Skin Lesion Segmentation Using Deep Learning with Auxiliary Task
Abstract
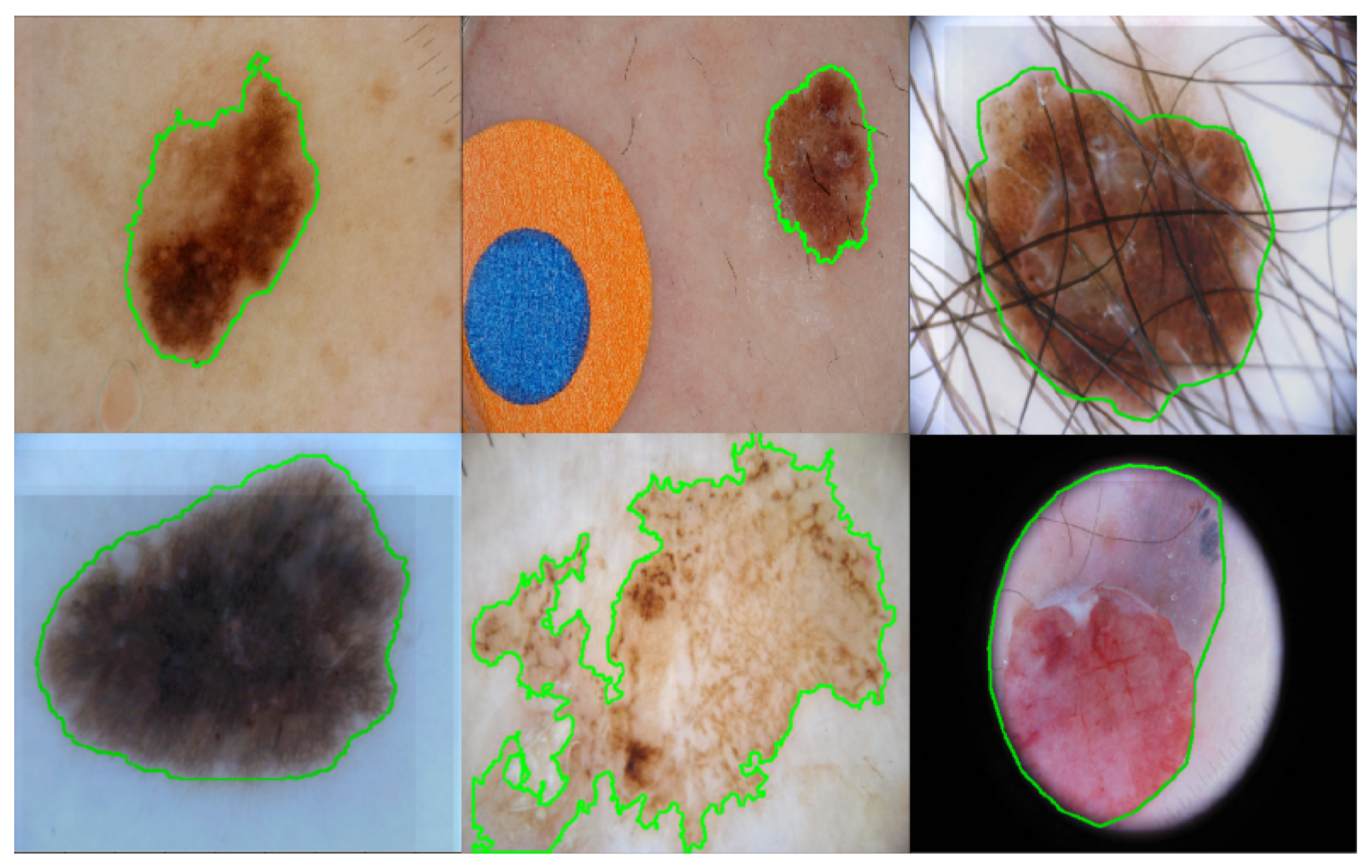
:1. Introduction
- Edge prediction is leveraged as an auxiliary task for the skin lesion segmentation task. The proposed method learns these two tasks simultaneously by two parallel branches (edge prediction and segmentation mask prediction). The edge prediction branch can guide the learned neural network to focus on the boundaries of the segmentation masks. Up to the authors’ knowledge, this is the first work that utilizes edge information to assist the skin lesion segmentation task. Note that the edge of a segmentation mask can be obtained automatically by applying some contour detection methods and hence no extra labeling effort is required for the proposed method.
- A cross-connection layer (CCL) module and a multi-scale feature aggregation (MSFA) module are proposed in this paper. The interaction of different tasks is realized by the CCL module. During the training process, the CCL module can implicitly guide the learning of the two tasks jointly, and hence boost each task’s performance in turn. Meanwhile, the MSFA module can make use of multi-scale information. Typically, a prediction head is placed at the intermediate feature maps of each resolution for both the edge prediction and segmentation prediction branch. The weights for the feature maps of each resolution can be learned automatically during training.
2. Related Works
3. Methodology
3.1. CNN Backbone
3.2. Cross-Connection Layer (CCL)
3.3. Multi-Scale Feature Aggregation (MSFA)
- N: number of pixels;
- : target label for pixel n;
- : input pixel n;
- : model with neural network weights ;
- : weight for foreground pixels;
- : weight for background pixels;
4. Experimental Results
4.1. Implementation Details
4.2. Database
4.3. Evaluation Metrics
4.4. Parameter Setting of the Loss Function
4.5. Ablation Study
4.6. Comparison with State-of-The-Art Methods
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, S.; Frühauf, J.; Campbell, T.; Massone, C.; Schwantzer, G.; Soyer, H.P.; Hofmann-Wellenhof, R. Mobile teledermatology for skin tumour screening: Diagnostic accuracy of clinical and dermoscopic image tele-evaluation using cellular phones. Br. J. Dermatol. 2011, 164, 973–979. [Google Scholar] [CrossRef]
- Alves, J.; Moreira, D.; Alves, P.; Rosado, L.; Vasconcelos, M.J.M. Automatic focus assessment on dermoscopic images acquired with smartphones. Sensors 2019, 19, 4957. [Google Scholar] [CrossRef] [Green Version]
- Ngoo, A.; Finnane, A.; McMeniman, E.; Soyer, H.P.; Janda, M. Fighting melanoma with smartphones: A snapshot of where we are a decade after app stores opened their doors. Int. J. Med. Inform. 2018, 118, 99–112. [Google Scholar] [CrossRef] [Green Version]
- Stolz, W.; Riemann, A.; Cognetta, A.; Pillet, L.; Abmayr, W.; Holzel, D.; Bilek, P.; Nachbar, F.; Landthaler, M. Abcd rule of dermatoscopy-a new practical method for early recognition of malignant-melanoma. Eur. J. Dermatol. 1994, 4, 521–527. [Google Scholar]
- Hazen, B.P.; Bhatia, A.C.; Zaim, T.; Brodell, R.T. The clinical diagnosis of early malignant melanoma: Expansion of the ABCD criteria to improve diagnostic sensitivity. Dermatol. Online J. 1999, 5, 3. [Google Scholar] [PubMed]
- Argenziano, G.; Fabbrocini, G.; Carli, P.; De Giorgi, V.; Sammarco, E.; Delfino, M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: Comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch. Dermatol. 1998, 134, 1563–1570. [Google Scholar] [CrossRef] [Green Version]
- Pehamberger, H.; Steiner, A.; Wolff, K. In vivo epiluminescence microscopy of pigmented skin lesions. I. Pattern analysis of pigmented skin lesions. J. Am. Acad. Dermatol. 1987, 17, 571–583. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2016, 36, 994–1004. [Google Scholar] [CrossRef]
- Liu, L.; Mou, L.; Zhu, X.X.; Mandal, M. Automatic skin lesion classification based on mid-level feature learning. Comput. Med. Imaging Graph. 2020, 84, 101765. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.K.; Abdel-Nasser, M.; Rashwan, H.A.; Akram, F.; Pandey, N.; Lalande, A.; Presles, B.; Romani, S.; Puig, D. FCA-Net: Adversarial learning for skin lesion segmentation based on multi-scale features and factorized channel attention. IEEE Access 2019, 7, 130552–130565. [Google Scholar] [CrossRef]
- Yang, X.; Zeng, Z.; Yeo, S.Y.; Tan, C.; Tey, H.L.; Su, Y. A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv 2017, arXiv:1703.01025. [Google Scholar]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans. Med. Imaging 2020. [Google Scholar] [CrossRef] [Green Version]
- Humayun, J.; Malik, A.S.; Kamel, N. Multilevel thresholding for segmentation of pigmented skin lesions. In Proceedings of the 2011 IEEE International Conference on Imaging Systems and Techniques, Batu Ferringhi, Malaysia, 17–18 May 2011; pp. 310–314. [Google Scholar]
- Wong, A.; Scharcanski, J.; Fieguth, P. Automatic skin lesion segmentation via iterative stochastic region merging. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 929–936. [Google Scholar] [CrossRef]
- Riaz, F.; Naeem, S.; Nawaz, R.; Coimbra, M. Active contours based segmentation and lesion periphery analysis for characterization of skin lesions in dermoscopy images. IEEE J. Biomed. Health Inform. 2018, 23, 489–500. [Google Scholar] [CrossRef]
- Abbas, Q.; Fondón, I.; Sarmiento, A.; Celebi, M.E. An improved segmentation method for non-melanoma skin lesions using active contour model. In Proceedings of the International Conference Image Analysis and Recognition, Vilamoura, Portugal, 22–24 October 2014; pp. 193–200. [Google Scholar]
- Tang, J. A multi-direction GVF snake for the segmentation of skin cancer images. Pattern Recognit. 2009, 42, 1172–1179. [Google Scholar] [CrossRef]
- Jafari, M.H.; Samavi, S.; Soroushmehr, S.M.R.; Mohaghegh, H.; Karimi, N.; Najarian, K. Set of descriptors for skin cancer diagnosis using non-dermoscopic color images. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 2638–2642. [Google Scholar]
- Ali, A.R.; Couceiro, M.S.; Hassenian, A.E. Melanoma detection using fuzzy C-means clustering coupled with mathematical morphology. In Proceedings of the International Conference on Hybrid Intelligent Systems (HIS), Hawally, Kuwait, 14–16 December 2014; pp. 73–78. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Maninis, K.K.; Caelles, S.; Pont-Tuset, J.; Van Gool, L. Deep extreme cut: From extreme points to object segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 616–625. [Google Scholar]
- Awan, M.J.; Rahim, M.S.M.; Salim, N.; Mohammed, M.A.; Garcia-Zapirain, B.; Abdulkareem, K.H. Efficient Detection of Knee Anterior Cruciate Ligament from Magnetic Resonance Imaging Using Deep Learning Approach. Diagnostics 2021, 11, 105. [Google Scholar] [CrossRef]
- Jafari, M.H.; Karimi, N.; Nasr-Esfahani, E.; Samavi, S.; Soroushmehr, S.M.R.; Ward, K.; Najarian, K. Skin lesion segmentation in clinical images using deep learning. In Proceedings of the International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 337–342. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Berseth, M. ISIC 2017-Skin Lesion Analysis Towards Melanoma Detection. arXiv 2017, arXiv:1703.00523. [Google Scholar]
- Chang, H. Skin cancer reorganization and classification with deep neural network. arXiv 2017, arXiv:1703.00534. [Google Scholar]
- Liu, L.; Mou, L.; Zhu, X.X.; Mandal, M. Skin Lesion Segmentation Based on Improved U-net. In Proceedings of the 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE), Edmonton, AB, Canada, 5–8 May 2019; pp. 1–4. [Google Scholar]
- Abhishek, K.; Hamarneh, G.; Drew, M.S. Illumination-based Transformations Improve Skin Lesion Segmentation in Dermoscopic Images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 728–729. [Google Scholar]
- Yuan, Y. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv 2017, arXiv:1703.05165. [Google Scholar]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Ahn, E.; Kumar, A.; Feng, D.; Fulham, M. Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recognit. 2019, 85, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Sarker, M.M.K.; Rashwan, H.A.; Akram, F.; Banu, S.F.; Saleh, A.; Singh, V.K.; Chowdhury, F.U.; Abdulwahab, S.; Romani, S.; Radeva, P.; et al. SLSDeep: Skin lesion segmentation based on dilated residual and pyramid pooling networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 21–29.
- Cheng, T.; Wang, X.; Huang, L.; Liu, W. Boundary-preserving mask R-CNN. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 660–676. [Google Scholar]
- Kim, M.; Woo, S.; Kim, D.; Kweon, I.S. The devil is in the boundary: Exploiting boundary representation for basis-based instance segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 5–9 January 2021; pp. 929–938. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Schaefer, S.; McPhail, T.; Warren, J. Image deformation using moving least squares. In Proceedings of the ACM Transactions on Graphics (TOG), Boston, MA, USA, 30 July–3 August 2006; Volume 25, pp. 533–540. [Google Scholar]
- Wei, Z.; Song, H.; Chen, L.; Li, Q.; Han, G. Attention-based DenseUnet network with adversarial training for skin lesion segmentation. IEEE Access 2019, 7, 136616–136629. [Google Scholar] [CrossRef]
- Tu, W.; Liu, X.; Hu, W.; Pan, Z. Dense-residual network with adversarial learning for skin lesion segmentation. IEEE Access 2019, 7, 77037–77051. [Google Scholar] [CrossRef]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access 2019, 8, 4171–4181. [Google Scholar] [CrossRef]
- Ribeiro, V.; Avila, S.; Valle, E. Handling inter-annotator agreement for automated skin lesion segmentation. arXiv 2019, arXiv:1906.02415. [Google Scholar]






| Layer Name | Output Size | Output Channel Dimension | Operations |
|---|---|---|---|
| 64 | |||
| 256 | |||
| 512 | |||
| 1024 | |||
| F | 128 | PPM [26] | |
| 128 | |||
| 128 | |||
| 128 |
| ACC | DC | SEN | SP | JA | |
|---|---|---|---|---|---|
| 94.17 | 87.14 | 88.77 | 95.56 | 79.43 | |
| 94.32 | 87.13 | 88.76 | 96.51 | 79.46 | |
| 94.33 | 87.09 | 88.06 | 96.40 | 79.30 | |
| 94.11 | 86.78 | 89.25 | 93.39 | 79.01 |
| Method | ACC | DC | SEN | SP | JA |
|---|---|---|---|---|---|
| ResNet + PPM + Seg | 93.16 | 85.21 | 88.87 | 95.12 | 77.01 |
| ResNet + PPM + Seg + Edge | 93.54 | 85.66 | 87.11 | 96.61 | 77.58 |
| Proposed | 94.32 | 87.13 | 88.76 | 96.51 | 79.46 |
| Method | ACC | DC | SEN | SP | JA |
|---|---|---|---|---|---|
| Liu et al. [32] | 93.00 | 84.00 | 82.90 | 98.00 | 75.20 |
| Abhishek et al. [33] | 92.22 | 83.86 | 87.06 | 95.16 | 75.70 |
| Yuan et al. [34] | 93.40 | 84.90 | 82.50 | 97.50 | 76.50 |
| AI-Masni et al. [35] | 94.03 | 87.08 | 85.40 | 96.69 | 77.11 |
| Bi et al. [36] | 94.08 | 85.66 | 86.20 | 96.71 | 77.73 |
| Sarker et al. [37] | 93.60 | 87.80 | 81.60 | 98.30 | 78.20 |
| Proposed | 94.32 | 87.13 | 88.76 | 96.51 | 79.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Tsui, Y.Y.; Mandal, M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. J. Imaging 2021, 7, 67. https://doi.org/10.3390/jimaging7040067
Liu L, Tsui YY, Mandal M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. Journal of Imaging. 2021; 7(4):67. https://doi.org/10.3390/jimaging7040067
Chicago/Turabian StyleLiu, Lina, Ying Y. Tsui, and Mrinal Mandal. 2021. "Skin Lesion Segmentation Using Deep Learning with Auxiliary Task" Journal of Imaging 7, no. 4: 67. https://doi.org/10.3390/jimaging7040067
APA StyleLiu, L., Tsui, Y. Y., & Mandal, M. (2021). Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. Journal of Imaging, 7(4), 67. https://doi.org/10.3390/jimaging7040067







