Early Challenges in the Implementation of Automated CranialRebuild Freeware for Generation of Patient-Specific Cranial Implant Using Additive Manufacturing: A Pilot Project in Review
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Feedback from the Philippine Team
3.2. Feedback from the Ukrainian Team
4. Discussion
Limitations
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, B.; Anderson, D.B.; Chen, L.; Feng, S.; Zhou, H. Global, regional and national burden of traumatic brain injury and spinal cord injury, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. BMJ Open. 2023, 13, e075049. [Google Scholar] [CrossRef] [PubMed]
- Hatamleh, M.M. Contemporary Review on Craniectomy and Cranioplasty; Part 1: Decompressive Craniectomy. J. Craniofac Surg. 2022, 33, 838–841. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, P.J.; Kolias, A.G.; Timofeev, I.S.; Corteen, E.A.; Czosnyka, M.; Timothy, J.; Anderson, I.; Bulters, D.O.; Belli, A.; Eynon, C.A.; et al. Trial of Decompressive Craniectomy for Traumatic Intracranial Hypertension. N. Engl. J. Med. 2016, 375, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, C.; Viaroli, E.; Fricia, M.; Serchi, E.; Poli, T.; Servadei, F. Preliminary Results of a Prospective Study on Methods of Cranial Reconstruction. J. Oral. Maxillofac. Surg. 2015, 73, 2375–2378. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, C.; Kolias, A.G.; Roumy, L.G.; Fountas, K.; Adeleye, A.O. Cranioplasty following decompressive craniectomy. Front. Neurol. 2020, 10, 1357. [Google Scholar] [CrossRef] [PubMed]
- Ozoner, B. Cranioplasty Following Severe Traumatic Brain Injury: Role in Neurorecovery. Curr. Neurol. Neurosci. Rep. 2021, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Honeybul, S.; Ho, K.M. How “successful” is calvarial reconstruction using frozen autologous bone? Plast. Reconstr. Surg. 2012, 130, 1110–1117. [Google Scholar] [CrossRef]
- Binhammer, A.; Jakubowski, J.; Antonyshyn, O.; Binhammer, P. Comparative Cost-Effectiveness of Cranioplasty Implants. Plast. Surg. 2020, 28, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Gilardino, M.S.; Karunanayake, M.; Al-Humsi, T.; Izadpanah, A.; Al-Ajmi, H.; Marcoux, J.; Atkinson, J.; Farmer, J.P. A comparison and cost analysis of cranioplasty techniques: Autologous bone versus custom computer-generated implants. J. Craniofac Surg. 2015, 26, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Ernst, G.; Qeadan, F.; Carlson, A.P. Subcutaneous bone flap storage after emergency craniectomy: Cost-effectiveness and rate of resorption. J. Neurosurg. 2018, 129, L1604–L1610. [Google Scholar] [CrossRef] [PubMed]
- Bečulić, H.; Spahić, D.; Begagić, E.; Pugonja, R.; Skomorac, R.; Jusić, A.; Selimović, E.; Mašović, A.; Pojskić, M. Breaking Barriers in Cranioplasty: 3D Printing in Low and Middle-Income Settings-Insights from Zenica, Bosnia and Herzegovina. Medicina 2023, 59, 1732. [Google Scholar] [CrossRef]
- Ashraf, M.; Choudhary, N.; Kamboh, U.A.; Raza, M.A.; Sultan, K.A.; Ghulam, N.; Hussain, S.S.; Ashraf, N. Early experience with patient-specific low-cost 3D-printed polymethylmethacrylate cranioplasty implants in a lower-middle-income-country: Technical note and economic analysis. Surg. Neurol. Int. 2022, 13, 270. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.T.W.; Ling, J.M.; Dinesh, S.K. The feasibility of producing patient-specific acrylic cranioplasty implants with a low-cost 3D printer. J. Neurosurg. 2016, 124, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Lannon, M.; Algird, A.; Alsunbul, W.; Wang, B.H. Cost-Effective Cranioplasty Utilizing 3D Printed Molds: A Canadian Single-Center Experience. J. Can. Des. Sci. Neurol. 2022, 49, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, V.; McConaha, M.; Ghalsasi, O.; Xu, A.; Anand, S.; Forbes, J.; Cheng, J.; Aryal, M.R. Method of Designing Patient-Specific Cranioplasty Implants. US Patent US20230414367A1, 28 December 2023. [Google Scholar]
- Venugopal, V.; Ghalsasi, O.; McConaha, M.; Xu, A.; Forbes, J.; Anand, S. Image processing-based method for automatic design of patient-specific cranial implant for additive manufacturing. Proc. Manuf. 2021, 53, 375–386. [Google Scholar] [CrossRef]
- Xu, A.; Venugopal, V.; Aryal, M.R.; Alfawares, Y.; Matur, A.V.; Cheng, J.; Kosco, E.; McConaha, M.; Ghalsasi, O.; Lockett, D.; et al. Toward global availability of low-cost, patient-specific cranial implants: Creation and validation of automated CranialRebuild freeware application. Acta Neurochir. 2023, 165, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Wambani, J.; Korir, G.K.; Onditi, E.G.; Korir, I.K. A survey of computed tomography imaging techniques and patient dose in Kenya. East Afr. Med. J. 2010, 87, 400–407. [Google Scholar] [PubMed]
- Thickman, D.; Wang, G.; White, J.; Cutter, G. National Variation of Technical Factors in Computed Tomography of the Head. J. Comput. Assist. Tomogr. 2013, 37, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D. 2022—Geometry in Medical Imaging: DICOM and NIfTI Formats. Available online: https://discovery.ucl.ac.uk/id/eprint/10146893/1/geometry_medim.pdf (accessed on 14 April 2024).
- Morales-Gomez, J.A.; Garcia-Estrada, E.; Leos-Bortoni, J.E.; Delgado-Brito, M.; Flores-Huerta, L.E.; De La Cruz-Arriaga, A.A.; Torres-Diaz, L.J.; Martinez-Ponce de Leon, A.R. Cranioplasty with a low-cost customized polymethylmethacrylate implant using a desktop 3D printer. J. Neurosurg. 2018, 130, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, C.; Kolias, A.; Adelson, P.D.; Rubiano, A.M.; Viaroli, E.; Buki, A.; Cinalli, G.; Fountas, K.; Khan, T.; Signoretti, S.; et al. Consensus statement from the international consensus meeting on post-traumatic cranioplasty. Acta Neurochir. 2021, 163, 423–440. [Google Scholar] [CrossRef] [PubMed]

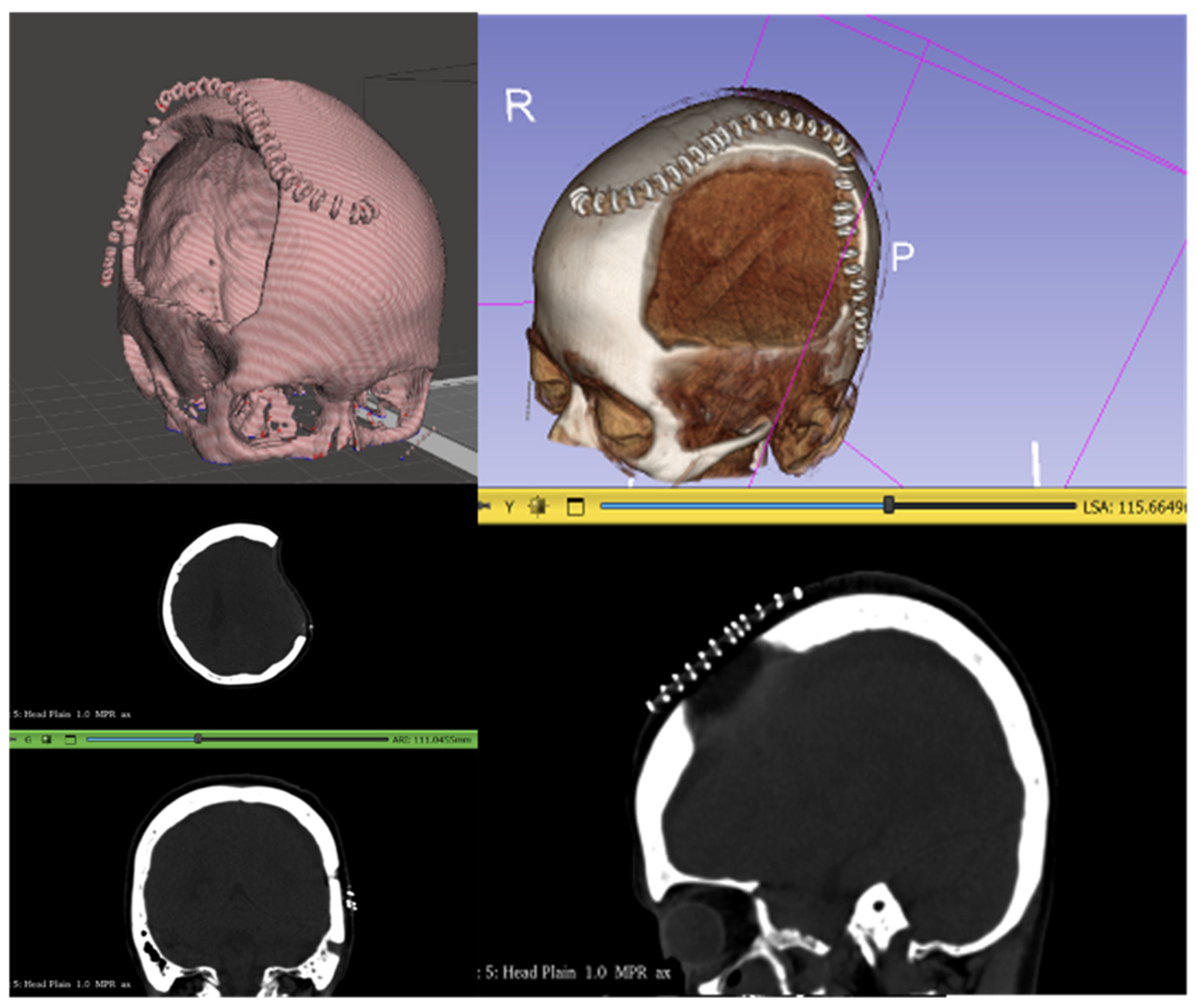
| Challenges | Brief Description | Short-Term Solution | Long-Term Solution |
|---|---|---|---|
| Variation in CT scan slice thickness. | The Philippine team used CT scans with a slice thickness of 142.89 mm, while CranialRebuild 1.0 was designed to accept a slice thickness ranging from 0.1 to 10 mm. | The Philippine team was advised to adjust for this requirement in slice thickness during CT scan acquisition. | The slice thickness requirement of 0.1–1 mm will be specified for the future use of CranialRebuild to not compromise the precision of the product. |
| Bone and soft tissue data storage in separate folders. | While the DICOM files used in our cadaveric testing stored bone and soft tissue data in a combined folder, the data used by the Philippine team were separated into two distinct folders. | The Philippine team had to re-export the bone CT data into a single DICOM folder, which could be input to CranialRebuild. | CranialRebuild will be updated to automatically extract the largest folder, which has been found to typically store the bone data regardless of whether the soft tissue data are also stored in that folder. |
| Inconsistency in array size in a single DICOM file. | The DICOM files used by the Philippine team matched the array size to the physical size of the skull in cross-section through each CT scan slice, causing arrays of different sizes. | This continues to pose a barrier to progress in testing at the time of publication of this paper. | CranialRebuild 2.0 will be engineered to allow the user to select the size of their largest array, and subsequently resize all other arrays to ensure size consistency before further processing. |
| Misidentification of surgical staples as bone. | Some CT scans included surgical staples overlying the DHC and were detected by CranialRebuild as bone, which impacted the software’s determination of the midsagittal plane. | The recommendation was made to acquire CT scans after staple removal (typically 2 weeks after DHC) but before cranioplasty (2–3 months after DHC). | This specific timing of the CT scan should be recommended in the future use of CranialRebuild 2.0. |
| Suggestion/Challenge | CranialRebuild Team Comments | Priority |
|---|---|---|
| Inconsistency in array size in a single DICOM file. | Effort required to address: Medium. Ability to improve software: Highly valuable as the Philippine team encountered the same issue. | 5 |
| Add ability to visualize implant fitted onto DHC site in virtual modeling before 3D printing. |
Effort required to address: Medium. Ability to improve software: Highly valuable for 3D model visualization. | 4 |
| Add option to align the implant with one edge of the DHC site and trim any overlap with the bone. | Effort required to address: High. Ability to improve software: Valuable in the long run, as some neurosurgeons might prefer this to our original scaling method. | 2 |
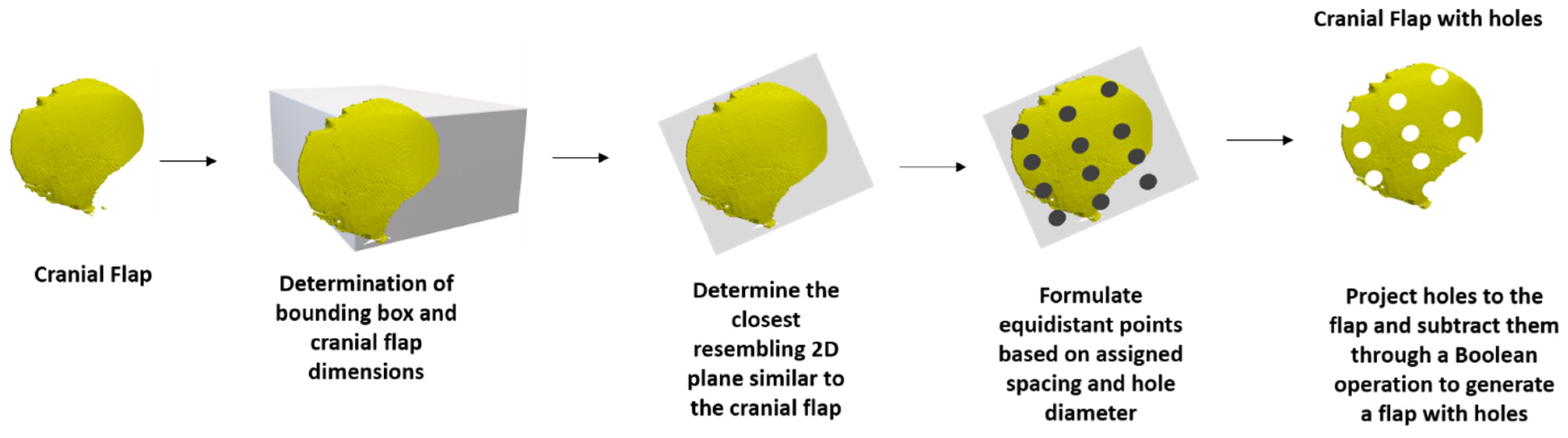
| Add the detail of including a grid of outlet holes onto the designed implant. | Effort required to address: High. Ability to improve software: Valuable as it is more in line with current clinically used implants and would allow future users the option of directly printing implants rather than molds. | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strelko, O.; Aryal, M.R.; Zack, A.; Alfawares, Y.; Remenyi, R.; Bayan, I.K.; Briones, Y.L.; Holovenko, Y.; Maksymenko, M.; Sirko, A.; et al. Early Challenges in the Implementation of Automated CranialRebuild Freeware for Generation of Patient-Specific Cranial Implant Using Additive Manufacturing: A Pilot Project in Review. Biomimetics 2024, 9, 430. https://doi.org/10.3390/biomimetics9070430
Strelko O, Aryal MR, Zack A, Alfawares Y, Remenyi R, Bayan IK, Briones YL, Holovenko Y, Maksymenko M, Sirko A, et al. Early Challenges in the Implementation of Automated CranialRebuild Freeware for Generation of Patient-Specific Cranial Implant Using Additive Manufacturing: A Pilot Project in Review. Biomimetics. 2024; 9(7):430. https://doi.org/10.3390/biomimetics9070430
Chicago/Turabian StyleStrelko, Oleksandr, Manish Raj Aryal, Abigail Zack, Yara Alfawares, Roland Remenyi, Ian Kristopher Bayan, Yumi L. Briones, Yaroslav Holovenko, Maksym Maksymenko, Andrii Sirko, and et al. 2024. "Early Challenges in the Implementation of Automated CranialRebuild Freeware for Generation of Patient-Specific Cranial Implant Using Additive Manufacturing: A Pilot Project in Review" Biomimetics 9, no. 7: 430. https://doi.org/10.3390/biomimetics9070430
APA StyleStrelko, O., Aryal, M. R., Zack, A., Alfawares, Y., Remenyi, R., Bayan, I. K., Briones, Y. L., Holovenko, Y., Maksymenko, M., Sirko, A., Anand, S., & Forbes, J. A. (2024). Early Challenges in the Implementation of Automated CranialRebuild Freeware for Generation of Patient-Specific Cranial Implant Using Additive Manufacturing: A Pilot Project in Review. Biomimetics, 9(7), 430. https://doi.org/10.3390/biomimetics9070430







