Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients at Pre-Treatment Using CT Textural Features and Machine Learning: Comparison of Feature Selection Methods
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Protocol and Data Acquisition
2.2. Pathological Evaluation of Tumor Response
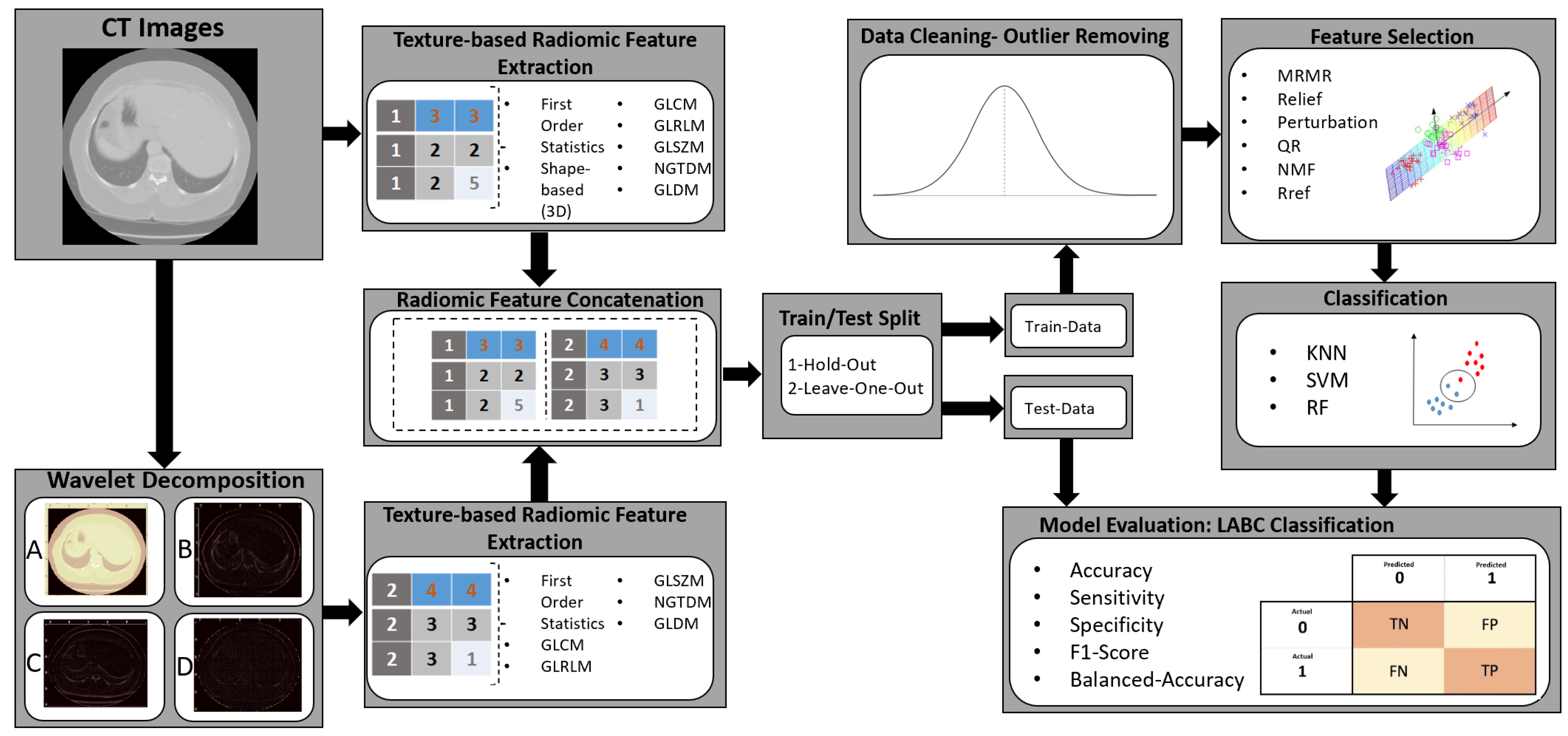
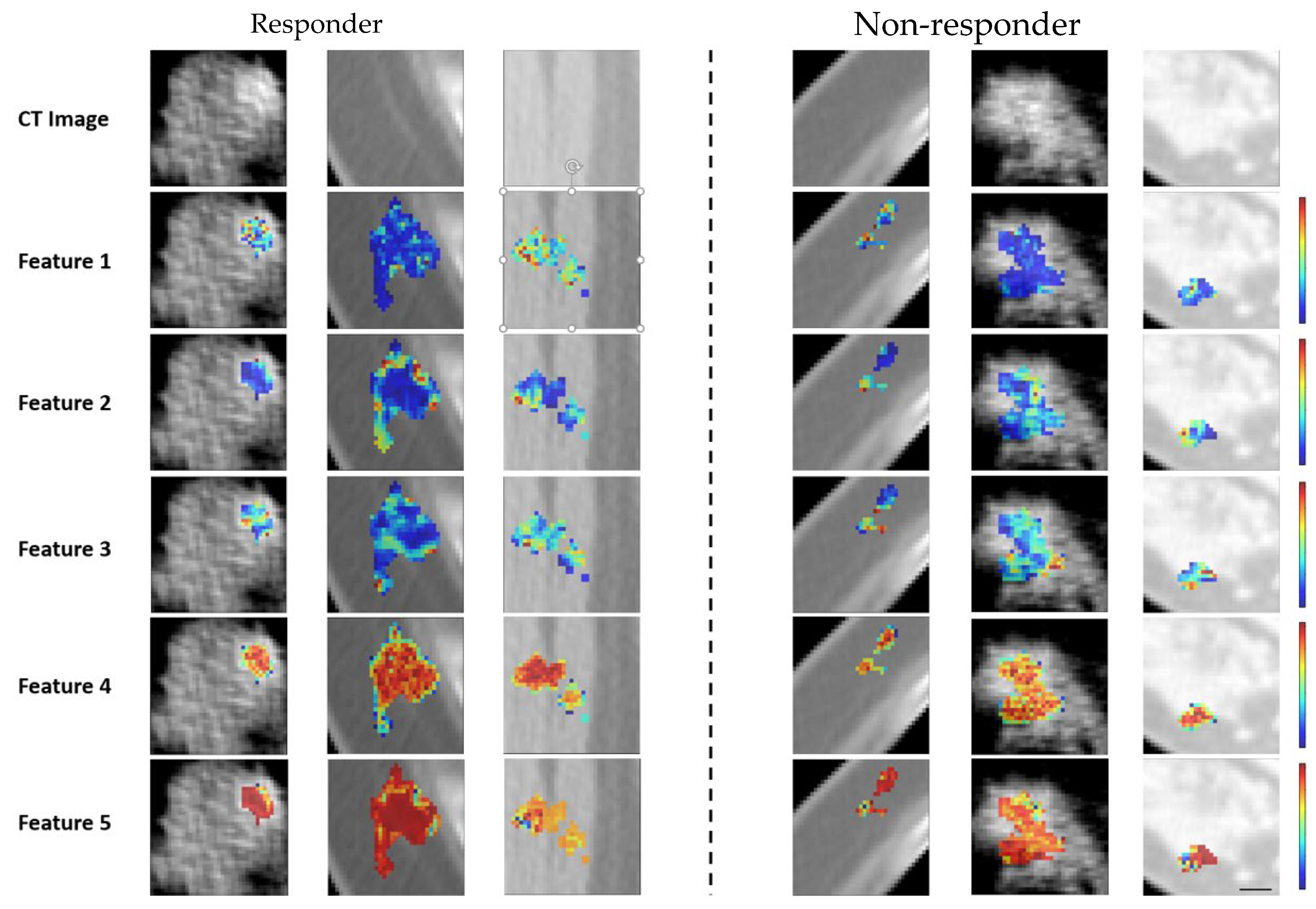
2.3. Feature Determination and Pre-Processing
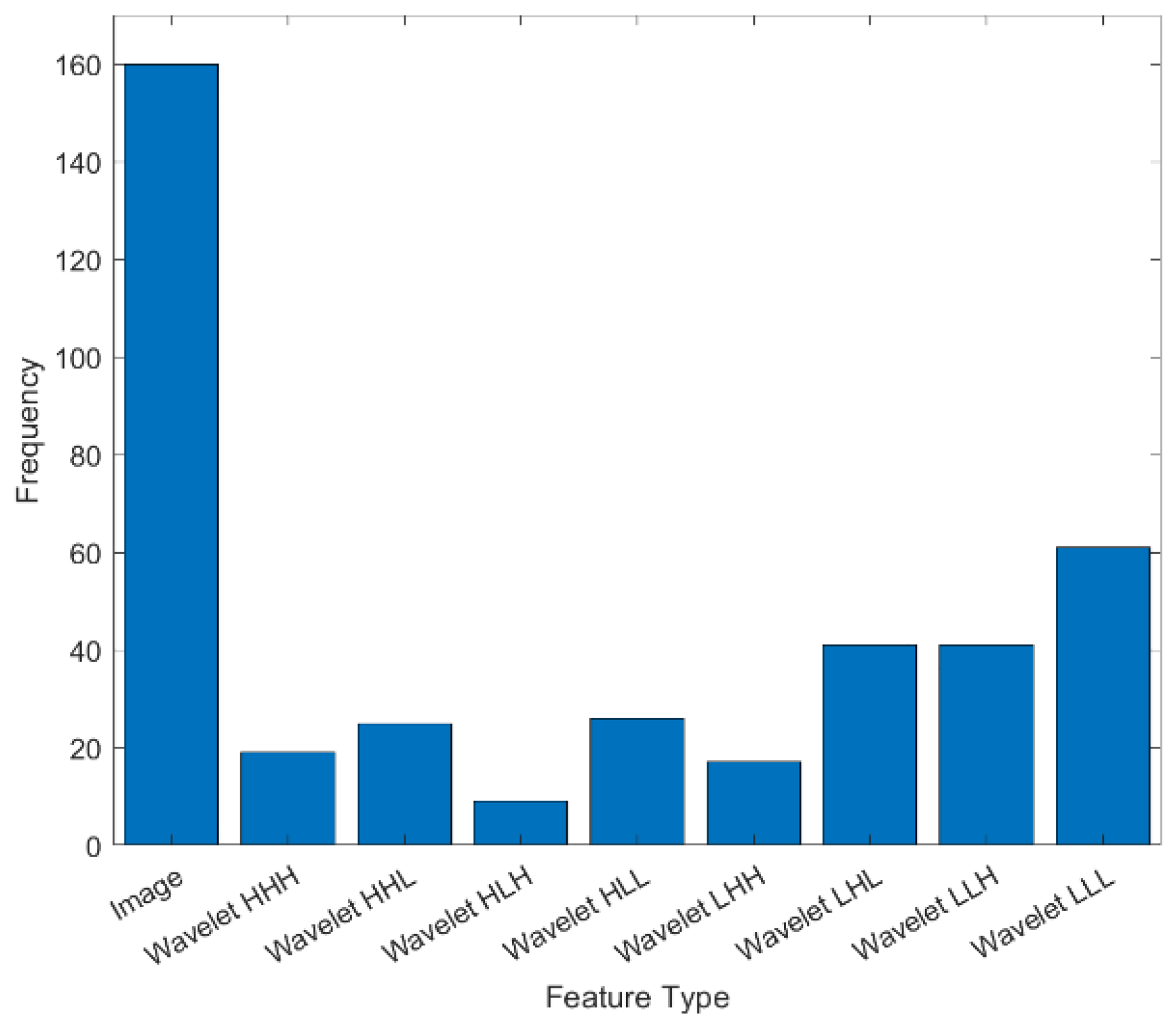
2.4. Feature Determination Using Wavelet Transform
2.5. Feature Selection
Feature Selection Techniques
2.6. Training Model
2.7. Response Prediction
2.8. Evaluation Metric
3. Statistical Analysis
4. Implementation of Method
5. Results
Feature Selection Techniques Comparison
6. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giordano, S.H. Update on locally advanced breast cancer. Oncologist 2003, 8, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Tryfonidis, K.; Senkus, E.; Cardoso, M.J.; Cardoso, F. Management of locally advanced breast cancer—Perspectives and future directions. Nat. Rev. Clin. Oncol. 2015, 12, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Gradishar, W.J.; Anderson, B.O.; Abraham, J.; Aft, R.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. Breast cancer, version 3.2018 featured updates to the NCCN guidelines. JNCCN J. Natl. Compr. Cancer Netw. 2019, 17, 118–126. [Google Scholar] [CrossRef]
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, 8–30. [Google Scholar] [CrossRef]
- Rajan, R.; Poniecka, A.; Smith, T.L.; Yang, Y.; Frye, D.; Pusztai, L.; Fiterman, D.J.; Gal-Gombos, E.; Whitman, G.; Rouzier, R.; et al. Change in tumour cellularity of breast carcinoma after neoadjuvant chemotherapy as a variable in the pathologic assessment of response. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2004, 100, 1365–1373. [Google Scholar]
- Ogston, K.N.; Miller, I.D.; Payne, S.; Hutcheon, A.W.; Sarkar, T.K.; Smith, I.; Schofield, A.; Heys, S.D. A new histological grading system to assess response of breast cancers to primary chemotherapy: Prognostic significance and survival. Breast 2003, 12, 320–327. [Google Scholar] [CrossRef]
- Sahoo, S.; Lester, S.C. Pathology of breast carcinomas after neoadjuvant chemotherapy: An overview with recommendations on specimen processing and reporting. Arch. Pathol. Lab. Med. 2009, 133, 633–642. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Kononenko, I. Machine learning for medical diagnosis: History, state of the art and perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef]
- Chang, J.; Ormerod, M.; Powles, T.J.; Allred, D.C.; Ashley, S.E.; Dowsett, M. Apoptosis and proliferation as predictors of chemotherapy response in patients with breast carcinoma. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2000, 89, 2145–2152. [Google Scholar] [CrossRef]
- Chang, J.; Powles, T.J.; Allred, D.C.; Ashley, S.E.; Clark, G.M.; Makris, A.; Assersohn, L.; Gregory, R.K.; Osborne, C.K.; Dowsett, M. Biologic markers as predictors of clinical outcome from systemic therapy for primary operable breast cancer. J. Clin. Oncol. 1999, 17, 3058–3063. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Mazouni, C.; Liedtke, C.; Kau, S.W.; Frye, D.; Green, M.; Gonzalez-Angulo, A.M.; Symmans, W.F.; Hortobagyi, G.N.; Pusztai, L. HER2 expression and efficacy of preoperative paclitaxel/FAC chemotherapy in breast cancer. Breast Cancer Res. Treat. 2008, 108, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Cerussi, A.; Hsiang, D.; Shah, N.; Mehta, R.; Durkin, A.; Butler, J.; Tromberg, B.J. Predicting response to breast cancer neoadjuvant chemotherapy using diffuse optical spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 4014–4019. [Google Scholar] [CrossRef]
- Jiang, S.; Pogue, B.W.; Kaufman, P.A.; Gui, J.; Jermyn, M.; Frazee, T.E.; Poplack, S.P.; DiFlorio-Alexander, R.; Wells, W.A.; Paulsen, K.D. Predicting breast tumour response to neoadjuvant chemotherapy with diffuse optical spectroscopic tomography prior to treatment. Clin. Cancer Res. 2014, 20, 6006–6015. [Google Scholar] [CrossRef]
- Sadeghi-Naini, A.; Falou, O.; Hudson, J.M.; Bailey, C.; Burns, P.N.; Yaffe, M.J.; Stanisz, G.J.; Kolios, M.C.; Czarnota, G.J. Imaging innovations for cancer therapy response monitoring. Imaging Med. 2012, 4, 311. [Google Scholar] [CrossRef]
- Tudorica, A.; Oh, K.Y.; Chui, S.Y.; Roy, N.; Troxell, M.L.; Naik, A.; Kemmer, K.A.; Chen, Y.; Holtorf, M.L.; Afzal, A.; et al. Early prediction and evaluation of breast cancer response to neoadjuvant chemotherapy using quantitative DCE-MRI. Transl. Oncol. 2016, 9, 8–17. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Pantel, K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res. 2015, 17, 136. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Thomssen, C.; Gnant, M. St. gallen 2013: Brief preliminary summary of the consensus discussion. Breast Care 2013, 8, 102–109. [Google Scholar] [CrossRef]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Tran, W.T.; Gangeh, M.J.; Sannachi, L.; Chin, L.; Watkins, E.; Bruni, S.G.; Rastegar, R.F.; Curpen, B.; Trudeau, M.; Gandhi, S.; et al. Predicting breast cancer response to neoadjuvant chemotherapy using pretreatment diffuse optical spectroscopic texture analysis. Br. J. Cancer 2017, 116, 1329–1339. [Google Scholar] [CrossRef]
- Tadayyon, H.; Sannachi, L.; Gangeh, M.J.; Kim, C.; Ghandi, S.; Trudeau, M.; Pritchard, K.; Tran, W.T.; Slodkowska, E.; Sadeghi-Naini, A.; et al. A priori prediction of neoadjuvant chemotherapy response and survival in breast cancer patients using quantitative ultrasound. Sci. Rep. 2017, 7, 45733. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Humbert, O.; Cochet, A.; Riedinger, J.M.; Berriolo-Riedinger, A.; Arnould, L.; Coudert, B.; Desmoulins, I.; Toubeau, M.; Dygai-Cochet, I.; Guiu, S.; et al. HER2-positive breast cancer: 18F-FDG PET for early prediction of response to trastuzumab plus taxane-based neoadjuvant chemotherapy. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1525–1533. [Google Scholar] [CrossRef]
- Juweid, M.E.; Cheson, B.D. Positron-emission tomography and assessment of cancer therapy. N. Engl. J. Med. 2006, 354, 496–507. [Google Scholar] [CrossRef]
- Sadeghi-Naini, A.; Vorauer, E.; Chin, L.; Falou, O.; Tran, W.T.; Wright, F.C.; Gandhi, S.; Yaffe, M.J.; Czarnota, G.J. Early detection of chemotherapy-refractory patients by monitoring textural alterations in diffuse optical spectroscopic images. Med. Phys. 2015, 42, 6130–6146. [Google Scholar] [CrossRef]
- Czarnota, G.J.; Kolios, M.C.; Abraham, J.; Portnoy, M.; Ottensmeyer, F.P.; Hunt, J.W.; Sherar, M.D. Ultrasound imaging of apoptosis: High-resolution non-invasive monitoring of programmed cell death in vitro, in situ and in vivo. Br. J. Cancer 1999, 81, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi-Naini, A.; Falou, O.; Tadayyon, H.; Al-Mahrouki, A.; Tran, W.; Papanicolau, N.; Kolios, M.C.; Czarnota, G.J. Conventional frequency ultrasonic biomarkers of cancer treatment response in vivo. Transl. Oncol. 2013, 6, 234-IN2. [Google Scholar] [CrossRef]
- Sadeghi-Naini, A.; Zhou, S.; Gangeh, M.J.; Jahedmotlagh, Z.; Falou, O.; Ranieri, S.; Azrif, M.; Giles, A.; Czarnota, G.J. Quantitative evaluation of cell death response in vitro and in vivo using conventional-frequency ultrasound. Oncoscience 2015, 2, 716. [Google Scholar] [CrossRef]
- Sadeghi-Naini, A.; Papanicolau, N.; Falou, O.; Tadayyon, H.; Lee, J.; Zubovits, J.; Sadeghian, A.; Karshafian, R.; Al-Mahrouki, A.; Giles, A.; et al. Low-frequency quantitative ultrasound imaging of cell death in vivo. Med. Phys. 2013, 40, 082901. [Google Scholar] [CrossRef]
- Sannachi, L.; Tadayyon, H.; Sadeghi-Naini, A.; Tran, W.; Gandhi, S.; Wright, F.; Oelze, M.; Czarnota, G. Non-invasive evaluation of breast cancer response to chemotherapy using quantitative ultrasonic backscatter parameters. Med. Image Anal. 2015, 20, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi-Naini, A.; Sannachi, L.; Tadayyon, H.; Tran, W.T.; Slodkowska, E.; Trudeau, M.; Gandhi, S.; Pritchard, K.; Kolios, M.C.; Czarnota, G.J. Chemotherapy-response monitoring of breast cancer patients using quantitative ultrasound-based intra-tumour heterogeneities. Sci. Rep. 2017, 7, 10352. [Google Scholar] [CrossRef]
- Makimoto, K.; Au, R.; Moslemi, A.; Hogg, J.C.; Bourbeau, J.; Tan, W.C.; Kirby, M. Comparison of Feature Selection Methods and Machine Learning Classifiers for Predicting Chronic Obstructive Pulmonary Disease Using Texture-Based CT Lung Radiomic Features. Acad. Radiol. 2023, 30, 900–910. [Google Scholar] [CrossRef]
- Moslemi, A.; Kontogianni, K.; Brock, J.; Wood, S.; Herth, F.; Kirby, M. Differentiating COPD and asthma using quantitative CT imaging and machine learning. Eur. Respir. J. 2022, 60, 2103078. [Google Scholar] [CrossRef] [PubMed]
- Moslemi, A.; Makimoto, K.; Tan, W.C.; Bourbeau, J.; Hogg, J.C.; Coxson, H.O.; Kirby, M. Quantitative CT lung imaging and machine learning improves prediction of emergency room visits and hospitalizations in COPD. Acad. Radiol. 2023, 30, 707–716. [Google Scholar] [CrossRef]
- O’Connell, A.M.; Karellas, A.; Vedantham, S. The potential role of dedicated 3D breast CT as a diagnostic tool: Review and early clinical examples. Breast J. 2014, 20, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Moghadas-Dastjerdi, H.; Sannachi, L.; Wright, F.C.; Gandhi, S.; Trudeau, M.E.; Sadeghi-Naini, A.; Czarnota, G.J. Prediction of chemotherapy response in breast cancer patients at pre-treatment using second derivative texture of CT images and machine learning. Transl. Oncol. 2021, 14, 101183. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, Y.; Mao, Y.; Wang, H.; Lv, M.; Su, X.; Wang, Y.; Li, Z.; Zhang, Z.; Bian, T.; et al. Development of MRI-Based Deep Learning Signature for Prediction of Axillary Response After NAC in Breast Cancer. Acad. Radiol. 2024, 31, 800–811. [Google Scholar] [CrossRef]
- Yongfeng, P.; Chuner, J.; Lei, W.; Fengqin, Y.; Zhimin, Y.; Zhenfu, F.; Haitao, J.; Yangming, J.; Fangzheng, W. The Usefulness of Pretreatment MR-Based Radiomics on Early Response of Neoadjuvant Chemotherapy in Patients with Locally Advanced Nasopharyngeal Carcinoma. Oncol. Res. 2021, 28, 605–613. [Google Scholar] [CrossRef]
- Yu, F.H.; Miao, S.M.; Li, C.Y.; Hang, J.; Deng, J.; Ye, X.H.; Liu, Y. Pretreatment ultrasound-based deep learning radiomics model for the early prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Eur. Radiol. 2023, 33, 5634–5644. [Google Scholar] [CrossRef]
- Oda, S.; Kuno, H.; Hiyama, T.; Sakashita, S.; Sasaki, T.; Kobayashi, T. Computed tomography-based radiomic analysis for predicting pathological response and prognosis after neoadjuvant chemotherapy in patients with locally advanced esophageal cancer. Abdom. Radiol. 2023, 48, 2503–2513. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Thibault, G.; Angulo, J.; Meyer, F. Advanced statistical matrices for texture characterization: Application to cell classification. IEEE Trans. Biomed. Eng. 2013, 61, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Moslemi, A. A tutorial-based survey on feature selection: Recent advancements on feature selection. Eng. Appl. Artif. Intell. 2023, 126, 107136. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Kira, K.; Rendell, L.A. A practical approach to feature selection. In Machine Learning Proceedings 1992; Morgan Kaufmann: Burlington, MA, USA, 1992; pp. 249–256. [Google Scholar]
- Afshar, M.; Usefi, H. High-dimensional feature selection for genomic datasets. Knowl. Based Syst. 2020, 206, 106370. [Google Scholar] [CrossRef]
- Ebrahimpour, M.K.; Zare, M.; Eftekharia, M.; Aghamollaei, G.H. Occam’s razor in dimension reduction: Using reduced row Echelon form for finding linear independent features in high dimensional microarray datasets. Eng. Appl. Artif. Intell. 2017, 62, 214–221. [Google Scholar] [CrossRef]
- Moslemi, A.; Ahmadian, A. Subspace learning for feature selection via rank revealing QR factorization: Fast feature selection. Expert Syst. Appl. 2024, 256, 124919. [Google Scholar] [CrossRef]
- Wang, S.; Pedrycz, W.; Zhu, Q.; Zhu, W. Subspace learning for unsupervised feature selection via matrix factorization. Pattern Recognit. 2015, 48, 10–19. [Google Scholar] [CrossRef]
- Fernández, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for learning from imbalanced data: Progress and challenges, marking the 15-year anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Bailey, C.; Siow, B.; Panagiotaki, E.; Hipwell, J.H.; Mertzanidou, T.; Owen, J.; Gazinska, P.; Pinder, S.E.; Alexander, D.C.; Hawkes, D.J. Microstructural models for diffusion MRI in breast cancer and surrounding stroma: An ex vivo study. NMR Biomed. 2017, 30, e3679. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, L.; Peng, W.; Wang, H.; Zhang, Y. Comparison of stretched-exponential and monoexponential model diffusion-weighted imaging in prostate cancer and normal tissues. J. Magn. Reson. Imaging 2015, 42, 1078–1085. [Google Scholar] [CrossRef]
- Bedair, R.; Priest, A.N.; Patterson, A.J.; McLean, M.A.; Graves, M.J.; Manavaki, R.; Gill, A.B.; Abeyakoon, O.; Griffiths, J.R.; Gilbert, F.J. Assessment of early treatment response to neoadjuvant chemotherapy in breast cancer using non-mono-exponential diffusion models: A feasibility study comparing the baseline and mid-treatment MRI examinations. Eur. Radiol. 2017, 27, 2726–2736. [Google Scholar] [CrossRef] [PubMed]
- Moghadas-Dastjerdi, H.; Ahmadzadeh, M.; Samani, A. Towards computer based lung disease diagnosis using accurate lung air segmentation of CT images in exhalation and inhalation phases. Expert Syst. Appl. 2017, 71, 396–403. [Google Scholar] [CrossRef]
- Saberi-Movahed, F.; Eftekhari, M.; Mohtashami, M. Supervised feature selection by constituting a basis for the original space of features and matrix factorization. Int. J. Mach. Learn. Cybern. 2020, 11, 1405–1421. [Google Scholar] [CrossRef]
- Blagus, R.; Lusa, L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinform. 2013, 14, 106. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Moghadas-Dastjerdi, H.; Sha-E-Tallat, H.R.; Sannachi, L.; Sadeghi-Naini, A.; Czarnota, G.J. A priori prediction of tumour response to neoadjuvant chemotherapy in breast cancer patients using quantitative CT and machine learning. Sci. Rep. 2020, 10, 10936. [Google Scholar] [CrossRef] [PubMed]
- Teruel, J.R.; Heldahl, M.G.; Goa, P.E.; Pickles, M.; Lundgren, S.; Bathen, T.F.; Gibbs, P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014, 27, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, J.; Wang, Y.; Xu, X.; Zhang, Y.; Zhang, Y.; Liu, G.; Cheng, J. Textural features of 18 F-FDG PET after two cycles of neoadjuvant chemotherapy can predict pCR in patients with locally advanced breast cancer. Ann. Nucl. Med. 2017, 31, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Gómez, O.V.; Herraiz, J.L.; Udías, J.M.; Haug, A.; Papp, L.; Cioni, D.; Neri, E. Analysis of cross-combinations of feature selection and machine-learning classification methods based on [18F] F-FDG PET/CT radiomic features for metabolic response prediction of metastatic breast cancer lesions. Cancers 2022, 14, 2922. [Google Scholar] [CrossRef]
- Sudjai, N.; Siriwanarangsun, P.; Lektrakul, N.; Saiviroonporn, P.; Maungsomboon, S.; Phimolsarnti, R.; Asavamongkolkul, A.; Chandhanayingyong, C. Robustness of Radiomic Features: Two-Dimensional versus Three-Dimensional MRI-Based Feature Reproducibility in Lipomatous Soft-Tissue Tumours. Diagnostics 2023, 13, 258. [Google Scholar] [CrossRef]
- Moslemi, A.; Osapoetra, L.O.; Dasgupta, A.; Alberico, D.; Trudeau, M.; Gandhi, S.; Eisen, A.; Wright, F.; Look-Hong, N.; Curpen, B.; et al. Apriori Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients using CT Imaging and Deep Learning: Transformer versus Transfer Learning. Front. Oncol. 2024, 14, 1359148. [Google Scholar] [CrossRef]
- Dasgupta, A.; Bhardwaj, D.; DiCenzo, D.; Fatima, K.; Osapoetra, L.O.; Quiaoit, K.; Saifuddin, M.; Brade, S.; Trudeau, M.; Gandhi, S.; et al. Radiomics in predicting recurrence for patients with locally advanced breast cancer using quantitative ultrasound. Oncotarget 2021, 12, 2437. [Google Scholar] [CrossRef]
- Bhardwaj, D.; Dasgupta, A.; DiCenzo, D.; Brade, S.; Fatima, K.; Quiaoit, K.; Trudeau, M.; Gandhi, S.; Eisen, A.; Wright, F.; et al. Early changes in quantitative ultrasound imaging parameters during neoadjuvant chemotherapy to predict recurrence in patients with locally advanced breast cancer. Cancers 2022, 14, 1247. [Google Scholar] [CrossRef]
- Pinsky, M.A. Introduction to Fourier Analysis and Wavelets; American Mathematical Society: Evanston, IL, USA, 2002; Volume 102. [Google Scholar]
- Brunton, S.L.; Kutz, J.N. Data-Driven Science and Engineering: Machine Learning, Dynamical Systems and Control; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar]
- Nason, G.P.; Silverman, B.W. The stationary wavelet transform and some statistical applications. In Wavelets and Statistics; Springer: New York, NY, USA, 1995; pp. 281–299. [Google Scholar]
- Wolf, L.; Shashua, A. Feature selection for unsupervised and supervised inference: The emergence of sparsity in a weight based approach. J. Mach. Learn. Res. 2005, 6, 1855–1887. [Google Scholar]
- Zhao, Z.; Liu, H. Semi-supervised feature selection via spectral analysis. In Proceedings of the SIAM International Conference on Data Mining (SDM), Minneapolis, MN, USA, 26–28 April 2007; pp. 641–646. [Google Scholar]
- Cai, D.; Zhang, C.; He, X. Unsupervised feature selection for multi-cluster data. In Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 24–28 July 2010; pp. 333–342. [Google Scholar]
- He, X.; Cai, D.; Niyogi, P. Laplacian score for feature selection. In Proceedings of the 19th International Conference on Neural Information Processing Systems (NIPS’05), Vancouver, BC, Canada, 5–8 December 2005; pp. 507–514. [Google Scholar]
- Baiti, R. Using mutual information for selecting features in supervised neural net learning. IEEE Trans. Neural Netw. Learn. Syst. 1994, 5, 537–550. [Google Scholar] [CrossRef]
- Lazar, C.; Taminau, J.; Meganck, S.; Steenhoff, D.; Coletta, A.; Molter, C.; de Schaetzen, V.; Duque, R.; Bersini, H.; Nowé, A. A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Trans Comput. Biol. Bioinform. 2012, 9, 1106–1119. [Google Scholar] [CrossRef]
- Forman, G. An extensive empirical study of feature selection metrics for text classification. JMLR 2003, 3, 1289–1305. [Google Scholar]
- Kwak, N.; Choi, C.-H. Input feature selection for classification problems. IEEE Trans. Neural Netw. Learn. Syst. 2002, 13, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Eldén, L.; Berry, M.W. Matrix methods in data mining and paern recognition. SIAM Rev. 2008, 50, 154. [Google Scholar]
- Halko, N.; Per-Gunnar, M.; Tropp, J.A. Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev. 2011, 53, 217–288. [Google Scholar] [CrossRef]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. JMLR 2003, 3, 1157–1182. [Google Scholar]
- Langley, P. Selection of relevant features in machine learning. In Proceedings of the AAAI Fall Symposium on Relevance, New Orleans, LA, USA, 4–6 November 1994; pp. 245–271. [Google Scholar]
- Blum, A.L.; Langley, P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997, 97, 245–271. [Google Scholar] [CrossRef]
- John, G.H.; Kohavi, R.; Pfleger, K. Irrelevant features and the subset selection problem. In Proceedings of the 11th International Conference on Machine Learning, New Brunswick, NJ, USA, 10–13 July 1994; pp. 121–129. [Google Scholar]
- Pudil, P.; Novovičová, J.; Kiler, J. Floating search methods in feature selection. Pattern Recognit. Lett. 1994, 15, 1119–1125. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, X.; Shi, Q.; Xu, X.-Q.; Hon-Chiu, E.; Leung, L.N.; Harris, J.D.; Iglehart, A.M.; Liu, J.S.; Wong, W.H. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinform. 2006, 7, 197. [Google Scholar] [CrossRef]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar] [CrossRef]
- Anaraki, J.R.; Usefi, H. A feature selection based on perturbation theory. Expert Syst. Appl. 2019, 127, 1–8. [Google Scholar] [CrossRef]
| Radiomics Features Type | Radiomics Features | |
|---|---|---|
| First Order Features: | Energy Total Energy Entropy Minimum 10th percentile 90th percentile Maximum Mean Median | Interquartile Range Range Mean Absolute Deviation (MAD) Robust Mean Absolute Deviation Root Mean Squared (RMS) Skewness Kurtosis Variance Uniformity |
| Shape Features: | Elongation Flatness Least Axis Length Major Axis Length Maximum 2D Diameter Column Maximum 2D Diameter Row Maximum 2D Diameter Slice | Maximum 3D Diameter Mesh Volume Minor Axis Length Sphericity Surface Area Surface Volume Ratio Voxel Volume |
| GLCM: | Autocorrelation Joint Average Cluster Prominence Cluster Shade Cluster Tendency Contrast Correlation Difference Average Difference Entropy Difference Variance Dissimilarity Joint Energy | Joint Entropy Homogeneity 1 Homogeneity 2 Informational Measure of Correlation (IMC) 1 Informational Measure of Correlation (IMC) 2 Inverse Difference Moment (IDM) Maximal Correlation Coefficient (MCC) Inverse Difference Moment Normalized (IDMN) Inverse Difference (ID) Inverse Difference Normalized (IDN) Inverse Variance Maximum Probability Sum Average Sum Variance Sum Entropy Sum of Squares |
| GLRLM: | Short Run Emphasis (SRE) Long Run Emphasis (LRE) Gray Level Non-Uniformity (GLN) Gray Level Non-Uniformity Normalized (GLNN) Run Length Non-Uniformity (RLN) Run Length Non-Uniformity Normalized (RLNN) Long Run Low Gray Level Emphasis (LRLGLE) Long Run High Gray Level Emphasis (LRHGLE) | Run Percentage (RP) Gray Level Variance (GLV) Run Variance (RV) Run Entropy (RE) Low Gray Level Run Emphasis (LGLRE) High Gray Level Run Emphasis (HGLRE) Short Run Low Gray Level Emphasis (SRLGLE) Short Run High Gray Level Emphasis (SRHGLE) Long Run Low Gray Level Emphasis (LRLGLE) Long Run High Gray Level Emphasis (LRHGLE) |
| GLSZM: | Small Area Emphasis (SAE) Large Area Emphasis (LAE) Gray Level Non-Uniformity (GLN) Gray Level Non-Uniformity Normalized (GLNN) Size-Zone Non-Uniformity (SZN) Size-Zone Non-Uniformity Normalized (SZNN) Zone Percentage (ZP) Gray Level Variance (GLV) | Zone Variance (ZV) Zone Entropy (ZE) Low Gray Level Zone Emphasis (LGLZE) High Gray Level Zone Emphasis (HGLZE) Small Area Low Gray Level Emphasis (SALGLE) Small Area High Gray Level Emphasis (SAHGLE) Large Area Low Gray Level Emphasis (LALGLE) Large Area High Gray Level Emphasis (LAHGLE) |
| GLDM: | Small Dependence Emphasis (SDE) Large Dependence Emphasis (LDE) Gray Level Non-Uniformity (GLN) Gray Level Non-Uniformity Normalized (GLNN) Dependence Non-Uniformity (DN) Dependence Non-Uniformity Normalized (DNN) Gray Level Variance (GLV) | Dependence Variance (DV) Dependence Entropy (DE) Dependence Percentage Low Gray Level Emphasis (LGLE) High Gray Level Emphasis (HGLE) Small Dependence Low Gray Level Emphasis (SDLGLE) Small Dependence High Gray Level Emphasis (SDHGLE) |
| NGLDM: | Coarseness Contrast Busyness Complexity Strength |
| Characteristics | Responders Mean (Std) | Non-Responders Mean (Std) |
|---|---|---|
| Age | 52 (11) | 54 (10) |
| Initial Tumor Size | 5.2 (2.5) cm | 5.6 (2.7) cm |
| Histology | Percentage (Count) | |
| IDC | 58 (70) | 23 (65) |
| ILC | 1 (1) | 4 (11) |
| IMC | 3 (3) | 2 (5) |
| Molecular Features | Percentage (Count) | |
| ER+ | 42 (51) | 29 (82) |
| PR+ | 37 (45) | 24 (68) |
| † HER2+ | 28 (34) | 9 (26) |
| ER-/PR-/HER2- | 22 (27) | 4 (11) |
| ER+/PR+/HER2+ | 15 (18) | 6 (17) |
| ER+/PR+/HER2- | 22 (27) | 20 (57) |
| ER-/PR-/HER2+ | 15 (18) | 4 (11) |
| Residual Tumor Size | 1.4 (2.4) cm | 6 (5.5) cm |
| Response | Percentage (Count) | |
| Responding Patients | 70 (82) | - |
| Non-responding Patients | - | 30 (35) |
| Classifier | FST | # Features | Spec | Sens | Acc | B-Acc | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Max | Mean | Max | Mean | Max | Mean | Max | ||||
| (%) | (%) | (%) | (%) | ||||||||
| Model 1: | KNN | mRMR | Top-5 | 77 | 86 | 51 | 60 | 72 | 77 | 62 | 76 |
| Model 2: | KNN | mRMR | Top-10 | 78 | 88 | 52 | 58 | 74 | 78 | 64 | 73 |
| Model 3: | KNN | mRMR | Top-5 | 80 | 90 | 56 | 63 | 77 | 79 | 68 | 77 |
| Classifier | FST | # Features | Spec (%) | Sens (%) | Acc (%) | B-Acc (%) | |
|---|---|---|---|---|---|---|---|
| Model 1: | KNN | mRMR | Top-5 | 69 | 58 | 68 | 66 |
| Model 2: | KNN | mRMR | Top-10 | 71 | 61 | 71 | 70 |
| Model 3: | KNN | mRMR | Top-5 | 76 | 62 | 75 | 72 |
| Technique/Metric | Specificity | Sensitivity | Accuracy | B-Accuracy |
|---|---|---|---|---|
| mRMR | 76 | 62 | 75 | 72 |
| Relief | 78 | 36 | 64 | 57 |
| PFS | 81 | 35 | 67 | 56 |
| Rref | 86 | 30 | 69 | 57 |
| QR | 82 | 39 | 70 | 60 |
| NMFFS | 71 | 35 | 60 | 54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moslemi, A.; Osapoetra, L.O.; Dasgupta, A.; Halstead, S.; Alberico, D.; Trudeau, M.; Gandhi, S.; Eisen, A.; Wright, F.; Look-Hong, N.; et al. Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients at Pre-Treatment Using CT Textural Features and Machine Learning: Comparison of Feature Selection Methods. Tomography 2025, 11, 33. https://doi.org/10.3390/tomography11030033
Moslemi A, Osapoetra LO, Dasgupta A, Halstead S, Alberico D, Trudeau M, Gandhi S, Eisen A, Wright F, Look-Hong N, et al. Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients at Pre-Treatment Using CT Textural Features and Machine Learning: Comparison of Feature Selection Methods. Tomography. 2025; 11(3):33. https://doi.org/10.3390/tomography11030033
Chicago/Turabian StyleMoslemi, Amir, Laurentius Oscar Osapoetra, Archya Dasgupta, Schontal Halstead, David Alberico, Maureen Trudeau, Sonal Gandhi, Andrea Eisen, Frances Wright, Nicole Look-Hong, and et al. 2025. "Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients at Pre-Treatment Using CT Textural Features and Machine Learning: Comparison of Feature Selection Methods" Tomography 11, no. 3: 33. https://doi.org/10.3390/tomography11030033
APA StyleMoslemi, A., Osapoetra, L. O., Dasgupta, A., Halstead, S., Alberico, D., Trudeau, M., Gandhi, S., Eisen, A., Wright, F., Look-Hong, N., Curpen, B., Kolios, M., & Czarnota, G. J. (2025). Prediction of Chemotherapy Response in Locally Advanced Breast Cancer Patients at Pre-Treatment Using CT Textural Features and Machine Learning: Comparison of Feature Selection Methods. Tomography, 11(3), 33. https://doi.org/10.3390/tomography11030033









