIP [15]. WRKY TFs exist only in plants and feature an extremely conserved WRKYGQK sequence in N-terminal, which can specifically recognize and bind to W-box (C/T)TGAC(T/C) elements [16]. WRKY is involved in regulating different biological processes in plants [17], including plant defense regulatory networks, developmental processes, and biosynthesis of defensive metabolites [18,19]. In addition, WRKY is also engaged in mediating the biosynthesis of flavonoids. MdWRKY11 increases the accumulation of flavonoids and anthocyanins in red-fleshed apples [20]. Duan et al. find that AtWRKY41 is an inhibitor to regulate the anthocyanin biosynthesis in Arabidopsis thaliana [21]. Furthermore, CsWRKY57like positively impacts the biosynthesis of methylated EGCG in the tea plant [22]. However, there are few studies on WRKY TFs regulating the biosynthesis of EGCG.
In this study, different concentrations of nano-Se fertilizers were sprayed on summer tea leaves to evaluate their effect on EGCG accumulation in leaves. The CsWRKY70, which was positively associated with the EGCG content in tea leaves, was identified using transcriptome sequencing. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to further identify and validate the expression levels of differentially expressed flavonoids biosynthetic related genes. The influence of CsWRKY70 on CsLAR and CsUGT84A gene promoters was further analyzed. These results will provide a theoretical foundation for elucidating the regulatory mechanism of EGCG biosynthesis in tea plants.
2. Materials and Methods
2.1. Plant Materials and Treatments
Camellia sinensis var. sinensis cv. ‘Bixiangzao’ (>10-year-old) were used as test materials, which were cultivated in the Tea Germplasm Repository of Hunan Agricultural University (Changsha, China). Nano-Se fertilizers were purchased from Baiwo Huitong Technology Co., LTD., (Beijing, China); their primary component was Se nanoparticles (1500 mg/L) with an average size of 50–78 nm [23]. Five concentrations of nano-selenium fertilizer were investigated, including the control check group (CK, sterile water), 100-fold dilution (D1, 15 mg/L), 200-fold dilution (D2, 7.5 mg/L), 300-fold dilution (D3, 5 mg/L), and 400-fold dilution (D4, 3.75 mg/L). Prior to foliar spraying, all of the experimental tea plants underwent consistent pruning. Tea leaves were sprayed with nano-Se fertilizers until the leaves were moist. The spraying was performed once every 7 days for a total of 3 times in summer (July and August). The experimental field is flat with uniform fertility. Each treatment consisted of two rows of tea plants, and each row was 30 m long and 1 m wide with 1.5 m between rows. The areas of each experimental treatment were approximately 60 m2. After new leaves started sprouting, the healthy and morphologically similar tea leaves with the criterion of one bud and two leaves from three random tea plants in each treatment were collected as three independent biological replicates. The pretreatments of the tea leaves for molecular biology experiments and biochemical determinations were the same as our published methods [24]. Briefly, fresh tea leaves were instantly partly frozen in liquid nitrogen and preserved at −80 °C for molecular biology research. In addition, some of the fresh tea leaves were also liquid-nitrogen-frozen, lyophilized, and then, mechanically crushed for subsequent biochemical determinations.
2.2. Determination of EGCG in Tea Leaves
The major catechin in tea leaves, EGCG, was detected using high-performance liquid chromatograph (HPLC), as described in a previous study [25].
2.3. RNA Isolation, cDNA Synthesis, and Sequensing
Total RNA was extracted using the RNA Prep Pure Plant Kit (Tiangen, Beijing, China). The integrity and concentration of RNA were conducted by 1% agarose gel and NanoDrop 2000C (Thermo Fisher Scientific, Waltham, MA, USA), respectively. The PrimeScriptTM II 1st Strand cDNA Synthesis Kit (Takara, Shiga, Japan) was employed to synthesize the cDNA libraries and then sequenced by the Novogene Bioinformatics Institute (Novogene, Beijing, China). The C. sinensis genome was used as the reference genome [26].
2.4. Quantitative Real-Time PCR (qRT-PCR)
For the qRT-PCR analysis, the reverse-transcribed first-strand cDNA template mentioned above was employed as the template. qRT-PCR was performed by using SYBR® Green Pro Taq HS premixed qPCR Kit (Accurate Biology, Changsha, China) as described previously [27]. The reference gene was β-actin. The reaction system was 20 µL, and three biological replicates were performed for qRT-PCR analysis. Table S3 lists the primers that were utilized in this investigation.
2.5. Gene Clone and Bioinformatics Analysis
The differentially expressed transcription factors (TFs) were screened through transcriptome data (Table S4) to obtain the CsWRKY70, and the CDS sequence of CsWRKY70 (TEA008808) was derived from the reference genome. The full-length sequence of CsWRKY70 was cloned based on our published methods [28], and the gene-specific primers of CsWRKY70 were listed in Table S3. Cloned CsWRKY70 sequence was verified by sequencing.
The molecular weight and isoelectric point of CsWRKY70 were analyzed by ProtParam and ProtScale tools in ExPASy (https://web.expasy.org/protparam/, accessed on 10 January 2022). Homology analysis of the CsWRKY70 protein sequence were carried out in NCBI (BLAST, http://www.ncbi.nih.gov/BLAST/, accessed on 10 January 2022), and the sequences of neighboring species were downloaded as well. The protein sequences of CsWRKY70 and the homologous sequences were analyzed by the NCBI Conserved Domains Database (CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 13 January 2022) and then edited using the Genedoc software (http://genedoc.software.informer.com/, accessed on 15 January 2022). Amino acid sequences of WRKY family of Arabidopsis were downloaded from TAIR (https://www.arabidopsis.org/, accessed on 20 January 2022). A phylogenetic tree was constructed by using the MUSCLE program in MEGA X with 1000 bootstrap replicates (http://www.megasoftware.net/, accessed on 30 January 2022).
2.6. Promoter Isolation and Cis-Acting Elements Analysis
Total DNA was extracted using the cetyl trimethylammonium bromide method from tea leaves. The upstream 2000 bp promoter sequences of CsLAR and CsUGT84A genes were obtained from the plant-care database (http://tpdb.shengxin.ren/, accessed on 16 August 2022). The primers were designed using Premier 5.0 and listed in Table S3. The promoters of CsLAR and CsUGT84A were isolated using high-fidelity enzyme 2X Pro Taq Master Mix (New England Biolabs, Ipswich, MA, USA) by PCR [28]. Cis-acting element analysis of promoters of CsLAR and CsUGT84A was used according to the plant-care database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 18 March 2022).
2.7. Subcellular Location of CsWRKY70 in Tobacco (Nicotiana benthamiana)
The online software Cell-PLOC 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 17 January 2022) was employed to predict the protein subcellular localization of CsWRKY70. To verify the subcellular localization of CsWRKY70, the CsWRKY70 was constructed into a transient expression vector containing the CAMV 35S promoter fused with a green fluorescent protein (GFP) to obtain CsWRKY70-GFP, and 35S-GFP empty was used as a negative control. Subsequently, both GFP-Empty and CsWRKY70-GFP vectors were transformed into Agrobacterium tumefaciens strain EHA105 (Weidi Biotechnology Co., Ltd., Shanghai, China), and the constructs were then injected into tobacco leaves from the lower epidermis. After incubation for 2–3 days, the GFP signals were observed under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
2.8. Transcriptional Activation Analysis
We first used a yeast system to analyze the transcriptional activity of CsWRKY70. The full length of the CsWRKY70 gene obtained a PGBKT7-CsWRKY70 expression vector. The transformed recombinant plasmids, positive control (pGBKT7-53+ PGADT7-T), negative control (PGBKT7-Empty), and the PGBKT7-CsWRKY70 were transformed into yeast cell AH109 (Weidi Biotechnology Co., Ltd., Shanghai, China), respectively. The detailed methods for yeast two-hybrid assay were performed as described [28]. To further verify the transcriptional activity of CsWRKY70, the transcriptional activity analysis was performed in tobacco. The full length of CsWRKY70 was constructed into a 35S promoter-driven pBD vector as an effector. Reporter vector generation and dual luciferase reporter assays were carried out as previously reported [28]. The ratio of Luciferase/Renilla luciferase (LUC/REN) of pBD-CsWRKY70 and pBD-Empty was detected to verify the transcriptional activity of CsWRKY70.
2.9. Protein Expression and Purification
To obtain the recombinant protein, a sequence of CsWRKY70 containing WRKYGQK domain (130–189 aa, a total of 186 bp) was constructed into the prokaryotic expression vector pGEX 4T (Amersham Biosciences, Piscataway, NJ, USA) to generate GST-CsWRKY70 plasmids. The recombinant plasmids of GST-Empty and GST-CsWRKY70 were transformed into Rosetta DE3 (Vazyme, Nanjing, China) and cultured at 37 °C to an OD600 of 0.6, and then, induced with IPTG (Scientific Research Special, USA). After ultrasonic comminution, the GST-Empty and GST-CsWRKY70 recombinant proteins were purified with a Glutathione Resin Kit (TaKaRa, Beijing, China). The protein eluent was collected and stained with SDS-PAEG and Coomassie Bright Blue to verify the expression and purification of CsWRKY70 protein.
2.10. Electrophoretic Mobility Shift Assay (EMSA)
The promoters of CsLAR and CsUGT84A contain W-box (C/T)TGAC(T/C) sequences, which could be recognized by WRKY TFs. Probes were designed to contain the core element TGAC. The probes were labeled with biotin at the 3′ end using DNA Labeling Kit (Thermo Scientific, Waltham, MA, USA). EMSA experiment was performed using the Light ShiftTM EMSA Optimization and Control Kit (Thermo Scientific, Waltham, MA, USA), as previously described [27]. Fluorescent Chemical Imaging System (ProteinSimple, San Jose, CA, USA) was used to detect the binding of the target protein to the probes.
2.11. Dual-Luciferase Reporter Assay System
The CDS sequence of CsWRKY70 was constructed into the pEAQ vector to generate CsWRKY70-pEAQ as an effector, and the promoters of CsLAR and CsUGT84A were constructed into the pGreen II 0800-LUC vectors as reporter plasmids [29]. All the plasmids were then transformed into Agrobacterium tumefaciens strain EHA105, respectively. The effector and reporter plasmids were injected into tobacco leaves at a ratio of 9:1. After incubation for 2~3 days, the LUC/REN ratios were detected by Dual-Luciferase Reporter Kit to evaluate the transcriptional effect of CsWRKY70 on the CsLAR and CsUGT84A promoters.
2.12. Statistical Analysis of Data
At least three different biological duplicates of each experiment were carried out independently. All the experimental data were recorded as mean ± standard error. Statistical analyses were compared using one-way ANOVA or Student’s t-tests. Significant differences were defined as p < 0.05 (*) and p < 0.01 (**).
3. Results
3.1. Effect of Nano-Se Fertilizer on EGCG Content in Tea Leaves
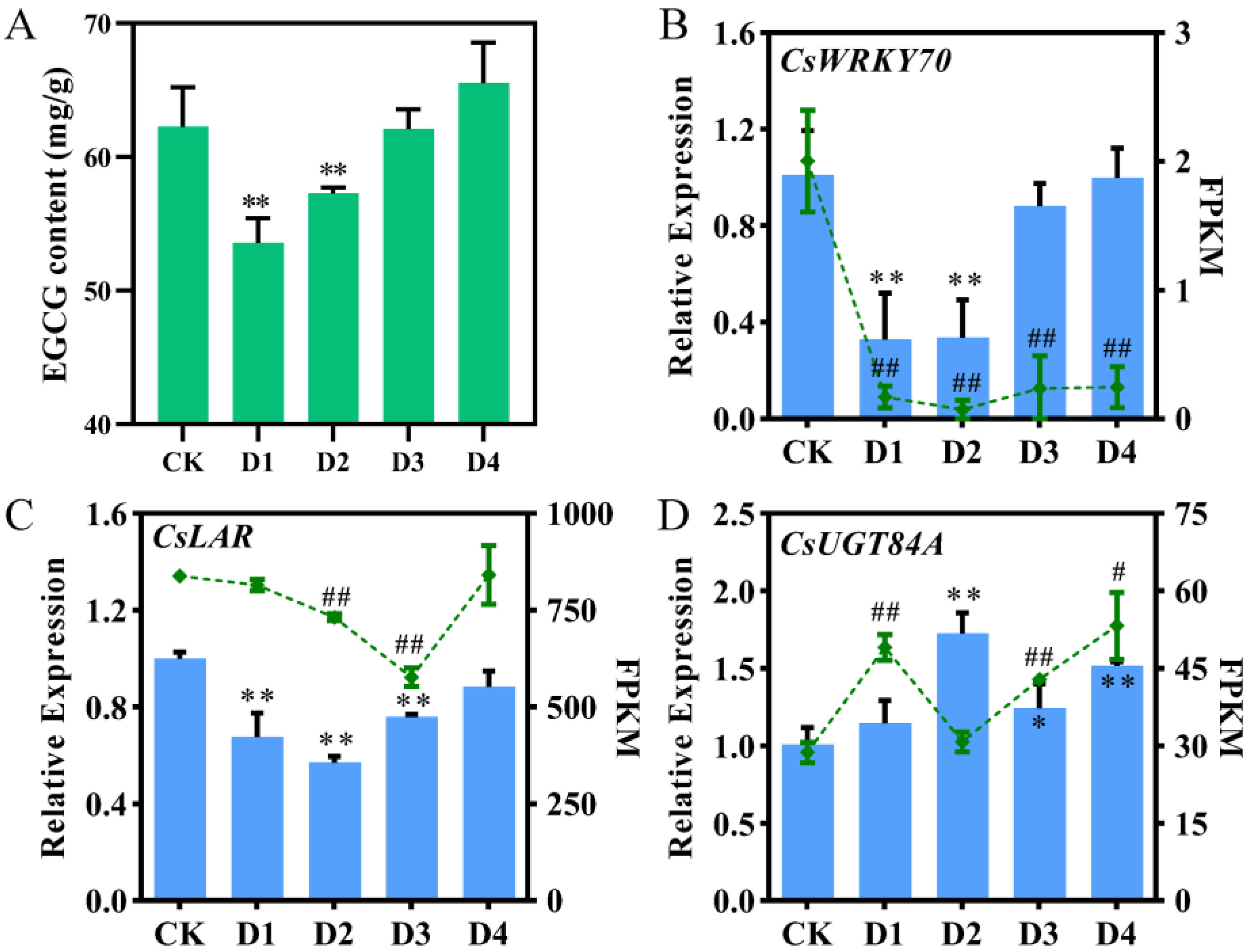
The EGCG content in tea leaves after foliar application of different concentrations of nano-Se was determined using HPLC (Figure 1A). The EGCG contents in the D1 (53.57 mg/g) and D2 (57.30 mg/g) groups were significantly decreased compared to the CK group (62.25 mg/g) (p < 0.01), while there is no significant difference in the D3 (62.08 mg/g) and D4 groups (65.55 mg/g) compared to the CK group. This result indicated that moderate and high concentrations of nano-Se could significantly decrease the EGCG content in summer tea leaves.
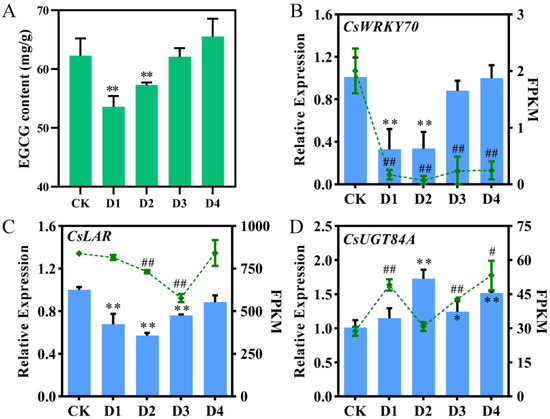
Figure 1.
EGCG content and expression levels of CsWRKY70, CsLAR, and CsUGT84A from qRT-PCR and RNA-seq. (A) EGCG content in tea leaves after foliar application of different concentrations of nano-Se. (B–D) Expression levels of CsWRKY70, CsLAR, and CsUGT84A from qRT-PCR (bar graphs) and RNA-seq (line graphs). FPKM represents fragments per kilobase of transcript per million mapped reads. * and ** represent significant differences (p < 0.05 and p < 0.01, respectively) compared to control in qRT-PCR data. # and ## represent significant differences (p < 0.05 and p < 0.01, respectively) compared to control in RNA-seq data. Data were analyzed by one-way ANOVA.
3.2. Expression Patterns of CsWRKY70, CsLAR, and CsUGT84A
Differentially expressed genes were obtained from our RNA-seq data (Figure S1). A WRKY TF with the same expression pattern as the EGCG content was identified and named CsWRKY70 (Figure 1B). Simultaneously, two flavonoid biosynthetic-related genes, CsLAR and CsUGT84A, were identified and cloned; they were positively and negatively correlated with the expression pattern of CsWRKY70, respectively (Figure 1C,D). To further verify the expression patterns of CsWRKY70, CsLAR, and CsUGT84A in tea leaves under foliar spraying of different concentrations of nano-Se, qRT-PCR was used. Results showed that nano-Se significantly decreased the CsWRKY70 expression in the D1 and D2 groups and showed no significant difference in the D3 and D4 groups compared to the CK (Figure 1B; p < 0.01), which was consistent with our RNA-seq data. Further, expression levels of CsLAR in nano-Se treated groups were all reduced compared to the CK group (Figure 1C; D1, D2, and D3, p < 0.01). In contrast, expression levels of CsUGT84A in nano-Se treated groups were increased compared to the CK group (Figure 1D; D2, D3, and D4, p < 0.01). The qRT-PCR results were basically consistent with our RNA-seq data, supporting the accuracy of the transcriptomic data.
3.3. Identification and Bioinformatics Analysis of CsWRKY70
The CDS sequence of the WRKY TF was cloned by PCR and named CsWRKY70. The open reading frame of CsWRKY70 was 891 bp and encoded a protein containing 296 amino acids. The predicted molecular weight and isoelectric point of CsWRKY70 were 33.39 KDa and 5.23, respectively.
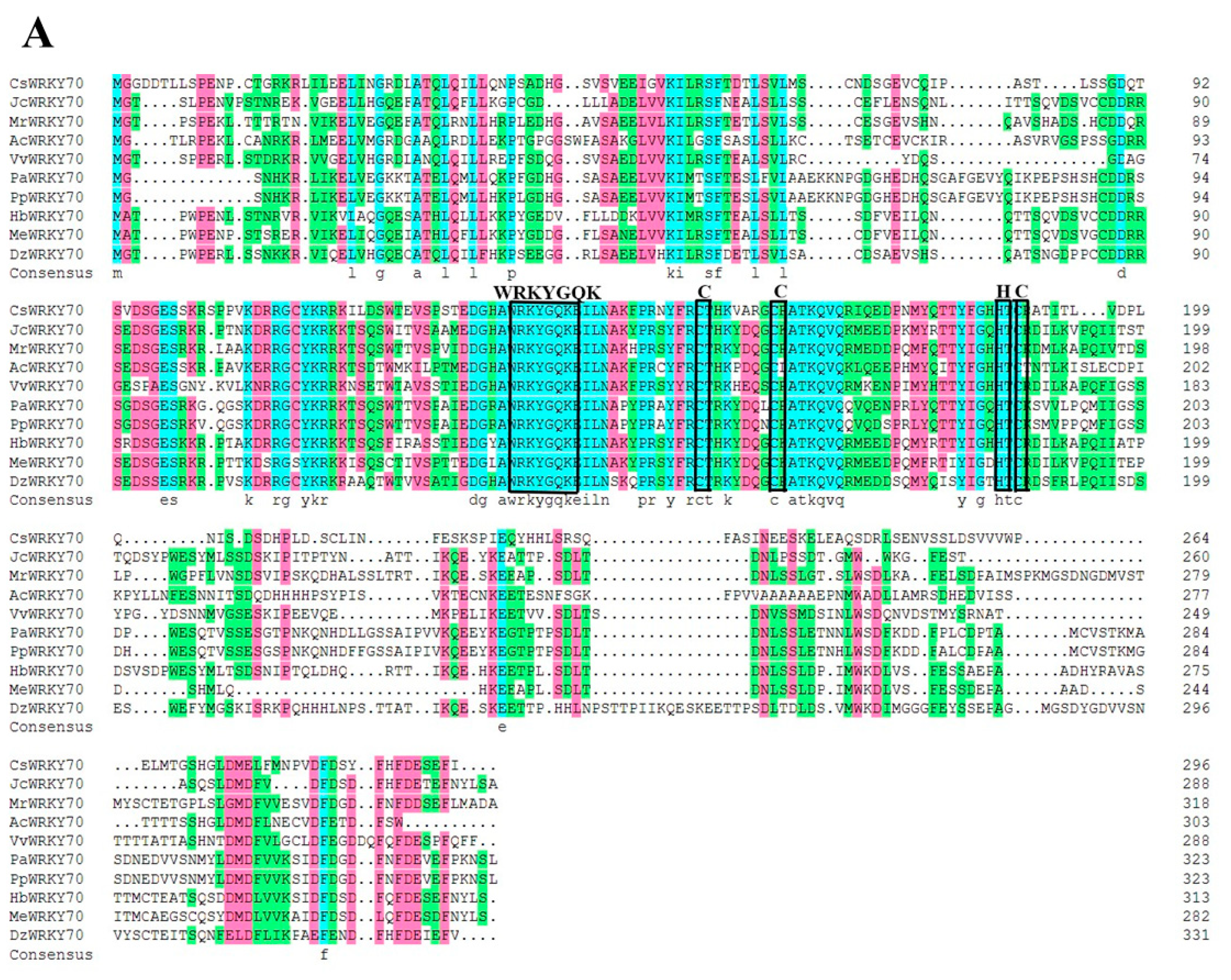
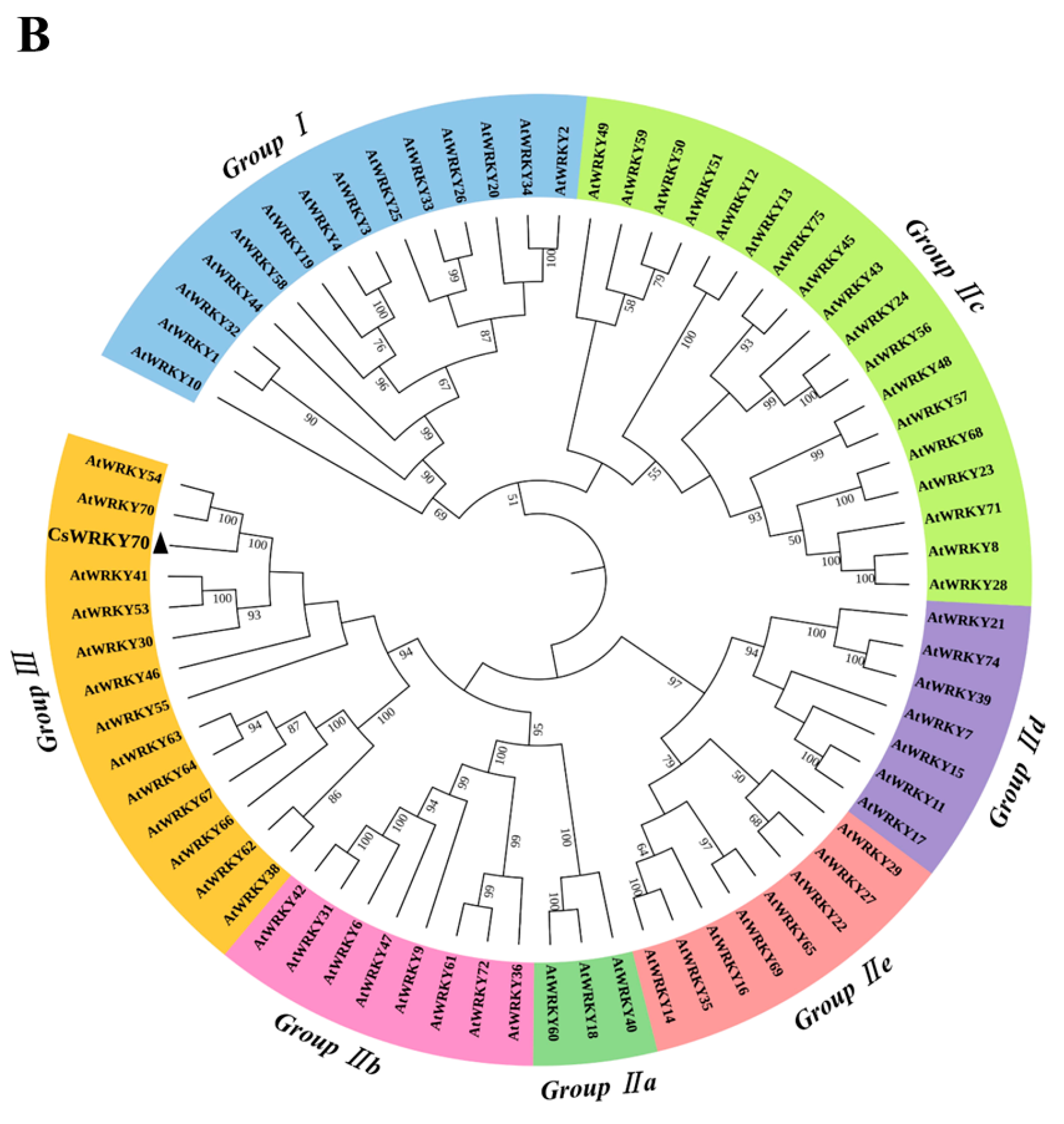
To further characterize the molecular features of CsWRKY70, multiple sequence alignments and phylogenetic analyses were carried out. Sequence alignment showed that CsWRKY70 has a typical WRKYGQK structure, which is an important characteristic of WRKY TFs. There is a WRKYGQK HeptaPeptide sequence at the N-terminal of CsWRKY70 and followed by a C2HC zinc finger structure (Figure 2A), indicating that CsWRKY70 belongs to the WRKY family. Further, a phylogenetic tree of amino acid sequences between CsWRKY70 and WRKY TFs from Arabidopsis thaliana was constructed to determine the phylogenetic characteristics of CsWRKY70. As shown in Figure 2B, WRKY TFs were divided into three groups: WRKY I, WRKY II, and WRKY III. There were five subgroups in the WRKY II group, including WRKY IIa, WRKY IIb, WRKY IIc, WRKY IId, and WRKY IIe. CsWRKY70 belonged to WRKY III group and was homologous to AtWRKY70 and AtWRKY54. It has been revealed that AtWRKY70 and AtWRKY54 are crucial in plant responses to biotic and abiotic stresses and in regulating plant tolerance to osmotic stresses [30]. According to these findings, CsWRKY70 could be critical for physiological and biochemical reactions, such as the biosynthesis of secondary metabolism in tea plants.
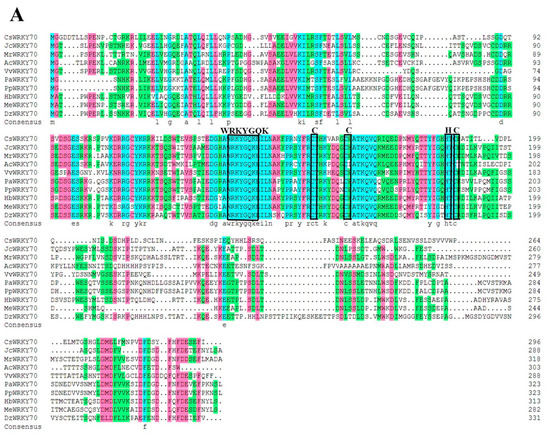
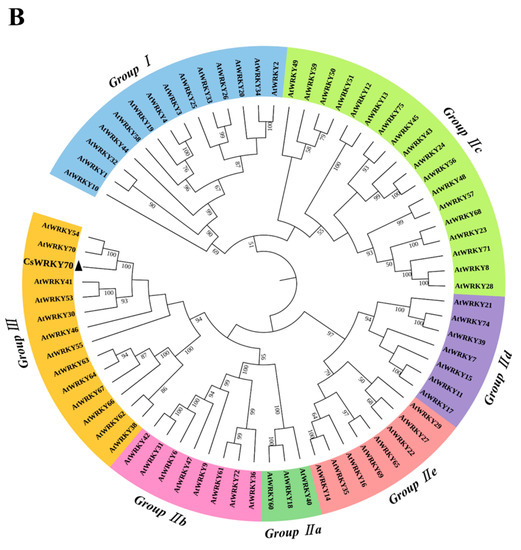
Figure 2.
Multiple sequences alignment and phylogenetic analyses of CsWRKY70. (A) Homology protein sequence alignment between CsWRKY70 and WRKY70 protein sequences of other species (Jatropha curcas, Myrica rubra, Actinidia chinensis, Vitis vinifera, Cerasus avium, Amygdalus persica, Hevea brasiliensis, Manihot esculenta, and Durio zibethinus). Identical or similar amino acids were indicated by blue, red, and green shading. The WRKY domain and C2HC zinc finger were indicated by black boxes. (B) Phylogenetic analysis of CsWRKY70 and Arabidopsis-derived WRKY TFs family. CsWRKY70 was marked with a black triangle.
3.4. Subcellular Localization of CsWRKY70
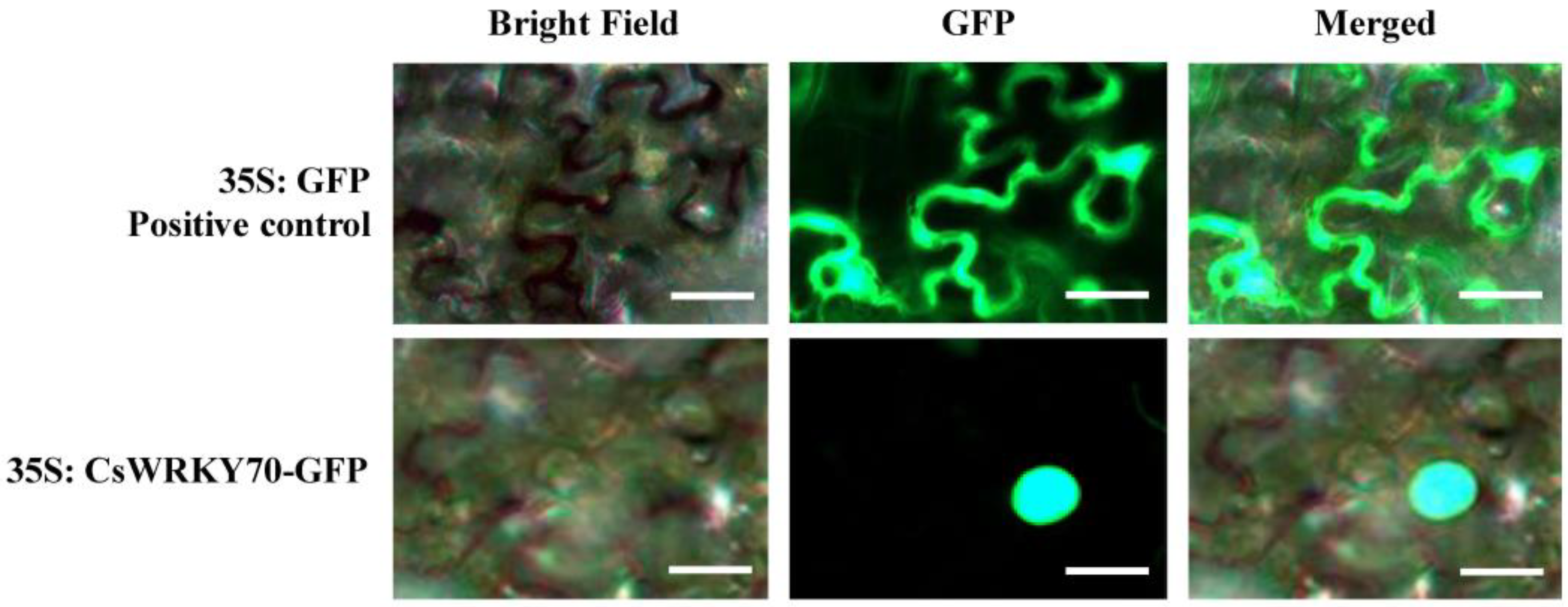
WRKY TFs are typical nuclear proteins with transcriptional activity [31]. CsWRKY70 was predicted to be a nuclear protein using the online software WoLF POSRT. We further verified this result by subcellular localization assays using tobacco. Results showed that CsWRKY70-GFP was localized in the nucleus (Figure 3), while the positive control of GFP-Empty was distributed in the whole cell, indicating that CsWRKY70 is a nuclear localization protein.
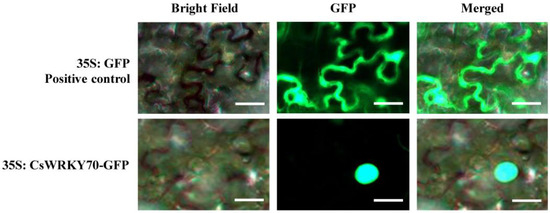
Figure 3.
Subcellular localization of CsWRKY70 via transient expression in tobacco leaves (Nicotiana benthamiana). The white bars denote 45 μm. GFP, green fluorescent protein.
3.5. Transcriptional Activity Analysis of CsWRKY70
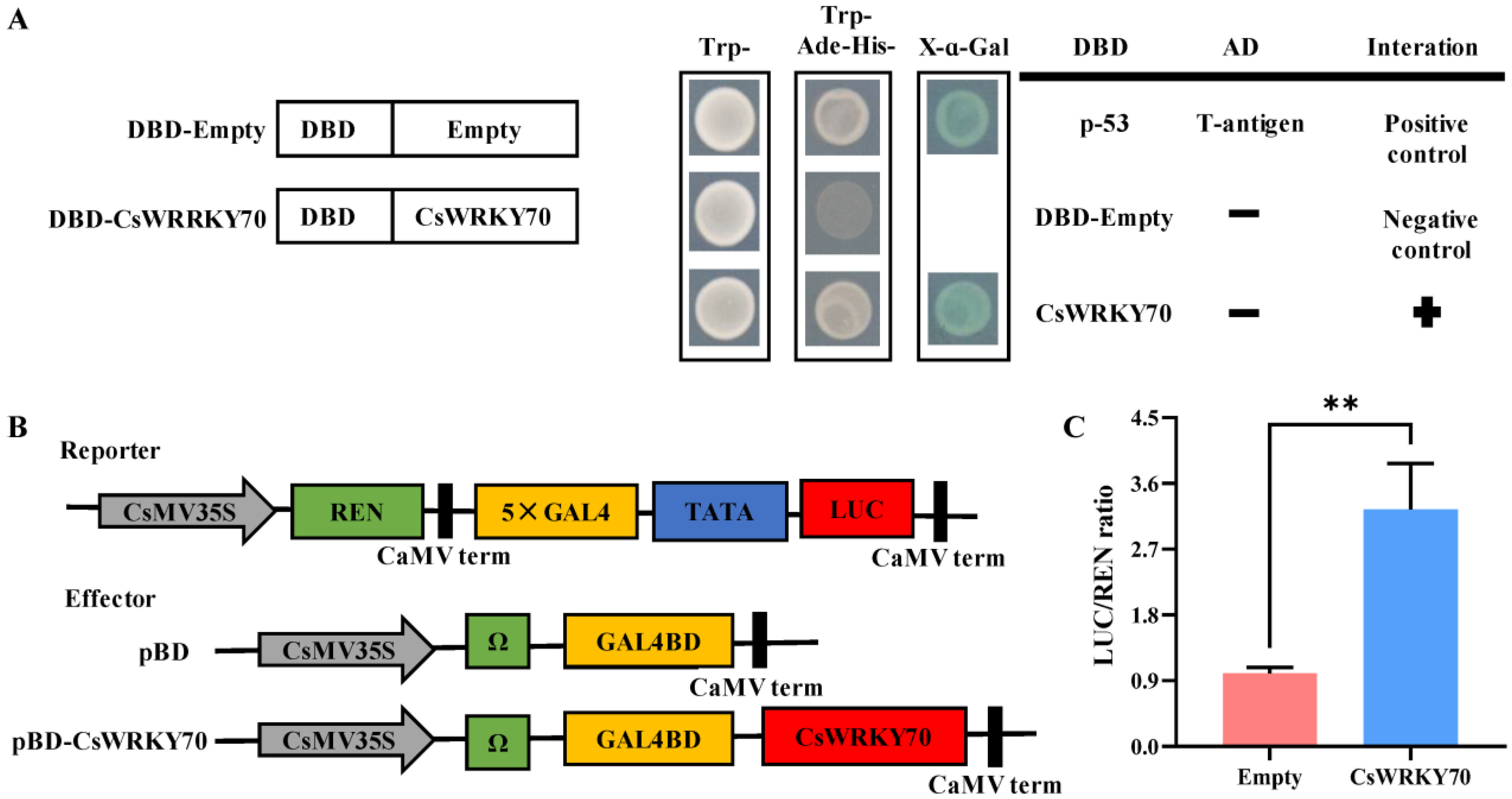
To evaluate the transcriptional activity of CsWRKY70, a GAL4-responsive reporter system was conducted in yeast. The yeast strains transformed with the positive control pGBKT7-53+pGADT7-T, negative control pGBKT7-Empty, and pGBKT7-CsWRKY70 plasmids were grown on the SD/-Trp medium plates, indicating that the yeast transformation was successful (Figure 4A). The transformed strains were transferred into the medium plates (SD/-Trp/-His/-Ade) for further cultivation. The positive control and pGBKT7-CsWRKY70 yeasts grew on the selected plates and exhibited α-galactosidase activity, suggesting that CsWRKY70 functions as a transcription activator with self-activating activity. We further verified the transcriptional regulatory activity of CsWRKY70 using tobacco. The pBD-Empty and pBD-CsWRKY70 were transformed into tobacco and detected by dual luciferase system. Results showed that the LUC/REN ratio of the pBD-CsWRKY70 was substantially higher than that of pBD-Empty (Figure 4B,C), suggesting that CsWRKY70 has transcriptional self-activation activity. The transcriptional activity assay in yeast and tobacco revealed that CsWRKY70 may act as a transcriptional activator.
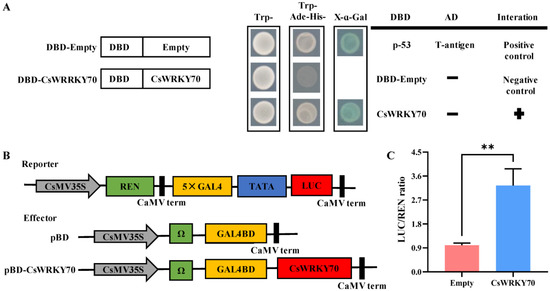
Figure 4.
Transcriptional activity characteristics of CsWRKY70 protein. (A) Transcriptional activity of CsWRKY70 in yeasts. Co-transformed yeasts were grown on selective media (SD medium without tryptophan, histidine, and adenine, SD/−Trp/−His/−Ade). The X-α-galactosidase test was performed to evaluate the transcription activation ability of CsWRKY70. (B) Transcriptional activity of CsWRKY70 in tobacco system (Nicotiana benthamiana). (C) The LUC/REN fluorescence intensity ratio was determined to evaluate the transcription activation ability of CsWRKY70. SD, synthetic defined medium; -Trp/-His/-Ade, medium lacking tryptophan, histidine, and adenine; LUC, Luciferase; REN, Renilla luciferase. ** represents significant differences (p < 0.01) compared to control. Data were analyzed by Student’s t-tests.
3.6. CsWRKY70 Binds to the Promoters of CsLAR and CsUGT84A
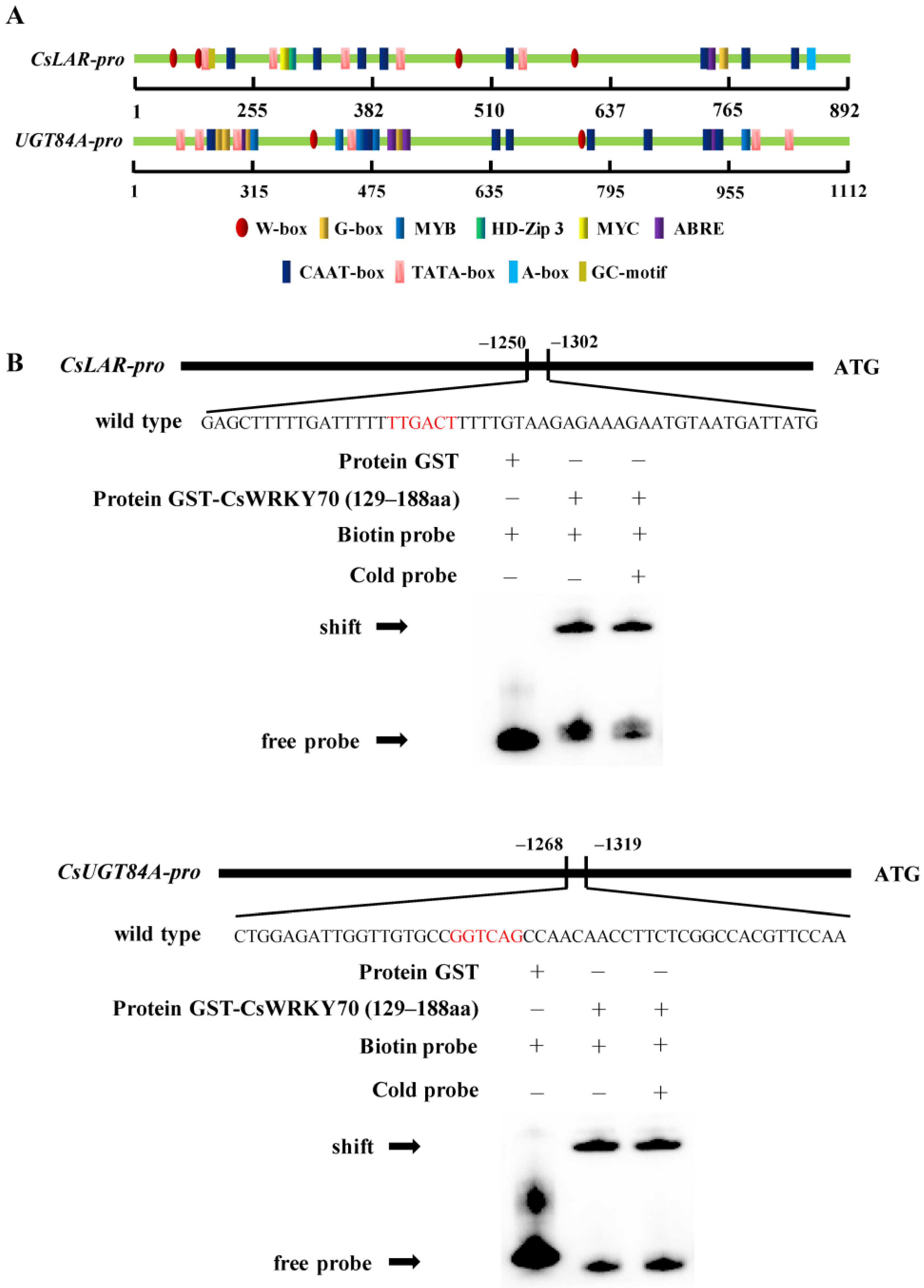
WRKY TFs specifically bind to the W-box elements on the promoter region of the target gene to affect gene expression [32]. Multiple W-box (5’-C/TTGACT/C-3’) sequences were found in CsLAR and CsUGT84A promoters (Figure 5A). The purified recombinant GST- CsWRKY70 protein (130–189 aa) was used to confirm the DNA–protein interactions between CsWRKY70 and the CsLAR and CsUGT84A promoters using EMSA. Results showed that the protein electrophoretic mobility shifts were observed in the presence of recombinant GST-CsWRKY70 protein and biotin-labeled probes of CsLAR and CsUGT84A promoters containing the W-box elements, while the GST-empty protein only the band for the free probe was observed (Figure 5B). These results indicated that the CsWRKY70 fusion protein could directly bind to the W-box motif on the promoters of CsLAR and CsUGT84A.
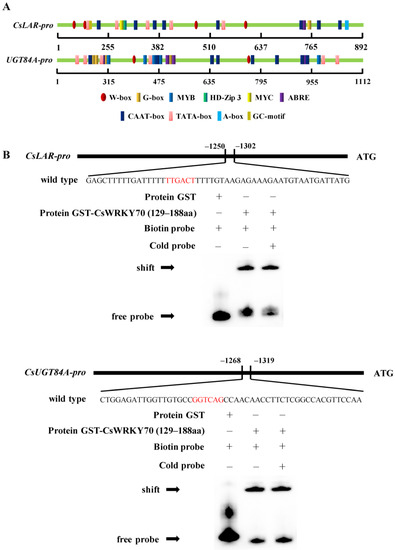
Figure 5.
Cis-elements analysis of CsLAR and CsUGT84A promoters and electrophoretic mobility assays (EMSA) of CsWRKY70 binding to CsLAR and CsUGT84A promoters. (A) Prediction of cis-elements on the CsLAR and CsUGT84A promoters. Ellipses represent W-box elements that can bind to WRKY TFs. (B) Recombinant GST-CsWRKY70 fusion protein binds directly to the promoters of CsLAR and CsUGT84A containing the W-box (T/CTGACT/C). The DNA−protein complexes were separated on 6% native polyacrylamide gels. The ‘−’ and ‘+’ indicate the absence and the presence of specific proteins, respectively.
3.7. Transcriptional Regulatory Effects of CsWRKY70 on CsLAR and CsUGT84A Promoters
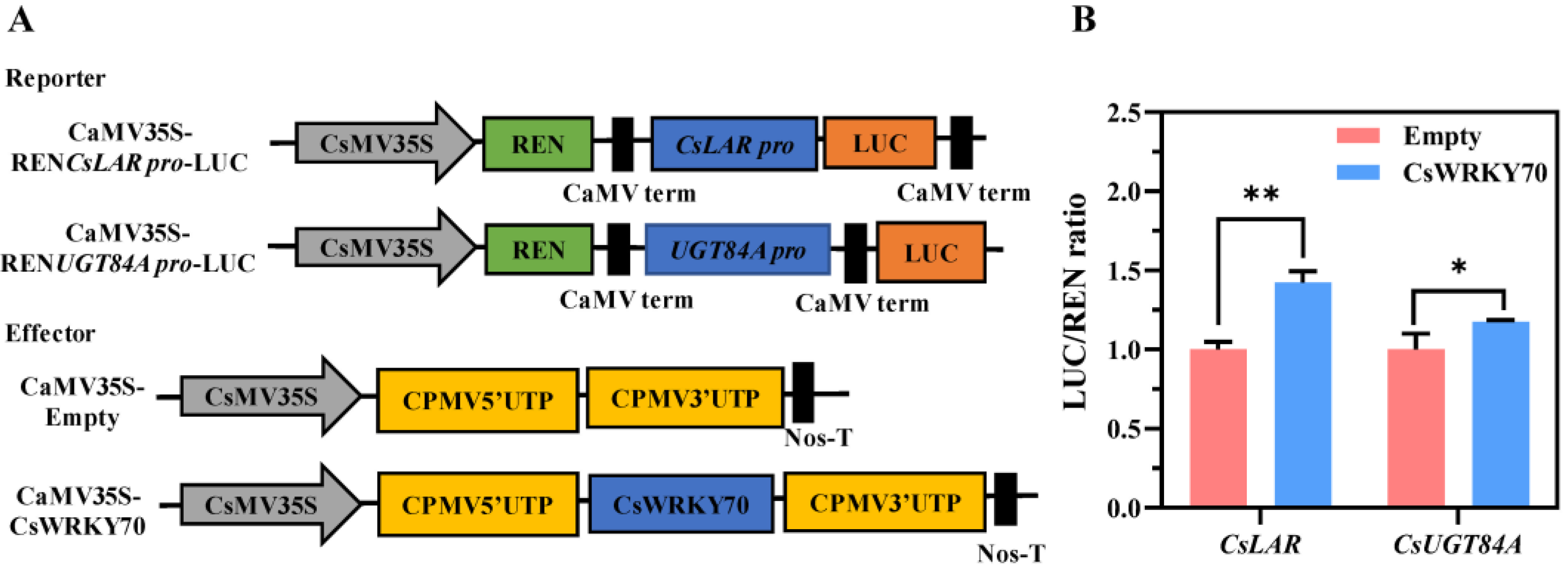
To further determine whether CsWRKY70 can promote the promoter activities of CsLAR and CsUGT84A, a dual luciferase reporter gene assay was performed in tobacco. As shown in Figure 6A, the pEAQ-Empty and pEAQ-CsWRKY70 vectors were used as effectors, and the CsLAR and CsUGT84A promoters were transformed into the pGreen II 0800-LUC vectors as reporters. Compared to the empty control, the LUC/REN ratios of co-transformations of CsWRKY70 and CsLAR pro-LUC and CsWRKY70 and CsUGT84A pro-LUC were remarkably increased (Figure 6B). These results indicated that CsWRKY70 may influence EGCG biosynthesis in tea leaves by regulating CsLAR and CsUGT84A expressions.
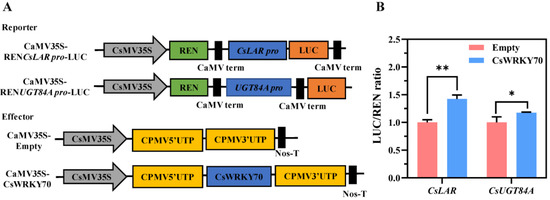
Figure 6.
The ability of CsWRKY70 to activate transcription of CsLAR and CsUGT84A promoters in tobacco leaves (Nicotiana benthamiana). (A) Schematic diagram of vector plasmids. (B) CsWRKY70 activates the promoter activities of CsLAR and CsUGT84A. * and ** represent significant differences (p < 0.05 and p < 0.01, respectively) compared to the empty control. Data were analyzed by Student’s t-tests.
4. Discussion
4.1. CsLAR and CsUGT84A Encode Key Enzymes for EGCG Biosynthesis
EGCG biosynthesis is regulated by several crucial enzymes. LAR has been demonstrated in association with flavonoid biosynthesis in multiple plants, including Vitis vinifera [33], Populus [34], and Lycium ruthenicum Murr [35]. Further, numerous studies have found that LAR is closely related to catechin biosynthesis in tea plants. Kumar et al. found that LAR is one of the key regulators involved in catechin biosynthesis during the development and seasonal variation among different tea plant cultivars [36]. LARs can also promote the biosynthesis of catechin monomers and inhibit their polymerization [37]. Furthermore, LAR gene expression is up-regulated in the pruned tea plants, which is highly correlated with changes in EGCG content in pruned tea plants. Additionally, glycosylation by UGT enzymes is commonly the last process for natural product biosynthesis in plants [38]. Two glycosyltransferases, UGT79B1 and UGT84A2, were found to be involved in anthocyanin biosynthesis in Arabidopsis [39]. In tea plants, it was found that the important precursor of EGCG biosynthesis, β-glucogallin is catalyzed by UGT enzymes from gallic acid and glucose urate diphosphate [40]. Therefore, CsLAR and CsUGT84A may be crucial genes for regulating catechin biosynthesis in tea plants, especially for the most abundant EGCG. In this study, we found that EGCG content is significantly decreased after foliar application of moderate and high concentrations of nano-Se, which is accompanied by the same expression patterns of CsLAR and CsUGT84A genes, indicating that CsLAR and CsUGT84A may be the target genes in response to the effect of nano-Se on EGCG content in tea leaves.
4.2. CsWRKY70 Belongs to the Group III of the WRKY Family and Is Positively Correlated with EGCG Content
WRKY TF family members are critical to plant growth and development, morphogenesis [41], hormone signal transduction [42,43], biological and abiotic stress [44], and secondary metabolite biosynthesis [45]. Our results showed that CsWRKY70 belongs to Group III of the WRKY TF family. Further, CsWRKY70 is homologous to AtWRKY70 and AtWRKY45 in Arabidopsis thaliana (Figure 2B). AtWRKY70 and AtWRKY54 are found to be involved in plant responses to biotic and abiotic stresses and regulate plant tolerance to osmotic stresses [46,47]. The WRKY TF family is also widely investigated in tea plants. Most of the identified WRKY genes in tea plants may be involved in response to drought, cold, ABA, or salt stresses [48]. Recently, WRKY TFs are involved in regulating phenylalanine metabolic pathways in various systems and play crucial roles in the flavonoid biosynthesis in plants. Several WRKY protein members were discovered to be involved in the biosynthesis of anthocyanins and proanthocyanidins in plants [49,50,51]. WRKY TFs are also found to associate with catechin and methylated catechin biosyntheses in tea plants [27,52]. These findings suggest that CsWRKY70 may exhibit the same biological functions in tea plants. In this study, we found that moderate and high concentrations of nano-Se dramatically down-regulated the CsWRKY70 expression in tea leaves. This finding is compatible with the variations in EGCG content after nano-Se treatments. Therefore, we speculated that CsWRKY70 may regulate EGCG biosynthesis affected by nano-Se.
4.3. CsWRKY70 Binds to Cis-Elements of CsLAR and CsUGT84A Promoters and Acts as a Transcriptional Activator
WRKY TFs usually bind to the W-box elements on the promoter region of the target gene [45]. Cis-elements analysis revealed that CsLAR and CsUGT84A promoters contain several W-box elements, indicating that there exist potential interactions between CsWRKY70 and CsLAR and CsUGT84A. Previous research indicated that CsWRKY57like activated the transcription of methylated EGCG biosynthesis-related genes (CCoAOMT, CsLAR, and CsDFR) by binding to the W-box elements [22]. This finding is similar to our result. In our results, CsWRKY70 is localized in the nucleus and processes transcriptional activation ability. However, no information is currently available concerning the regulatory effect of WRKY TFs on CsUGT84A. Further, we demonstrated that CsWRKY70 directly binds to the CsLAR and CsUGT84A promoters and activates the expressions of CsLAR and CsUGT84A. Therefore, CsWRKY70 may regulate EGCG biosynthesis under moderate and high levels of nano-Se treatments by affecting CsLAR and CsUGT84A gene expressions.
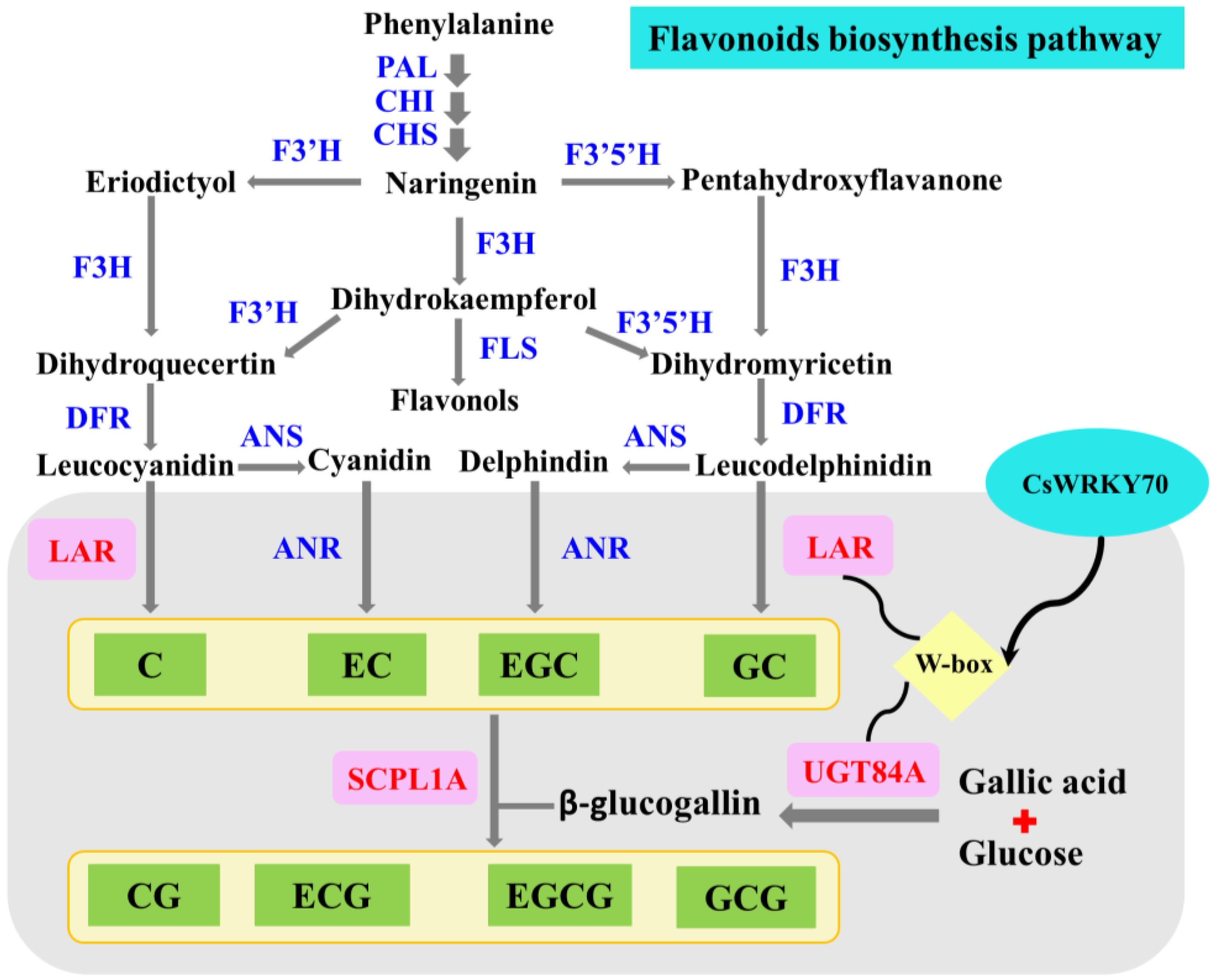
The transcriptional regulatory mechanism of CsWRKY70 on CsLAR and CsUGT84A in the flavonoids biosynthesis pathway is schematized in Figure 7. CsLAR catalyzes the formation of non-galloylated catechins from phenylalanine. CsUGT84A is involved in the formation of β-glucogallin. Further, non-galloylated catechins and β-glucogallin are catalyzed to galloylated catechins by the key enzyme SCPL1A [9]. Recently, CsMYB1 is found to regulate galloylated catechins biosyntheses, such as EGCG and ECG, by regulating CsANR and CsSCPL1A gene expressions in tea leaves [53]. Our results enriched the understanding with regard to how CsWRKY70 might regulate EGCG biosynthesis. In the future, we will explore the regulatory mechanisms of the two crucial gene families, UGT84A and SCPL1A, on the biosynthesis of galloylated catechins in tea plants more deeply.
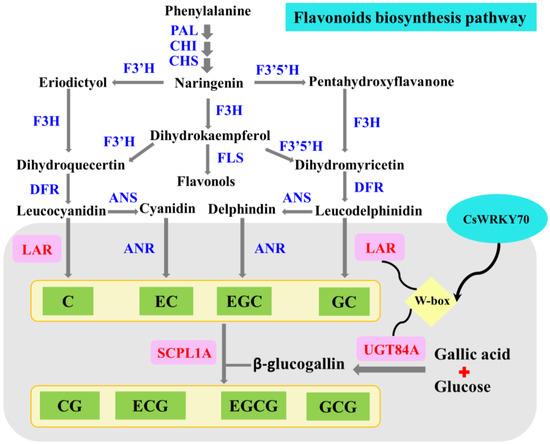
Figure 7.
Diagram depicting the mechanism of CsWRKY70 affecting EGCG biosynthesis by regulating CsANR and CsSCPL1A expressions in tea leaves after nano-Se foliar application. PAL, phenylalanine ammonia lyase; CHI, chalcone isomerase; CHS, chalcone synthase; F3′H, flavanone 3-hydroxylase; F3′5′H, flavonoid 3′, 5′-hydroxylase; F3H, flavanone-3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; LAR, leucoanthocyanidin reductase; SCPL1A, serine carboxypeptidase-like acyltransferase; UGT84A, uridine diphosphate glucuronosyltransferase; C, catechin; EC, epicatechin; EGC, epigallocatechin; GC, gallocatechin; CG, catechin gallate; ECG, epicatechin gallate; EGCG, epigallocatechin gallate; GCG, gallocatechin gallate.
5. Conclusions
This work demonstrated that nano-Se fertilizers can significantly decrease the EGCG content in summer tea leaves. Transcriptome sequencing data suggested that expressions of a WRKY TF (CsWRKY70) and catechin biosynthetic-related genes (CsANR and CsSCPL1A) were positively correlated with EGCG content. CsWRKY70 was further isolated from tea leaves and found to be a transcriptional activator. CsWRKY70 is a nuclear localization protein, which can specifically bind to cis-acting elements (W-box) on the promoters of CsLAR and CsUGT84A. These results indicated that CsWRKY70 affects EGCG biosynthesis by regulating CsANR and CsSCPL1A expressions in tea leaves after nano-Se foliar application. Our results enrich the knowledge of the molecular regulatory mechanism of WRKY TFs involved in catechin biosynthesis and provide a theoretical basis for molecular breeding and innovation of tea cultivars.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9010120/s1, Table S1: contents of major chemical components in tea leaves after foliar spraying of nano-Se (%). Table S2: contents of catechins in tea leaves after foliar spraying of nano-Se (mg/g). Table S3: list of primers used in this study. Table S4: differentially expressed transcription factors after foliar application of different concentrations of nano-Se. Figure S1: gene expression levels of differentially expressed catechin biosynthesis-related genes from transcriptome.
Author Contributions
Conceptualization, X.S., K.W. and M.Z.; methodology, Q.L., H.L. and S.B.; software, X.S. and X.H.; validation, X.S. and X.H.; formal analysis, J.L., M.Z. and Q.L.; investigation, X.S.; resources, K.W.; data curation, X.S. and X.H.; writing—original draft preparation, X.S. and X.H.; writing—review and editing, X.S., K.W., M.Z., J.L. and X.H.; visualization, X.H. and H.L.; supervision, H.L., S.B., Q.L. and K.W.; project administration, J.L., M.Z. and K.W.; funding acquisition, J.L., M.Z. and K.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Key Research and Development Program of China (No. 2022YFD2101102), National Natural Science Foundation of China (32072629, U19A2030), Natural Science Foundation of Hunan Province for Outstanding Young Scholars (2022JJ20028), Yunnan Province Key Research and Development Program of China (202202AE090030), and the Changsha City Outstanding Innovative Youth Training Program (No. kq2107015).
Data Availability Statement
All data are available in the manuscript or Tables S1–S4 and Figure S1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu, Y.Q.; Zhang, Y.N.; Chen, J.X.; Wang, F.; Du, Q.Z.; Yin, J.F. Quantitative analyses of the bitterness and astringency of catechins from green tea. Food Chem. 2018, 258, 16–24. [Google Scholar] [CrossRef]
- Rasheed, N.O.A.; Ahmed, L.A.; Abdallah, D.M.; El-Sayeh, B.M. Paradoxical cardiotoxicity of intraperitoneally-injected epigallocatechin gallate preparation in diabetic mice. Sci. Rep. 2018, 8, 7880. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.X.; Xu, S.; Yu, L.J.; Zhou, Y.F.; Zhou, Y.; Liu, Z.H. Eurotium cristatum Fermented loose dark tea ameliorates cigarette smoke-induced lung injury by MAPK pathway and enhances hepatic metabolic detoxification by PXR/AhR pathway in mice. Oxid. Med. Cell. Longev. 2021, 2021, 6635080. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Xu, W.; Guo, T.; Pan, H.; Zou, H.; Jiang, L.; Li, C.; Gao, S. (-)-Epigallocatechin-3-gallate inhibits the life cycle of pseudorabies virus in vitro and protects mice against fatal infection. Front. Cell. Infect. Microbiol. 2020, 10, 616895. [Google Scholar] [CrossRef]
- Gong, A.-D.; Lian, S.-B.; Wu, N.-N.; Zhou, Y.-J.; Zhao, S.-Q.; Zhang, L.-M.; Cheng, L.; Yuan, H.-Y. Integrated transcriptomics and metabolomics analysis of catechins, caffeine and theanine biosynthesis in tea plant (Camellia sinensis) over the course of seasons. BMC Plant Biol. 2020, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.Q.; Zou, C.; Zhang, Y.H.; Du, Q.Z.; Yin, J.F.; Shi, J.; Xue, S.; Xu, Y.Q. Improving the taste of autumn green tea with tannase. Food Chem. 2019, 277, 432–437. [Google Scholar] [CrossRef]
- Huang, X.; Yu, S.; Chen, S.; Lin, H.; Luo, Y.; Li, J.; Zhu, M.; Wang, K. Complementary transcriptomic and metabolomics analysis reveal the molecular mechanisms of EGCG3″Me biosynthesis in Camellia sinensis. Sci. Hortic. 2022, 304, 111340. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, L.; Liu, L.; Yang, Q.; Lu, Z.; Nie, Z.; Wang, Y.; Xia, T. Purification and characterization of a novel galloyltransferase involved in catechin galloylation in the tea plant (Camellia sinensis). J. Biol. Chem. 2012, 287, 44406–44417. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.Z.; Li, P.; She, G.; Xia, E.; Benedito, V.A.; Wan, X.C.; Zhao, J. Genome-wide analysis of serine carboxypeptidase-like acyltransferase gene family for evolution and characterization of enzymes involved in the biosynthesis of galloylated catechins in the tea plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 848. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef]
- Zhang, X.; Song, J.Y.; Hu, Y.L.; Xu, J.; Xu, Z.C.; Ji, A.J.; Luo, H.M.; Chen, S.L. Research progress of the regulation on active compound biosynthesis by the bHLH transcription factors in plants. Yao Xue Xue Bao 2014, 49, 435–442. [Google Scholar]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Dadalto, S.P.; Goncalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Jin, W.; Zhou, Q.; Wei, Y.; Yang, J.; Hao, F.; Cheng, Z.; Guo, H.; Liu, W. NtWRKY-R1, a novel transcription factor, integrates iaa and ja signal pathway under topping damage stress in Nicotiana tabacum. Front. Plant Sci. 2017, 8, 2263. [Google Scholar] [CrossRef]
- Zhao, N.; He, M.; Li, L.; Cui, S.; Hou, M.; Wang, L.; Mu, G.; Liu, L.; Yang, X. Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS ONE 2020, 15, e0231396. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Zhang, T.; Jiang, S.; Xu, H.; Wang, Y.; Zhang, Z.; Wang, C.; Chen, X. Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 2018, 66, 7076–7086. [Google Scholar] [CrossRef]
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Huang, X.X.; Song, X.F.; Wen, B.B.; Xie, N.C.; Wang, K.B.; Huang, J.A.; Liu, Z.H. Identification of a WRKY transcriptional activator from Camellia sinensis that regulates methylated EGCG biosynthesis. Hortic. Res. 2022, 9, uhac024. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, C.; Zou, N.; Wu, Y.; Zhang, J.; An, Q.; Li, J.-Q.; Pan, C. Nanoselenium foliar application enhances biosynthesis of tea leaves in metabolic cycles and associated responsive pathways. Environ. Pollut. 2021, 273, 116503. [Google Scholar] [CrossRef]
- Huang, X.; Tang, Q.; Li, Q.; Lin, H.; Li, J.; Zhu, M.; Liu, Z.; Wang, K. Integrative analysis of transcriptome and metabolome reveals the mechanism of foliar application of Bacillus amyloliquefaciens to improve summer tea quality (Camellia sinensis). Plant Physiol. Biochem. 2022, 185, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Z.; Zhou, F.; Ran, L.S.; Li, Y.L.; Tan, B.; Wang, K.B.; Huang, J.A.; Liu, Z.H. Metabolic Profiling and Gene Expression Analyses of Purple-Leaf Formation in Tea Cultivars (Camellia sinensis var. sinensis and var. assamica). Front. Plant Sci. 2021, 12, 606962. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef]
- Luo, Y.; Yu, S.S.; Li, J.; Li, Q.; Wang, K.B.; Huang, J.N.; Liu, Z.H. Molecular characterization of WRKY transcription factors that act as negative regulators of O-methylated catechin biosynthesis in tea plants (Camellia sinensis L.). J. Agric. Food Chem. 2018, 66, 11234–11243. [Google Scholar] [CrossRef]
- Huang, X.X.; Ou, S.Q.; Li, Q.; Luo, Y.; Lin, H.Y.; Li, J.; Zhu, M.Z.; Wang, K.B. The R2R3 Transcription factor CsMYB59 regulates polyphenol oxidase gene CsPPO1 in tea plants (Camellia sinensis). Front. Plant Sci. 2021, 12, 39951. [Google Scholar] [CrossRef]
- Fan, Z.Q.; Ba, L.J.; Shan, W.; Xiao, Y.Y.; Lu, W.J.; Kuang, J.F.; Chen, J.Y. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef]
- Yang, Y.; Chi, Y.J.; Wang, Z.; Zhou, Y.; Fan, B.F.; Chen, Z.X. Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. J. Exp. Bot. 2016, 67, 4727–4742. [Google Scholar] [CrossRef]
- Kang, G.J.; Yan, D.; Chen, X.L.; Li, Y.; Yang, L.F.; Zeng, R.Z. Molecular characterization and functional analysis of a novel WRKY transcription factor HbWRKY83 possibly involved in rubber production of Hevea brasiliensis. Plant Physiol. Biochem. 2020, 155, 483–493. [Google Scholar] [CrossRef]
- Bi, C.W.; Xu, Y.Q.; Ye, Q.L.; Yin, T.M.; Ye, N. Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. Peerj 2016, 4, e2437. [Google Scholar] [CrossRef]
- Wen, P.F.; Ji, W.; Gao, M.Y.; Niu, T.Q.; Xing, Y.F.; Niu, X.Y. Accumulation of flavanols and expression of leucoanthocyanidin reductase induced by postharvest UV-C irradiation in grape berry. Genet. Mol. Res. 2015, 14, 7687–7695. [Google Scholar] [CrossRef]
- Ullah, C.; Unsicker, S.B.; Reichelt, M.; Gershenzon, J.; Hammerbacher, A. Accumulation of catechin and proanthocyanidins in black poplar stems after infection by Plectosphaerella populi: Hormonal regulation, biosynthesis and antifungal activity. Front. Plant Sci. 2019, 10, 1441. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, Y. Characterization of proanthocyanin-related leucoanthocyanidin reductase and anthocyanidin reductase genes in Lycium ruthenicum Murr. J. Chin. Pharm. Sci. 2014, 23, 369–377. [Google Scholar] [CrossRef]
- Kumar, A.; Chawla, V.; Sharma, E.; Mahajan, P.; Shankar, R.; Yadav, S.K. Comparative transcriptome analysis of chinary, assamica and cambod tea (Camellia sinensis) types during development and seasonal variation using RNA-seq technology. Sci. Rep. 2016, 6, 37244. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Jiang, X.; Dai, X.; Xu, L.; Li, T.; Xing, D.; Li, Y.; Li, M.; Gao, L.; et al. Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 2018, 247, 139–154. [Google Scholar] [CrossRef]
- Gilbert, M.K.; Bland, J.M.; Shockey, J.M.; Cao, H.; Hinchliffe, D.J.; Fang, D.D.; Naoumkina, M. A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of Ligon lintless-2 mutant of cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e75268. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, P.C.; Liu, P.P.; Song, X.W.; Guo, F.; Li, Y.Y.; Ni, D.J.; Jiang, C.J. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019, 272, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, Y.; Zhang, Q.; Ren, S.; Shen, Y.; Qin, L.; Xing, Y. Genome-wide analysis of the expression of WRKY family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLoS ONE 2016, 11, e0154312. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Yan, D.; Chen, X.; Yang, L.; Zeng, R. HbWRKY82, a novel IIc WRKY transcription factor from Hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in Arabidopsis. Physiol. Plant 2021, 171, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C.; Pattanaik, S.; Patra, B.; Yuan, L. Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genom. 2014, 15, 502. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Wu, F.; Hao, Q.; Ji, K. Transcriptome-wide identification of WRKY transcription factors and their expression profiles under different types of biological and abiotic stress in Pinus massoniana Lamb. Genes 2020, 11, 1386. [Google Scholar] [CrossRef]
- Wei, W.; Cheng, M.N.; Ba, L.J.; Zeng, R.X.; Luo, D.L.; Qin, Y.H.; Liu, Z.L.; Kuang, J.F.; Lu, W.J.; Chen, J.Y.; et al. Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1. Int. J. Mol. Sci. 2019, 20, 1890. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Törönen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 2013, 200, 457–472. [Google Scholar] [CrossRef]
- Wang, P.; Yue, C.; Chen, D.; Zheng, Y.; Zhang, Q.; Yang, J.; Ye, N. Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 2019, 41, 17–33. [Google Scholar] [CrossRef]
- Verweij, W.; Spelt, C.E.; Bliek, M.; de Vries, M.; Wit, N.; Faraco, M.; Koes, R.; Quattrocchio, F.M. Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell 2016, 28, 786–803. [Google Scholar] [CrossRef]
- Chen, P.; Liu, Q.Z. Genome-wide characterization of the WRKY gene family in cultivated strawberry (Fragaria x ananassa Duch.) and the importance of several group III members in continuous cropping. Sci. Rep. 2019, 9, 8423. [Google Scholar] [CrossRef]
- Amato, A.; Cavallini, E.; Zenoni, S.; Finezzo, L.; Begheldo, M.; Ruperti, B.; Tornielli, G.B. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front Plant Sci. 2017, 7, 1979. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhang, X.; He, Y.; Li, L.; Wang, Y.; Hong, G.; Xu, P. Transcriptomic analysis reveals the molecular adaptation of three major secondary metabolic pathways to multiple macronutrient starvation in tea (Camellia sinensis). Genes 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Fu, J.; Xu, Y.; Shen, Y.; Zhang, Y.; Ye, Z.; Tong, W.; Zeng, X.; Yang, J.; Tang, D.; et al. CsMYB1 integrates the regulation of trichome development and catechins biosynthesis in tea plant domestication. New Phytol 2022, 234, 902–917. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).