The Constantly Evolving Role of Medical Image Processing in Oncology: From Traditional Medical Image Processing to Imaging Biomarkers and Radiomics
Abstract
:1. Introduction
2. Traditional Image Analysis: The First Efforts towards CAD Systems
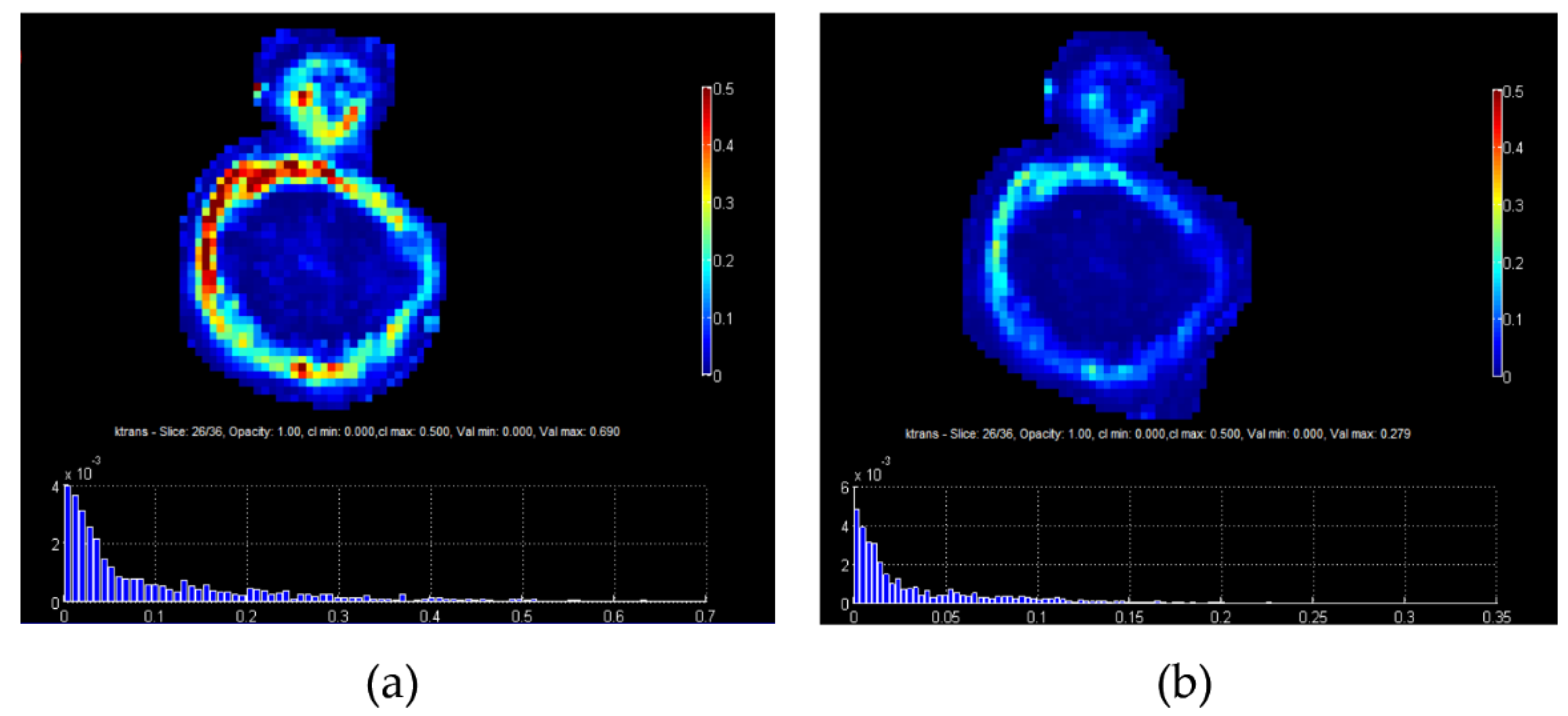
3. Quantitative Imaging Based on Models
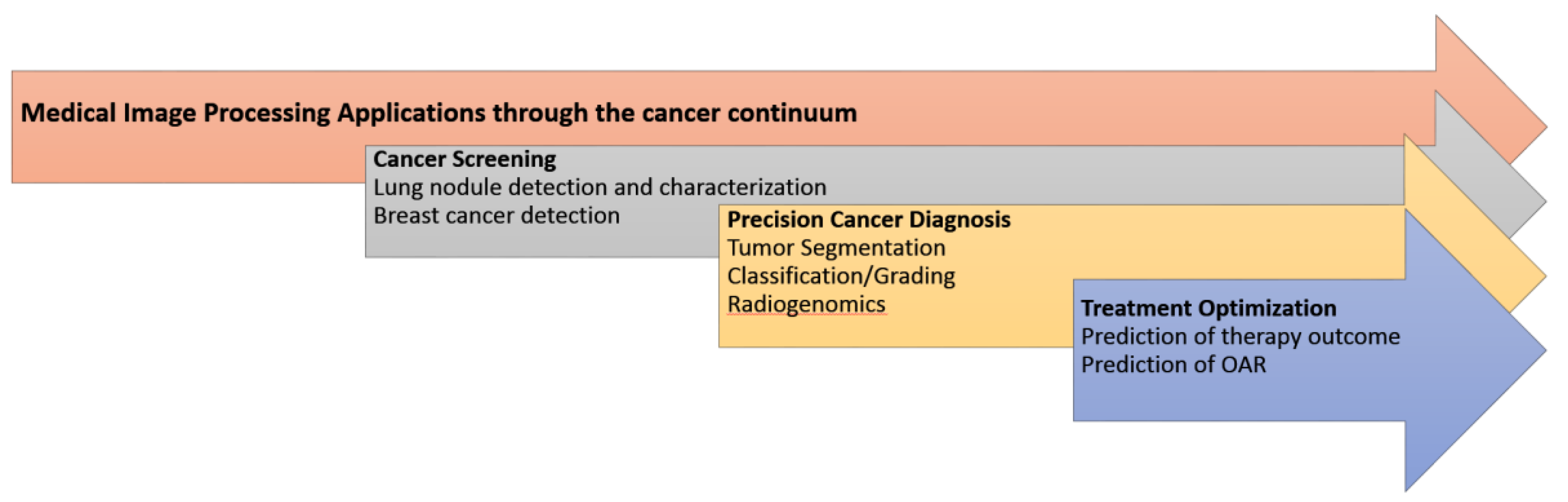
4. Radiomics and Deep Learning Approaches in Oncology through the Cancer Continuum
4.1. Cancer Screening
4.2. Precision Cancer Diagnosis
4.3. Treatment Optimization
5. Radiomics Limitations Regarding Clinical Translation
6. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Oakden-Rayner, L.; Carneiro, G.; Bessen, T.; Nascimento, J.C.; Bradley, A.P.; Palmer, L.J. Precision Radiology: Predicting longevity using feature engineering and deep learning methods in a radiomics framework. Sci. Rep. 2017, 7, 1648. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.-L.; Hong, G.-B. Quantitative image analysis for evaluation of tumor response in clinical oncology. Chronic Dis. Transl. Med. 2018, 4, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.; Chapiro, J.; Frangakis, C.; De Lin, M.; Schlachter, T.R.; Schernthaner, R.E.; Wang, Z.; Savic, L.J.; Tacher, V.; Kamel, I.R.; et al. Uveal melanoma metastatic to the liver: The role of quantitative volumetric contrast-enhanced MR imaging in the assessment of early tumor response after transarterialchemo. Transl. Oncol. 2014, 7, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Aykan, N.F.; Özatlı, T. Objective response rate assessment in oncology: Current situation and future expectations. World J. Clin. Oncol. 2020, 11, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Froelich, M.F.; Petersen, E.L.; Heinemann, V.; Nörenberg, D.; Hesse, N.; Gesenhues, A.B.; Modest, D.P.; Sommer, W.H.; Hofmann, F.O.; Stintzing, S.; et al. Impact of Size and Location of Metastases on Early Tumor Shrinkage and Depth of Response in Patients With Metastatic Colorectal Cancer: Subgroup Findings of the Randomized, Open-Label Phase 3 Trial FIRE-3/AIO KRK-0306. Clin. Colorectal Cancer 2020, 19, 291–300.e5. [Google Scholar] [CrossRef] [PubMed]
- Sasieni, P. Evaluation of the UK breast screening programmes. Ann. Oncol. 2003, 14, 1206–1208. [Google Scholar] [CrossRef]
- Marias, K.; Behrenbruch, C.; Parbhoo, S.; Seifalian, A.; Brady, M. A registration framework for the comparison of mammogram sequences. IEEE Trans. Med. Imaging 2005, 24, 782–790. [Google Scholar] [CrossRef]
- Funovics, M.; Schamp, S.; Lackner, B.; Wunderbaldinger, P.; Lechner, G.; Wolf, G. Computerassistierte diagnose in der mammographie: Das R2 imagechecker- system in der detektion spikulierter lasionen. Wien. Med. Wochenschr. 1998, 148, 321–324. [Google Scholar]
- Manikis, G.C.; Nikiforaki, K.; Lagoudaki, E.; de Bree, E.; Maris, T.G.; Marias, K.; Karantanas, A.H. Differentiating low from high-grade soft tissue sarcomas using post-processed imaging parameters derived from multiple DWI models. Eur. J. Radiol. 2021, 138, 109660. [Google Scholar] [CrossRef]
- Messina, C.; Bignone, R.; Bruno, A.; Bruno, A.; Bruno, F.; Calandri, M.; Caruso, D.; Coppolino, P.; De Robertis, R.; Gentili, F.; et al. Diffusion-Weighted Imaging in Oncology: An Update. Cancers 2020, 12, 1493. [Google Scholar] [CrossRef]
- Fliedner, F.P.; Engel, T.B.; El-Ali, H.H.; Hansen, A.E.; Kjaer, A. Diffusion weighted magnetic resonance imaging (DW-MRI) as a non-invasive, tissue cellularity marker to monitor cancer treatment response. BMC Cancer 2020, 20, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz-Hansen, T.; Rostrup, E.; Larsson, H.B.W.; Søndergaard, L.; Ring, P.; Henriksen, O. Measurement of the arterial concentration of Gd-DTPA using MRI: A step toward quantitative perfusion imaging. Magn. Reson. Med. 1996, 36, 225–231. [Google Scholar] [CrossRef]
- Woolf, D.K.; Taylor, N.J.; Makris, A.; Tunariu, N.; Collins, D.J.; Li, S.P.; Ah-See, M.-L.; Beresford, M.; Padhani, A.R. Arterial input functions in dynamic contrast-enhanced magnetic resonance imaging: Which model performs best when assessing breast cancer response? Br. J. Radiol. 2016, 89, 20150961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef]
- Liu, L.; Chen, J.; Fieguth, P.; Zhao, G.; Chellappa, R.; Pietikäinen, M. From BoW to CNN: Two Decades of Texture Representation for Texture Classification. Int. J. Comput. Vis. 2019, 127, 74–109. [Google Scholar] [CrossRef] [Green Version]
- Svoboda, E. Artificial intelligence is improving the detection of lung cancer. Nature 2020, 587, S20–S22. [Google Scholar] [CrossRef] [PubMed]
- Binczyk, F.; Prazuch, W.; Bozek, P.; Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res. 2021, 10, 1186–1199. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Salim, M.; Wåhlin, E.; Dembrower, K.; Azavedo, E.; Foukakis, T.; Liu, Y.; Smith, K.; Eklund, M.; Strand, F. External Evaluation of 3 Commercial Artificial Intelligence Algorithms for Independent Assessment of Screening Mammograms. JAMA Oncol. 2020, 6, 1581. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A Deep Learning Approach for Brain Tumor Classification and Segmentation Using a Multiscale Convolutional Neural Network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, L.-F.; Zhang, X.; Han, Y.; Nan, H.-Y.; Hu, Y.-C.; Hu, B.; Yan, S.-L.; Zhang, J.; Cheng, D.-L.; et al. Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front. Neurosci. 2018, 12. [Google Scholar] [CrossRef] [Green Version]
- Trivizakis, E.; Papadakis, G.Z.; Souglakos, I.; Papanikolaou, N.; Koumakis, L.; Spandidos, D.A.; Tsatsakis, A.; Karantanas, A.H.; Marias, K. Artificial intelligence radiogenomics for advancing precision and effectiveness in oncologic care (Review). Int. J. Oncol. 2020, 57, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Nam, Y.; Lee, Y.S.; Kim, J.; Ahn, K.-J.; Jang, J.; Shin, N.-Y.; Kim, B.-S.; Jeon, S.-S. IDH1 mutation prediction using MR-based radiomics in glioblastoma: Comparison between manual and fully automated deep learning-based approach of tumor segmentation. Eur. J. Radiol. 2020, 128, 109031. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Tang, F.; Huang, X.; Yang, K.; Zhong, T.; Hu, R.; Liu, S.; Yuan, X.; Zhang, Y. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur. Radiol. 2019, 29, 1961–1967. [Google Scholar] [CrossRef]
- Vulchi, M.; El Adoui, M.; Braman, N.; Turk, P.; Etesami, M.; Drisis, S.; Plecha, D.; Benjelloun, M.; Madabhushi, A.; Abraham, J. Development and external validation of a deep learning model for predicting response to HER2-targeted neoadjuvant therapy from pretreatment breast MRI. J. Clin. Oncol. 2019, 37, 593. [Google Scholar] [CrossRef]
- Spadarella, G.; Calareso, G.; Garanzini, E.; Ugga, L.; Cuocolo, A.; Cuocolo, R. MRI based radiomics in nasopharyngeal cancer: Systematic review and perspectives using radiomic quality score (RQS) assessment. Eur. J. Radiol. 2021, 140, 109744. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, D.; Kim, H.S.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Shin, J.H.; Kim, J.H. Quality of science and reporting of radiomics in oncologic studies: Room for improvement according to radiomics quality score and TRIPOD statement. Eur. Radiol. 2020, 30, 523–536. [Google Scholar] [CrossRef]
- Stanzione, A.; Gambardella, M.; Cuocolo, R.; Ponsiglione, A.; Romeo, V.; Imbriaco, M. Prostate MRI radiomics: A systematic review and radiomic quality score assessment. Eur. J. Radiol. 2020, 129, 109095. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Li, L.; Hou, W.; Ma, X.; Tian, R. Current status and quality of radiomics studies in lymphoma: A systematic review. Eur. Radiol. 2020, 30, 6228–6240. [Google Scholar] [CrossRef]
- McNitt-Gray, M.; Napel, S.; Jaggi, A.; Mattonen, S.A.; Hadjiiski, L.; Muzi, M.; Goldgof, D.; Balagurunathan, Y.; Pierce, L.A.; Kinahan, P.E.; et al. Standardization in Quantitative Imaging: A Multicenter Comparison of Radiomic Features from Different Software Packages on Digital Reference Objects and Patient Data Sets. Tomography 2020, 6, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Capobianco, E.; Dominietto, M. From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health. J. Pers. Med. 2020, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Rundo, L.; Militello, C.; Vitabile, S.; Russo, G.; Sala, E.; Gilardi, M.C. A Survey on Nature-Inspired Medical Image Analysis: A Step Further in Biomedical Data Integration. Fundam. Inform. 2019, 171, 345–365. [Google Scholar] [CrossRef]
- Zhu, H.-T.; Zhang, X.-Y.; Shi, Y.-J.; Li, X.-T.; Sun, Y.-S. A Deep Learning Model to Predict the Response to Neoadjuvant Chemoradiotherapy by the Pretreatment Apparent Diffusion Coefficient Images of Locally Advanced Rectal Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Chaddad, A.; Daniel, P.; Sabri, S.; Desrosiers, C.; Abdulkarim, B. Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma. Cancers 2019, 11, 1148. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marias, K. The Constantly Evolving Role of Medical Image Processing in Oncology: From Traditional Medical Image Processing to Imaging Biomarkers and Radiomics. J. Imaging 2021, 7, 124. https://doi.org/10.3390/jimaging7080124
Marias K. The Constantly Evolving Role of Medical Image Processing in Oncology: From Traditional Medical Image Processing to Imaging Biomarkers and Radiomics. Journal of Imaging. 2021; 7(8):124. https://doi.org/10.3390/jimaging7080124
Chicago/Turabian StyleMarias, Kostas. 2021. "The Constantly Evolving Role of Medical Image Processing in Oncology: From Traditional Medical Image Processing to Imaging Biomarkers and Radiomics" Journal of Imaging 7, no. 8: 124. https://doi.org/10.3390/jimaging7080124
APA StyleMarias, K. (2021). The Constantly Evolving Role of Medical Image Processing in Oncology: From Traditional Medical Image Processing to Imaging Biomarkers and Radiomics. Journal of Imaging, 7(8), 124. https://doi.org/10.3390/jimaging7080124





