Transfer Learning in Magnetic Resonance Brain Imaging: A Systematic Review
Abstract
:1. Introduction
2. Transfer Learning
Related Approaches
3. Methods
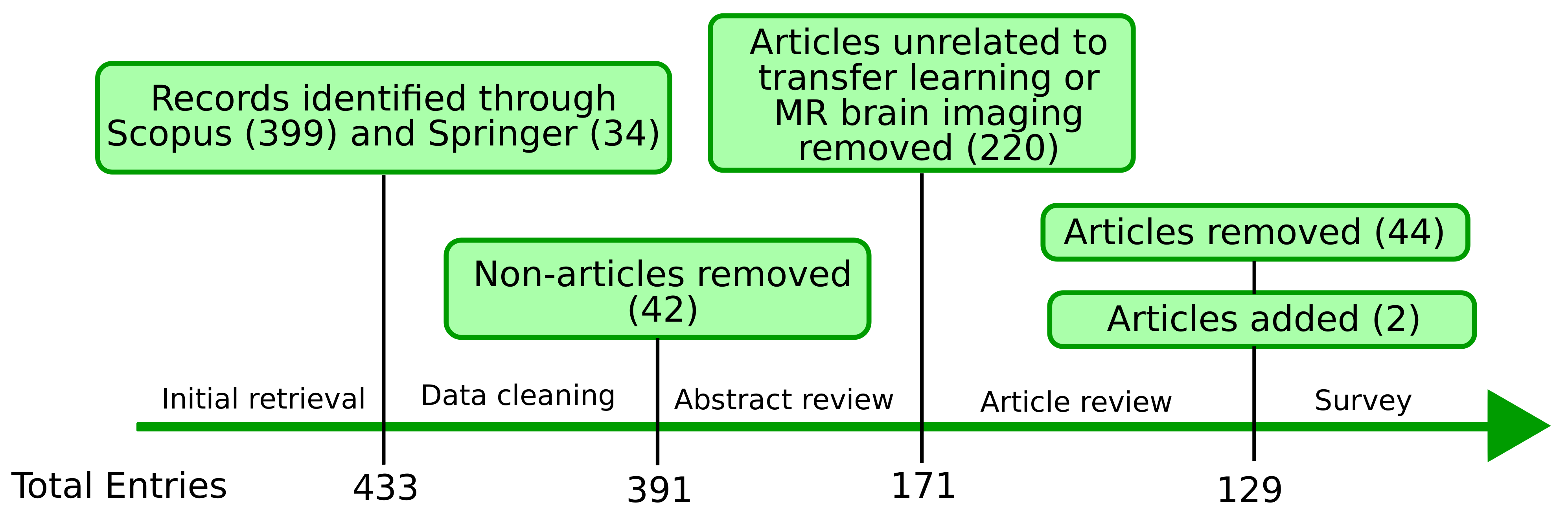
3.1. Search Strategy
3.2. Study Selection
3.3. Data Collection
3.4. Screening Process
4. Results
4.1. Applications
4.2. Machine Learning Approaches
4.3. Transfer Learning Approaches
4.3.1. Instance-Based Approaches
4.3.2. Feature-Based Approaches
4.3.3. Parameter-Based Approaches
4.4. Transfer Learning Approaches Relevant to MR Brain Imaging
- Brain MRI-specificity
- Privacy
- Unseen Target Domains
- Unlabeled Data
4.5. Tackling Transfer Learning Issues
5. Discussion
5.1. Research Directions of Transfer Learning in Brain MRI
5.2. Knowledge Gaps
5.3. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lerch, J.P.; van der Kouwe, A.J.; Raznahan, A.; Paus, T.; Johansen-Berg, H.; Miller, K.L.; Smith, S.M.; Fischl, B.; Sotiropoulos, S.N. Studying neuroanatomy using MRI. Nat. Neurosci. 2017, 20, 314–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glover, G.H. Overview of functional magnetic resonance imaging. Neurosurg. Clin. 2011, 22, 133–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.K. Diffusion Mri; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Cao, X.; Fan, J.; Dong, P.; Ahmad, S.; Yap, P.T.; Shen, D. Image registration using machine and deep learning. In Handbook of Medical Image Computing and Computer Assisted Intervention; Elsevier: Amsterdam, The Netherlands, 2020; pp. 319–342. [Google Scholar]
- Falahati, F.; Westman, E.; Simmons, A. Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. J. Alzheimer’s Dis. 2014, 41, 685–708. [Google Scholar] [CrossRef]
- Jovicich, J.; Czanner, S.; Greve, D.; Haley, E.; van Der Kouwe, A.; Gollub, R.; Kennedy, D.; Schmitt, F.; Brown, G.; MacFall, J.; et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006, 30, 436–443. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Calhoun, V.D.; Arias-Vasquez, A.; Zwiers, M.P.; Gupta, C.N.; Franke, B.; Turner, J.A. Exploration of scanning effects in multi-site structural MRI studies. J. Neurosci. Methods 2014, 230, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Day, O.; Khoshgoftaar, T.M. A survey on heterogeneous transfer learning. J. Big Data 2017, 4, 29. [Google Scholar] [CrossRef]
- Woodworth, R.S.; Thorndike, E. The influence of improvement in one mental function upon the efficiency of other functions (I). Psychol. Rev. 1901, 8, 247. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Cheplygina, V.; de Bruijne, M.; Pluim, J.P. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 2019, 54, 280–296. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Kiik, M.; Peek, N.; Curcin, V.; Marshall, I.J.; Rudd, A.G.; Wang, Y.; Douiri, A.; Wolfe, C.D.; Bray, B. A systematic review of machine learning models for predicting outcomes of stroke with structured data. PLoS ONE 2020, 15, e0234722. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Isaza, G.; Guyot, R.; Tabares-Soto, R. A systematic review of the application of machine learning in the detection and classification of transposable elements. PeerJ 2019, 7, e8311. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; Liu, D.; Philpotts, L.L.; Turner, D.P.; Houle, T.T.; Chen, L.; Zhang, M.; Yang, J.; Zhang, W.; Deng, H. Current state of science in machine learning methods for automatic infant pain evaluation using facial expression information: Study protocol of a systematic review and meta-analysis. BMJ Open 2019, 9, e030482. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Gao, J.; Ngo, H.; Li, K.; Zhang, A. On handling negative transfer and imbalanced distributions in multiple source transfer learning. Stat. Anal. Data Min. ASA Data Sci. J. 2014, 7, 254–271. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, Z.; Póczos, B.; Carbonell, J. Characterizing and avoiding negative transfer. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 11293–11302. [Google Scholar]
- Leen, G.; Peltonen, J.; Kaski, S. Focused multi-task learning in a Gaussian process framework. Mach. Learn. 2012, 89, 157–182. [Google Scholar] [CrossRef]
- Weninger, L.; Liu, Q.; Merhof, D. Multi-task Learning for Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2019; pp. 327–337. [Google Scholar]
- Zhou, C.; Ding, C.; Wang, X.; Lu, Z.; Tao, D. One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans. Image Process. 2020, 29, 4516–4529. [Google Scholar] [CrossRef] [Green Version]
- Nalepa, J.; Marcinkiewicz, M.; Kawulok, M. Data augmentation for brain-tumor segmentation: A review. Front. Comput. Neurosci. 2019, 13, 83. [Google Scholar] [CrossRef] [Green Version]
- Zhao, A.; Balakrishnan, G.; Durand, F.; Guttag, J.V.; Dalca, A.V. Data augmentation using learned transformations for one-shot medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 8543–8553. [Google Scholar]
- Liu, Q.; Dou, Q.; Heng, P.A. Shape-aware Meta-learning for Generalizing Prostate MRI Segmentation to Unseen Domains. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 475–485. [Google Scholar]
- Finn, C.; Abbeel, P.; Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. arXiv 2017, arXiv:1703.03400. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, I.; Nomura, Y.; Hanaoka, S.; Miki, S.; Hayashi, N.; Abe, O.; Masutani, Y. Managing Computer-Assisted Detection System Based on Transfer Learning with Negative Transfer Inhibition. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 695–704. [Google Scholar]
- Huang, Y.L.; Hsieh, W.T.; Yang, H.C.; Lee, C.C. Conditional Domain Adversarial Transfer for Robust Cross-Site ADHD Classification Using Functional MRI. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1190–1194. [Google Scholar]
- Martyn, P.; McPhilemy, G.; Nabulsi, L.; Martyn, F.; McDonald, C.; Cannon, D.; Schukat, M. Using Magnetic Resonance Imaging to Distinguish a Healthy Brain from a Bipolar Brain: A Transfer Learning Approach; AICS: Galway, Ireland, 6 December 2019. [Google Scholar]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Deep Learning Techniques for Automatic Detection of Embryonic Neurodevelopmental Disorders. Diagnostics 2020, 10, 27. [Google Scholar] [CrossRef] [Green Version]
- Si, X.; Zhang, X.; Zhou, Y.; Sun, Y.; Jin, W.; Yin, S.; Zhao, X.; Li, Q.; Ming, D. Automated Detection of Juvenile Myoclonic Epilepsy using CNN based Transfer Learning in Diffusion MRI. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1679–1682. [Google Scholar]
- Chougule, T.; Shinde, S.; Santosh, V.; Saini, J.; Ingalhalikar, M. On Validating Multimodal MRI Based Stratification of IDH Genotype in High Grade Gliomas Using CNNs and Its Comparison to Radiomics. In International Workshop on Radiomics and Radiogenomics in Neuro-Oncology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 53–60. [Google Scholar]
- Eitel, F.; Soehler, E.; Bellmann-Strobl, J.; Brandt, A.U.; Ruprecht, K.; Giess, R.M.; Kuchling, J.; Asseyer, S.; Weygandt, M.; Haynes, J.D.; et al. Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional MRI using layer-wise relevance propagation. Neuroimage Clin. 2019, 24, 102003. [Google Scholar] [CrossRef]
- Samani, Z.R.; Alappatt, J.A.; Parker, D.; Ismail, A.A.O.; Verma, R. QC-Automator: Deep learning-based automated quality control for diffusion mr images. Front. Neurosci. 2019, 13, 1456. [Google Scholar] [CrossRef] [Green Version]
- Dong, Q.; Zhang, J.; Li, Q.; Wang, J.; Leporé, N.; Thompson, P.M.; Caselli, R.J.; Ye, J.; Wang, Y. Alzheimer’s Disease Neuroimaging Initiative. Integrating Convolutional Neural Networks and Multi-Task Dictionary Learning for Cognitive Decline Prediction with Longitudinal Images. J. Alzheimer’s Dis. 2020, 75, 971–992. [Google Scholar] [CrossRef]
- Moradi, E.; Khundrakpam, B.; Lewis, J.D.; Evans, A.C.; Tohka, J. Predicting symptom severity in autism spectrum disorder based on cortical thickness measures in agglomerative data. Neuroimage 2017, 144, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Li, J.; Chen, K.; Wu, T.; Ye, J.; Wu, X.; Yao, L. A transfer learning approach for network modeling. IIE Trans. 2012, 44, 915–931. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.S.; Yoon, H.; Eschbacher, J.M.; Baxter, L.C.; Dueck, A.C.; Nespodzany, A.; Smith, K.A.; Nakaji, P.; Xu, Y.; Wang, L.; et al. Accurate patient-specific machine learning models of glioblastoma invasion using transfer learning. Am. J. Neuroradiol. 2019, 40, 418–425. [Google Scholar] [CrossRef]
- Hermessi, H.; Mourali, O.; Zagrouba, E. Convolutional neural network-based multimodal image fusion via similarity learning in the shearlet domain. Neural Comput. Appl. 2018, 30, 2029–2045. [Google Scholar] [CrossRef]
- Naselaris, T.; Kay, K.N.; Nishimoto, S.; Gallant, J.L. Encoding and decoding in fMRI. Neuroimage 2011, 56, 400–410. [Google Scholar] [CrossRef] [Green Version]
- Dinsdale, N.K.; Jenkinson, M.; Namburete, A.I. Unlearning Scanner Bias for MRI Harmonisation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 369–378. [Google Scholar]
- Cheng, B.; Liu, M.; Zhang, D.; Munsell, B.C.; Shen, D. Domain transfer learning for MCI conversion prediction. IEEE Trans. Biomed. Eng. 2015, 62, 1805–1817. [Google Scholar] [CrossRef] [Green Version]
- Cheng, B.; Liu, M.; Suk, H.I.; Shen, D.; Zhang, D.; Alzheimer’s Disease Neuroimaging Initiative. Multimodal manifold-regularized transfer learning for MCI conversion prediction. Brain Imaging Behav. 2015, 9, 913–926. [Google Scholar] [CrossRef] [Green Version]
- Goetz, M.; Weber, C.; Binczyk, F.; Polanska, J.; Tarnawski, R.; Bobek-Billewicz, B.; Koethe, U.; Kleesiek, J.; Stieltjes, B.; Maier-Hein, K.H. DALSA: Domain adaptation for supervised learning from sparsely annotated MR images. IEEE Trans. Med. Imaging 2015, 35, 184–196. [Google Scholar] [CrossRef]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; De Bruijne, M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans. Med. Imaging 2014, 34, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, W.; Fan, W.; Chen, X.; Wu, D. Alzheimer’s Disease Brain Network Classification Using Improved Transfer Feature Learning with Joint Distribution Adaptation. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2959–2963. [Google Scholar]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; De Bruijne, M. A transfer-learning approach to image segmentation across scanners by maximizing distribution similarity. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2013; pp. 49–56. [Google Scholar]
- Van Opbroek, A.; Vernooij, M.W.; Ikram, M.A.; de Bruijne, M. Weighting training images by maximizing distribution similarity for supervised segmentation across scanners. Med. Image Anal. 2015, 24, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Van Opbroek, A.; Achterberg, H.C.; Vernooij, M.W.; De Bruijne, M. Transfer learning for image segmentation by combining image weighting and kernel learning. IEEE Trans. Med. Imaging 2018, 38, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Prastawa, M.; Saha, A.; Awate, S.P.; Irimia, A.; Chambers, M.C.; Vespa, P.M.; Van Horn, J.D.; Pascucci, V.; Gerig, G. Modeling 4D changes in pathological anatomy using domain adaptation: Analysis of TBI imaging using a tumor database. In International Workshop on Multimodal Brain Image Analysis; Springer: Berlin/Heidelberg, Germany, 2013; pp. 31–39. [Google Scholar]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; de Bruijne, M. Supervised image segmentation across scanner protocols: A transfer learning approach. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2012; pp. 160–167. [Google Scholar]
- Tan, X.; Liu, Y.; Li, Y.; Wang, P.; Zeng, X.; Yan, F.; Li, X. Localized instance fusion of MRI data of Alzheimer’s disease for classification based on instance transfer ensemble learning. Biomed. Eng. Online 2018, 17, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.; He, W.; Xu, Y.; Xiong, G.; Cai, J. Feature selection and transfer learning for Alzheimer’s disease clinical diagnosis. Appl. Sci. 2018, 8, 1372. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gu, Y.; Dvornek, N.; Staib, L.; Ventola, P.; Duncan, J.S. Multi-site fmri analysis using privacy-preserving federated learning and domain adaptation: Abide results. arXiv 2020, arXiv:2001.05647. [Google Scholar] [CrossRef]
- Wachinger, C.; Reuter, M.; Alzheimer’s Disease Neuroimaging Initiative. Domain adaptation for Alzheimer’s disease diagnostics. Neuroimage 2016, 139, 470–479. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.B.; Gu, I.Y.H.; Berger, M.S.; Pallud, J.; Southwell, D.; Widhalm, G.; Roux, A.; Vecchio, T.G.; Jakola, A.S. Domain Mapping and Deep Learning from Multiple MRI Clinical Datasets for Prediction of Molecular Subtypes in Low Grade Gliomas. Brain Sci. 2020, 10, 463. [Google Scholar] [CrossRef]
- Tokuoka, Y.; Suzuki, S.; Sugawara, Y. An Inductive Transfer Learning Approach using Cycle-consistent Adversarial Domain Adaptation with Application to Brain Tumor Segmentation. In Proceedings of the 2019 6th International Conference on Biomedical and Bioinformatics Engineering, Shanghai, China, 13–15 November 2019; pp. 44–48. [Google Scholar]
- Li, W.; Zhao, Y.; Chen, X.; Xiao, Y.; Qin, Y. Detecting Alzheimer’s disease on small dataset: A knowledge transfer perspective. IEEE J. Biomed. Health Inform. 2018, 23, 1234–1242. [Google Scholar] [CrossRef]
- Wang, B.; Prastawa, M.; Irimia, A.; Saha, A.; Liu, W.; Goh, S.M.; Vespa, P.M.; Van Horn, J.D.; Gerig, G. Modeling 4D pathological changes by leveraging normative models. Comput. Vis. Image Underst. 2016, 151, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Van Opbroek, A.; Achterberg, H.C.; de Bruijne, M. Feature-space transformation improves supervised segmentation across scanners. In Medical Learning Meets Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2015; pp. 85–93. [Google Scholar]
- Van Opbroek, A.; Achterberg, H.C.; Vernooij, M.W.; Ikram, M.A.; de Bruijne, M.; Alzheimer’s Disease Neuroimaging Initiative. Transfer learning by feature-space transformation: A method for Hippocampus segmentation across scanners. Neuroimage Clin. 2018, 20, 466–475. [Google Scholar] [CrossRef]
- Qin, Y.; Li, Y.; Liu, Z.; Ye, C. Knowledge Transfer Between Datasets for Learning-Based Tissue Microstructure Estimation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1530–1533. [Google Scholar]
- Mansoor, A.; Linguraru, M.G. Communal Domain Learning for Registration in Drifted Image Spaces. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 479–488. [Google Scholar]
- Zhou, S.; Cox, C.R.; Lu, H. Improving whole-brain neural decoding of fmri with domain adaptation. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 265–273. [Google Scholar]
- Van Tulder, G.; de Bruijne, M. Representation learning for cross-modality classification. In Medical Computer Vision and Bayesian and Graphical Models for Biomedical Imaging; Springer: Berlin/Heidelberg, Germany, 2016; pp. 126–136. [Google Scholar]
- Gao, Y.; Zhang, Y.; Cao, Z.; Guo, X.; Zhang, J. Decoding Brain States from fMRI Signals by using Unsupervised Domain Adaptation. IEEE J. Biomed. Health Inform. 2019, 24, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wan, P.; Zhang, D. Transport-Based Joint Distribution Alignment for Multi-site Autism Spectrum Disorder Diagnosis Using Resting-State fMRI. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 444–453. [Google Scholar]
- Cheng, B.; Liu, M.; Shen, D.; Li, Z.; Zhang, D.; Alzheimer’s Disease Neuroimaging Initiative. Multi-domain transfer learning for early diagnosis of Alzheimer’s disease. Neuroinformatics 2017, 15, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.; Liu, M.; Zhang, D.; Shen, D.; Alzheimer’s Disease Neuroimaging Initiative. Robust multi-label transfer feature learning for early diagnosis of Alzheimer’s disease. Brain Imaging Behav. 2019, 13, 138–153. [Google Scholar] [CrossRef]
- Van Tulder, G.; de Bruijne, M. Learning cross-modality representations from multi-modal images. IEEE Trans. Med. Imaging 2018, 38, 638–648. [Google Scholar] [CrossRef]
- Li, Z.; Ogino, M. Augmented Radiology: Patient-Wise Feature Transfer Model for Glioma Grading. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 23–30. [Google Scholar]
- Ackaouy, A.; Courty, N.; Vallée, E.; Commowick, O.; Barillot, C.; Galassi, F. Unsupervised domain adaptation with optimal transport in multi-site segmentation of multiple sclerosis lesions from MRI data. Front. Comput. Neurosci. 2020, 14, 19. [Google Scholar] [CrossRef] [Green Version]
- Hofer, C.; Kwitt, R.; Höller, Y.; Trinka, E.; Uhl, A. Simple domain adaptation for cross-dataset analyses of brain MRI data. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 441–445. [Google Scholar]
- Cai, X.L.; Xie, D.J.; Madsen, K.H.; Wang, Y.M.; Bögemann, S.A.; Cheung, E.F.; Møller, A.; Chan, R.C. Generalizability of machine learning for classification of schizophrenia based on resting-state functional MRI data. Hum. Brain Mapp. 2020, 41, 172–184. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, R.; Ledig, C.; Rueckert, D. Manifold alignment and transfer learning for classification of Alzheimer’s disease. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2014; pp. 77–84. [Google Scholar]
- Wang, M.; Zhang, D.; Huang, J.; Yap, P.T.; Shen, D.; Liu, M. Identifying autism spectrum disorder with multi-site fMRI via low-rank domain adaptation. IEEE Trans. Med. Imaging 2019, 39, 644–655. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, M.; Pan, Y.; Shen, D. Unsupervised Conditional Consensus Adversarial Network for Brain Disease Identification with Structural MRI. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 391–399. [Google Scholar]
- Shen, Y.; Gao, M. Brain tumor segmentation on MRI with missing modalities. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; pp. 417–428. [Google Scholar]
- Robinson, R.; Dou, Q.; de Castro, D.C.; Kamnitsas, K.; de Groot, M.; Summers, R.M.; Rueckert, D.; Glocker, B. Image-level Harmonization of Multi-Site Data using Image-and-Spatial Transformer Networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 710–719. [Google Scholar]
- Guan, H.; Yang, E.; Yap, P.T.; Shen, D.; Liu, M. Attention-Guided Deep Domain Adaptation for Brain Dementia Identification with Multi-site Neuroimaging Data. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 31–40. [Google Scholar]
- Mahapatra, D.; Ge, Z. Training data independent image registration using generative adversarial networks and domain adaptation. Pattern Recognit. 2020, 100, 107109. [Google Scholar] [CrossRef]
- Orbes-Arteaga, M.; Varsavsky, T.; Sudre, C.H.; Eaton-Rosen, Z.; Haddow, L.J.; Sørensen, L.; Nielsen, M.; Pai, A.; Ourselin, S.; Modat, M.; et al. Multi-domain adaptation in brain MRI through paired consistency and adversarial learning. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Springer: Berlin/Heidelberg, Germany, 2019; pp. 54–62. [Google Scholar]
- Kamnitsas, K.; Baumgartner, C.; Ledig, C.; Newcombe, V.; Simpson, J.; Kane, A.; Menon, D.; Nori, A.; Criminisi, A.; Rueckert, D.; et al. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In Proceedings of the International Conference on Information Processing in Medical Imaging, Boone, NC, USA, 25–30 June 2017; pp. 597–609. [Google Scholar]
- Varsavsky, T.; Orbes-Arteaga, M.; Sudre, C.H.; Graham, M.S.; Nachev, P.; Cardoso, M.J. Test-time unsupervised domain adaptation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 428–436. [Google Scholar]
- Shanis, Z.; Gerber, S.; Gao, M.; Enquobahrie, A. Intramodality Domain Adaptation Using Self Ensembling and Adversarial Training. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Springer: Berlin/Heidelberg, Germany, 2019; pp. 28–36. [Google Scholar]
- Orbes-Arteainst, M.; Cardoso, J.; Sørensen, L.; Igel, C.; Ourselin, S.; Modat, M.; Nielsen, M.; Pai, A. Knowledge distillation for semi-supervised domain adaptation. In OR 2.0 Context-Aware Operating Theaters and Machine Learning in Clinical Neuroimaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 68–76. [Google Scholar]
- Grimm, F.; Edl, F.; Kerscher, S.R.; Nieselt, K.; Gugel, I.; Schuhmann, M.U. Semantic segmentation of cerebrospinal fluid and brain volume with a convolutional neural network in pediatric hydrocephalus—Transfer learning from existing algorithms. Acta Neurochir. 2020, 162, 2463–2474. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Hsu, Y.C.; Yang, L.Y.; Tung, Y.H.; Luo, W.B.; Liu, C.M.; Hwang, T.J.; Hwu, H.G.; Tseng, W.Y.I. Generalization of diffusion magnetic resonance imaging–based brain age prediction model through transfer learning. NeuroImage 2020, 217, 116831. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Chung, Y.C.; Kim, K.W.; Kim, W.S.; Oh, I.S. Classification and visualization of Alzheimer’s disease using volumetric convolutional neural network and transfer learning. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Abrol, A.; Bhattarai, M.; Fedorov, A.; Du, Y.; Plis, S.; Calhoun, V.; Alzheimer’s Disease Neuroimaging Initiative. Deep residual learning for neuroimaging: An application to predict progression to alzheimer’s disease. J. Neurosci. Methods 2020, 339, 108701. [Google Scholar] [CrossRef] [Green Version]
- Alex, V.; Vaidhya, K.; Thirunavukkarasu, S.; Kesavadas, C.; Krishnamurthi, G. Semisupervised learning using denoising autoencoders for brain lesion detection and segmentation. J. Med. Imaging 2017, 4, 041311. [Google Scholar] [CrossRef]
- Cui, S.; Mao, L.; Jiang, J.; Liu, C.; Xiong, S. Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network. J. Healthc. Eng. 2018, 2018, 4940593. [Google Scholar] [CrossRef]
- Gao, F.; Yoon, H.; Xu, Y.; Goradia, D.; Luo, J.; Wu, T.; Su, Y. Alzheimer’s Disease Neuroimaging Initiative. AD-NET: Age-adjust neural network for improved MCI to AD conversion prediction. Neuroimage Clin. 2020, 27, 102290. [Google Scholar] [CrossRef]
- Han, Y.; Yoo, J.; Kim, H.H.; Shin, H.J.; Sung, K.; Ye, J.C. Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn. Reson. Med. 2018, 80, 1189–1205. [Google Scholar] [CrossRef] [Green Version]
- Amin, J.; Sharif, M.; Yasmin, M.; Saba, T.; Anjum, M.A.; Fernandes, S.L. A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J. Med. Syst. 2019, 43, 326. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Content-based brain tumor retrieval for MR images using transfer learning. IEEE Access 2019, 7, 17809–17822. [Google Scholar] [CrossRef]
- Xu, Y.; Géraud, T.; Bloch, I. From neonatal to adult brain MR image segmentation in a few seconds using 3D-like fully convolutional network and transfer learning. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 4417–4421. [Google Scholar]
- Ladefoged, C.N.; Hansen, A.E.; Henriksen, O.M.; Bruun, F.J.; Eikenes, L.; Øen, S.K.; Karlberg, A.; Højgaard, L.; Law, I.; Andersen, F.L. AI-driven attenuation correction for brain PET/MRI: Clinical evaluation of a dementia cohort and importance of the training group size. NeuroImage 2020, 222, 117221. [Google Scholar] [CrossRef] [PubMed]
- Dar, S.U.H.; Özbey, M.; Çatlı, A.B.; Çukur, T. A Transfer-Learning Approach for Accelerated MRI Using Deep Neural Networks. Magn. Reson. Med. 2020, 84, 663–685. [Google Scholar] [CrossRef] [Green Version]
- Aderghal, K.; Khvostikov, A.; Krylov, A.; Benois-Pineau, J.; Afdel, K.; Catheline, G. Classification of Alzheimer disease on imaging modalities with deep CNNs using cross-modal transfer learning. In Proceedings of the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, 18–21 June 2018; pp. 345–350. [Google Scholar]
- Bashyam, V.M.; Erus, G.; Doshi, J.; Habes, M.; Nasralah, I.; Truelove-Hill, M.; Srinivasan, D.; Mamourian, L.; Pomponio, R.; Fan, Y.; et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain 2020, 143, 2312–2324. [Google Scholar] [CrossRef]
- Xu, Y.; Géraud, T.; Puybareau, É.; Bloch, I.; Chazalon, J. White matter hyperintensities segmentation in a few seconds using fully convolutional network and transfer learning. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2017; pp. 501–514. [Google Scholar]
- Han, X. MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepak, S.; Ameer, P. Retrieval of brain MRI with tumor using contrastive loss based similarity on GoogLeNet encodings. Comput. Biol. Med. 2020, 125, 103993. [Google Scholar] [CrossRef]
- Li, H.; Parikh, N.A.; He, L. A novel transfer learning approach to enhance deep neural network classification of brain functional connectomes. Front. Neurosci. 2018, 12, 491. [Google Scholar] [CrossRef]
- Alkassar, S.; Abdullah, M.A.; Jebur, B.A. Automatic Brain Tumour Segmentation using fully Convolution Network and Transfer Learning. In Proceedings of the 2019 2nd International Conference on Electrical, Communication, Computer, Power and Control Engineering (ICECCPCE), Mosul, Iraq, 13–14 February 2019; pp. 188–192. [Google Scholar]
- Afridi, M.J.; Ross, A.; Shapiro, E.M. L-CNN: Exploiting labeling latency in a cnn learning framework. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 2156–2161. [Google Scholar]
- Chen, K.T.; Schürer, M.; Ouyang, J.; Koran, M.E.I.; Davidzon, G.; Mormino, E.; Tiepolt, S.; Hoffmann, K.T.; Sabri, O.; Zaharchuk, G.; et al. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2998–3007. [Google Scholar] [CrossRef]
- Mahmood, U.; Rahman, M.M.; Fedorov, A.; Lewis, N.; Fu, Z.; Calhoun, V.D.; Plis, S.M. Whole MILC: Generalizing learned dynamics across tasks, datasets, and populations. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 407–417. [Google Scholar]
- Yang, Y.; Yan, L.F.; Zhang, X.; Han, Y.; Nan, H.Y.; Hu, Y.C.; Hu, B.; Yan, S.L.; Zhang, J.; Cheng, D.L.; et al. Glioma grading on conventional MR images: A deep learning study with transfer learning. Front. Neurosci. 2018, 12, 804. [Google Scholar] [CrossRef] [Green Version]
- Pravitasari, A.A.; Iriawan, N.; Almuhayar, M.; Azmi, T.; Fithriasari, K.; Purnami, S.W.; Ferriastuti, W. UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation. Telkomnika 2020, 18, 1310–1318. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, H.; Zhou, Y.; Bian, W.; Zhang, T.; Zou, X. Gibbs-ringing artifact suppression with knowledge transfer from natural images to MR images. Multimed. Tools Appl. 2020, 79, 33711–33733. [Google Scholar] [CrossRef]
- Coupé, P.; Mansencal, B.; Clément, M.; Giraud, R.; de Senneville, B.D.; Ta, V.T.; Lepetit, V.; Manjon, J.V. AssemblyNet: A novel deep decision-making process for whole brain MRI segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 466–474. [Google Scholar]
- Zhou, Z.; Sodha, V.; Siddiquee, M.M.R.; Feng, R.; Tajbakhsh, N.; Gotway, M.B.; Liang, J. Models genesis: Generic autodidactic models for 3d medical image analysis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 384–393. [Google Scholar]
- Tao, X.; Li, Y.; Zhou, W.; Ma, K.; Zheng, Y. Revisiting Rubik’s cube: Self-supervised learning with volume-wise transformation for 3D medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 238–248. [Google Scholar]
- Liu, Y.; Pan, Y.; Yang, W.; Ning, Z.; Yue, L.; Liu, M.; Shen, D. Joint Neuroimage Synthesis and Representation Learning for Conversion Prediction of Subjective Cognitive Decline. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 583–592. [Google Scholar]
- Ataloglou, D.; Dimou, A.; Zarpalas, D.; Daras, P. Fast and precise hippocampus segmentation through deep convolutional neural network ensembles and transfer learning. Neuroinformatics 2019, 17, 563–582. [Google Scholar] [CrossRef]
- Afridi, M.J.; Ross, A.; Shapiro, E.M. On automated source selection for transfer learning in convolutional neural networks. Pattern Recognit. 2018, 73, 65–75. [Google Scholar] [CrossRef]
- Kouw, W.M.; Ørting, S.N.; Petersen, J.; Pedersen, K.S.; de Bruijne, M. A cross-center smoothness prior for variational Bayesian brain tissue segmentation. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; pp. 360–371. [Google Scholar]
- Kuzina, A.; Egorov, E.; Burnaev, E. Bayesian generative models for knowledge transfer in mri semantic segmentation problems. Front. Neurosci. 2019, 13, 844. [Google Scholar] [CrossRef] [Green Version]
- Wee, C.Y.; Liu, C.; Lee, A.; Poh, J.S.; Ji, H.; Qiu, A.; Alzheimer’s Disease Neuroimaging Initiative. Cortical graph neural network for AD and MCI diagnosis and transfer learning across populations. Neuroimage Clin. 2019, 23, 101929. [Google Scholar] [CrossRef]
- Fei, X.; Wang, J.; Ying, S.; Hu, Z.; Shi, J. Projective parameter transfer based sparse multiple empirical kernel learning Machine for diagnosis of brain disease. Neurocomputing 2020, 413, 271–283. [Google Scholar] [CrossRef]
- Velioglu, B.; Vural, F.T.Y. Transfer learning for brain decoding using deep architectures. In Proceedings of the 2017 IEEE 16th International Conference on Cognitive Informatics & Cognitive Computing (ICCI* CC), Oxford, UK, 26–28 July 2017; pp. 65–70. [Google Scholar]
- Li, W.; Zhang, L.; Qiao, L.; Shen, D. Toward a Better Estimation of Functional Brain Network for Mild Cognitive Impairment Identification: A Transfer Learning View. IEEE J. Biomed. Health Inform. 2019, 24, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Jónsson, B.A.; Bjornsdottir, G.; Thorgeirsson, T.; Ellingsen, L.M.; Walters, G.B.; Gudbjartsson, D.; Stefansson, H.; Stefansson, K.; Ulfarsson, M. Brain age prediction using deep learning uncovers associated sequence variants. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ghafoorian, M.; Mehrtash, A.; Kapur, T.; Karssemeijer, N.; Marchiori, E.; Pesteie, M.; Guttmann, C.R.; de Leeuw, F.E.; Tempany, C.M.; Van Ginneken, B.; et al. Transfer learning for domain adaptation in mri: Application in brain lesion segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; pp. 516–524. [Google Scholar]
- Valverde, S.; Salem, M.; Cabezas, M.; Pareto, D.; Vilanova, J.C.; Ramió-Torrentà, L.; Rovira, À.; Salvi, J.; Oliver, A.; Lladó, X. One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. NeuroImage Clin. 2019, 21, 101638. [Google Scholar] [CrossRef]
- Shirokikh, B.; Zakazov, I.; Chernyavskiy, A.; Fedulova, I.; Belyaev, M. First U-Net Layers Contain More Domain Specific Information Than The Last Ones. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 117–126. [Google Scholar]
- Gao, Y.; Zhang, Y.; Wang, H.; Guo, X.; Zhang, J. Decoding Behavior Tasks From Brain Activity Using Deep Transfer Learning. IEEE Access 2019, 7, 43222–43232. [Google Scholar] [CrossRef]
- Naser, M.A.; Deen, M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 2020, 121, 103758. [Google Scholar] [CrossRef]
- Vakli, P.; Deák-Meszlényi, R.J.; Hermann, P.; Vidnyánszky, Z. Transfer learning improves resting-state functional connectivity pattern analysis using convolutional neural networks. GigaScience 2018, 7, giy130. [Google Scholar] [CrossRef]
- Jiang, H.; Guo, J.; Du, H.; Xu, J.; Qiu, B. Transfer learning on T1-weighted images for brain age estimation. Math. Biosci. Eng. 2019, 16, 4382–4398. [Google Scholar] [CrossRef]
- Kushibar, K.; Valverde, S.; González-Villà, S.; Bernal, J.; Cabezas, M.; Oliver, A.; Lladó, X. Supervised domain adaptation for automatic sub-cortical brain structure segmentation with minimal user interaction. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kollia, I.; Stafylopatis, A.G.; Kollias, S. Predicting Parkinson’s disease using latent information extracted from deep neural networks. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Menikdiwela, M.; Nguyen, C.; Shaw, M. Deep Learning on Brain Cortical Thickness Data for Disease Classification. In Proceedings of the 2018 Digital Image Computing: Techniques and Applications (DICTA), Canberra, Australia, 10–13 December 2018; pp. 1–5. [Google Scholar]
- Kaur, B.; Lemaître, P.; Mehta, R.; Sepahvand, N.M.; Precup, D.; Arnold, D.; Arbel, T. Improving Pathological Structure Segmentation via Transfer Learning Across Diseases. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Springer: Berlin/Heidelberg, Germany, 2019; pp. 90–98. [Google Scholar]
- Liu, R.; Hall, L.O.; Goldgof, D.B.; Zhou, M.; Gatenby, R.A.; Ahmed, K.B. Exploring deep features from brain tumor magnetic resonance images via transfer learning. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 235–242. [Google Scholar]
- Stawiaski, J. A pretrained densenet encoder for brain tumor segmentation. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2018; pp. 105–115. [Google Scholar]
- Mahapatra, D.; Ge, Z. Training data independent image registration with gans using transfer learning and segmentation information. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 709–713. [Google Scholar]
- Wang, L.; Li, S.; Meng, M.; Chen, G.; Zhu, M.; Bian, Z.; Lyu, Q.; Zeng, D.; Ma, J. Task-oriented Deep Network for Ischemic Stroke Segmentation in Unenhanced CT Imaging. In Proceedings of the 2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Manchester, UK, 26 October–2 November 2019; pp. 1–3. [Google Scholar]
- Wang, S.; Shen, Y.; Chen, W.; Xiao, T.; Hu, J. Automatic recognition of mild cognitive impairment from mri images using expedited convolutional neural networks. In Proceedings of the International Conference on Artificial Neural Networks, Alghero, Italy, 11–15 September 2017; pp. 373–380. [Google Scholar]
- Guy-Fernand, K.N.; Zhao, J.; Sabuni, F.M.; Wang, J. Classification of Brain Tumor Leveraging Goal-Driven Visual Attention with the Support of Transfer Learning. In Proceedings of the 2020 Information Communication Technologies Conference (ICTC), Nanjing, China, 29–31 May 2020; pp. 328–332. [Google Scholar]
- Khan, N.M.; Abraham, N.; Hon, M. Transfer learning with intelligent training data selection for prediction of Alzheimer’s disease. IEEE Access 2019, 7, 72726–72735. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.; Zhang, Q.N. Multiclass classification of initial stages of Alzheimer’s Disease through Neuroimaging modalities and Convolutional Neural Networks. In Proceedings of the 2020 IEEE 5th Information Technology and Mechatronics Engineering Conference (ITOEC), Chongqing, China, 12–14 June 2020; pp. 51–56. [Google Scholar]
- Castro, A.P.; Fernandez-Blanco, E.; Pazos, A.; Munteanu, C.R. Automatic assessment of Alzheimer’s disease diagnosis based on deep learning techniques. Comput. Biol. Med. 2020, 120, 103764. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Vijay, A.; Veeranjaneyulu, N. Brain tumor detection using deep features in the latent space. J. Homepage 2020, 25, 259–265. [Google Scholar] [CrossRef]
- Kang, L.; Jiang, J.; Huang, J.; Zhang, T. Identifying early mild cognitive impairment by multi-modality mri-based deep learning. Front. Aging Neurosci. 2020, 12, 206. [Google Scholar] [CrossRef]
- Thomas, A.W.; Müller, K.R.; Samek, W. Deep transfer learning for whole-brain FMRI analyses. In OR 2.0 Context-Aware Operating Theaters and Machine Learning in Clinical Neuroimaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 59–67. [Google Scholar]
- Ebrahimi-Ghahnavieh, A.; Luo, S.; Chiong, R. Transfer Learning for Alzheimer’s Disease Detection on MRI Images. In Proceedings of the 2019 IEEE International Conference on Industry 4.0, Artificial Intelligence, and Communications Technology (IAICT), Bali, Indonesia, 1–3 July 2019; pp. 133–138. [Google Scholar]
- Zheng, J.; Xia, K.; Zheng, Q.; Qian, P. A smart brain MR image completion method guided by synthetic-CT-based multimodal registration. J. Ambient. Intell. Humaniz. Comput. 2019. [Google Scholar] [CrossRef]
- Svanera, M.; Savardi, M.; Benini, S.; Signoroni, A.; Raz, G.; Hendler, T.; Muckli, L.; Goebel, R.; Valente, G. Transfer learning of deep neural network representations for fMRI decoding. J. Neurosci. Methods 2019, 328, 108319. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Luo, Q.; Gong, W.; Lu, W. Transfer Learning Models on Brain Age Prediction. In Proceedings of the Third International Symposium on Image Computing and Digital Medicine, Xi’an, China, 24–26 August 2019; pp. 278–282. [Google Scholar]
- Han, W.; Qin, L.; Bay, C.; Chen, X.; Yu, K.H.; Miskin, N.; Li, A.; Xu, X.; Young, G. Deep Transfer Learning and Radiomics Feature Prediction of Survival of Patients with High-Grade Gliomas. Am. J. Neuroradiol. 2020, 41, 40–48. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Carass, A.; Zuo, L.; Dewey, B.E.; Prince, J.L. Self domain adapted network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 437–446. [Google Scholar]
- Yang, Y.; Li, X.; Wang, P.; Xia, Y.; Ye, Q. Multi-Source Transfer Learning via Ensemble Approach for Initial Diagnosis of Alzheimer’s Disease. IEEE J. Transl. Eng. Health Med. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27 June–1 July 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–1 July 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Dwork, C.; Roth, A. The algorithmic foundations of differential privacy. Found. Trends Theor. Comput. Sci. 2014, 9, 211–407. [Google Scholar] [CrossRef]
- Petersen, R.C.; Aisen, P.; Beckett, L.A.; Donohue, M.; Gamst, A.; Harvey, D.J.; Jack, C.; Jagust, W.; Shaw, L.; Toga, A.; et al. Alzheimer’s disease neuroimaging initiative (ADNI): Clinical characterization. Neurology 2010, 74, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659–667. [Google Scholar] [CrossRef]
- Di Martino, A.; O’connor, D.; Chen, B.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Balsters, J.H.; Baxter, L.; Beggiato, A.; Bernaerts, S.; et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data 2017, 4, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Van Essen, D.C.; Smith, S.M.; Barch, D.M.; Behrens, T.E.; Yacoub, E.; Ugurbil, K.; Consortium, W.M.H. The WU-Minn human connectome project: An overview. Neuroimage 2013, 80, 62–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrol, A.; Fu, Z.; Salman, M.; Silva, R.; Du, Y.; Plis, S.; Calhoun, V. Deep learning encodes robust discriminative neuroimaging representations to outperform standard machine learning. Nat. Commun. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum learning. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 41–48. [Google Scholar]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]


| Type | Properties | Approach Example |
|---|---|---|
| Unsupervised | Transforming T1- and T2-weighted images into the same feature space with adversarial training. | |
| Transductive | Learning a feature mapping from T1- to T2-weighted images while optimizing to segment tumors in T2-weighted images. | |
| Inductive | Optimizing a classifier on a natural images dataset, and fine-tuning certain parameters for tumor segmentation. | |
| Optimizing a lesion segmentation algorithm in T2-weighted images, and re-optimizing certain parameters on FLAIR images. | ||
| Optimizing a lesion segmentation algorithm in T2-weighted images, and re-optimizing certain parameters in the same images for anatomical segmentation. |
| Scopus | Springer Website |
|---|---|
| (TITLE-ABS-KEY ((“transfer learning” OR “knowledge transfer” OR “domain adaptation”) AND (mri OR “magnetic resonance” OR “diffusion imaging” OR “diffusion weighted imaging” OR “arterial spin labeling” OR “susceptibility mapping” OR bold OR “blood oxygenation level dependent” OR “blood oxygen level dependent”))) | (“transfer learning” OR “knowledge transfer” OR “domain adaptation”) AND (mri OR “magnetic resonance” OR “diffusion imaging” OR “diffusion weighted imaging” OR “arterial spin labeling” OR “susceptibility mapping” OR “T1” OR “T2”) |
| Task (Total) | % of Studies | Application |
|---|---|---|
| Classification (68) | 52.71% | Alzheimer’s diagnostics/prognostics (31), Tumor (10), fMRI decoding (6), Autism spectrum disorder (5), Injected cells (2), Parkinson (2), Schizophrenia (2), Sex (2), Aneurysm [27], Attention deficit hyperactivity disorder [28], Bipolar disorder [29], Embryonic neurodevelopmental disorders [30], Epilepsy [31], IDH mutation [32], Multiple sclerosis [33], Quality control [34] |
| Segmentation (45) | 34.88% | Tumor (16), Anatomical (15), Lesion (14) |
| Regression (12) | 9.30% | Age (8), Alzheimer’s disease progression [35], Autism symptom severity [36], Brain connectivity in Alzheimer’s disease [37], Tumor cell density [38] |
| Others (15) | 11.63% | Reconstruction (5), Registration (4), Image translation (3), CBIR (2), Image fusion [39] |
| Type | % of Studies | Subtype | Subsubtype | Approaches |
|---|---|---|---|---|
| Instance (16) | 11.63% | Fixed | 6 (4.65%) | |
| Optimized | Unsupervised | 5 (3.88%) | ||
| Supervised | 5 (3.88%) | |||
| Feature (38) | 29.46% | Asymmetric | 7 (5.43%) | |
| Symmetric | Direct | 17 (13.18%) | ||
| Indirect | 14 (10.85%) | |||
| Parameter (87) | 65.89% | Prior sharing | 50 (38.76%) | |
| Parameter sharing | One model | 21 (16.28%) | ||
| Multiple | 16 (12.40%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valverde, J.M.; Imani, V.; Abdollahzadeh, A.; De Feo, R.; Prakash, M.; Ciszek, R.; Tohka, J. Transfer Learning in Magnetic Resonance Brain Imaging: A Systematic Review. J. Imaging 2021, 7, 66. https://doi.org/10.3390/jimaging7040066
Valverde JM, Imani V, Abdollahzadeh A, De Feo R, Prakash M, Ciszek R, Tohka J. Transfer Learning in Magnetic Resonance Brain Imaging: A Systematic Review. Journal of Imaging. 2021; 7(4):66. https://doi.org/10.3390/jimaging7040066
Chicago/Turabian StyleValverde, Juan Miguel, Vandad Imani, Ali Abdollahzadeh, Riccardo De Feo, Mithilesh Prakash, Robert Ciszek, and Jussi Tohka. 2021. "Transfer Learning in Magnetic Resonance Brain Imaging: A Systematic Review" Journal of Imaging 7, no. 4: 66. https://doi.org/10.3390/jimaging7040066
APA StyleValverde, J. M., Imani, V., Abdollahzadeh, A., De Feo, R., Prakash, M., Ciszek, R., & Tohka, J. (2021). Transfer Learning in Magnetic Resonance Brain Imaging: A Systematic Review. Journal of Imaging, 7(4), 66. https://doi.org/10.3390/jimaging7040066







