A Comparative Study of Phase I and II Hepatic Microsomal Biotransformation of Phenol in Three Species of Salmonidae: Hydroquinone, Catechol, and Phenylglucuronide Formation
Abstract
1. Introduction
2. Methods
2.1. Chemicals
2.2. Standard and Sample Preparation and Handling
2.3. Animals
2.4. Microsomal Characterization
2.5. Phase I Microsomal Incubations
2.6. Phase II Microsomal Incubations
2.7. Metabolite Identification and Quantification
2.8. Data Analysis
3. Results
3.1. Microsome Characterization and Assay Optimization
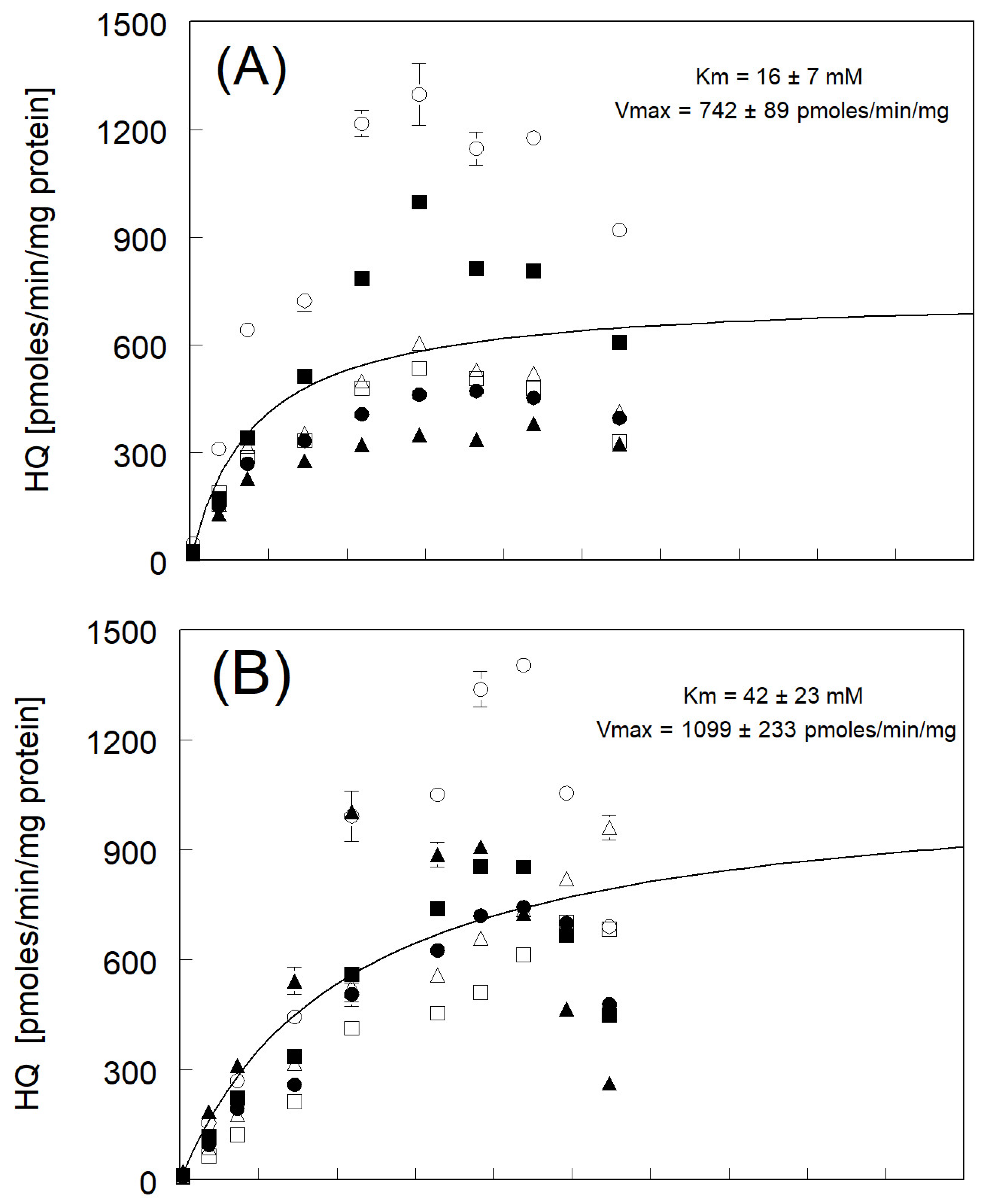
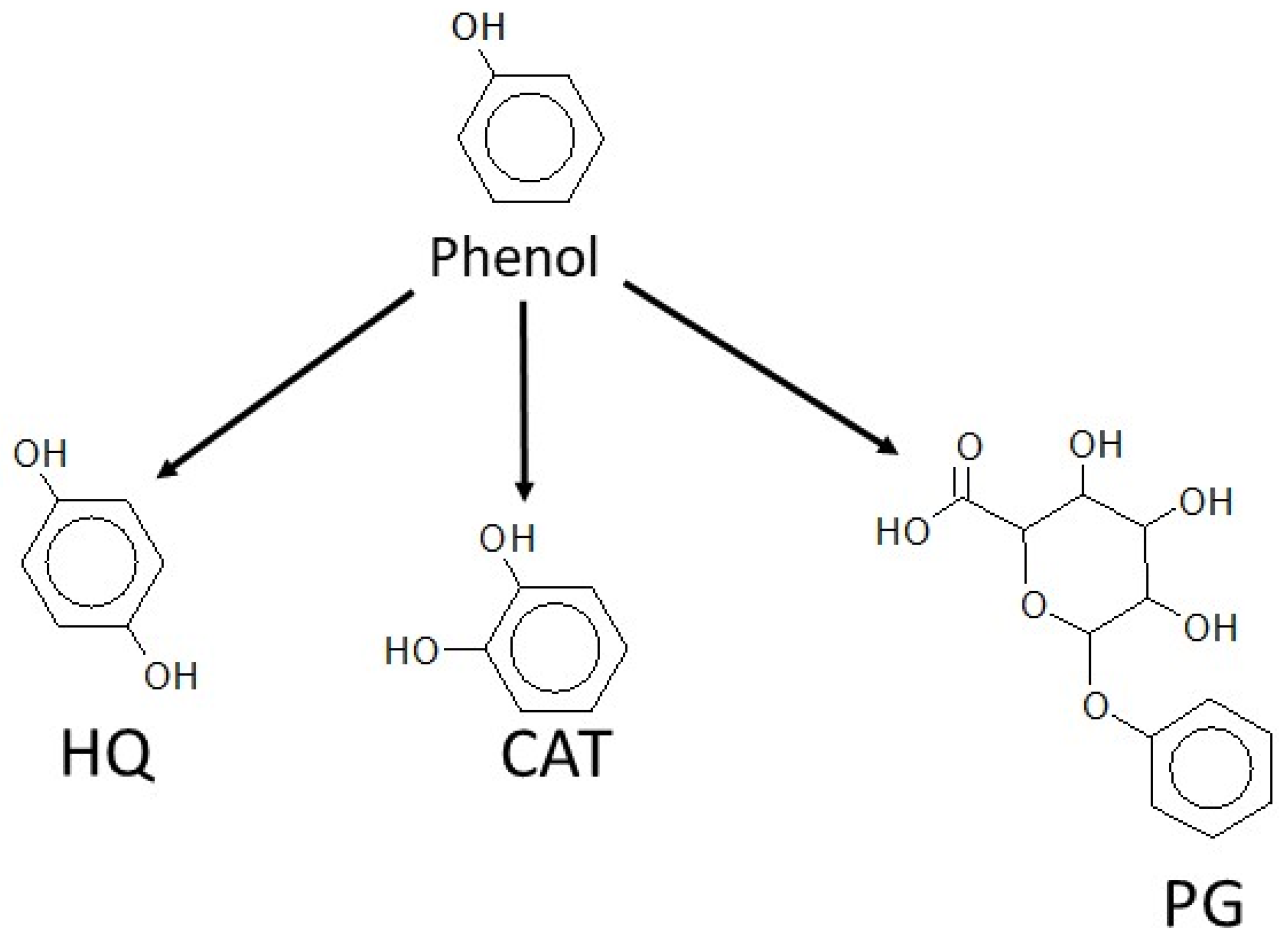
3.2. Hydroquinone Formation
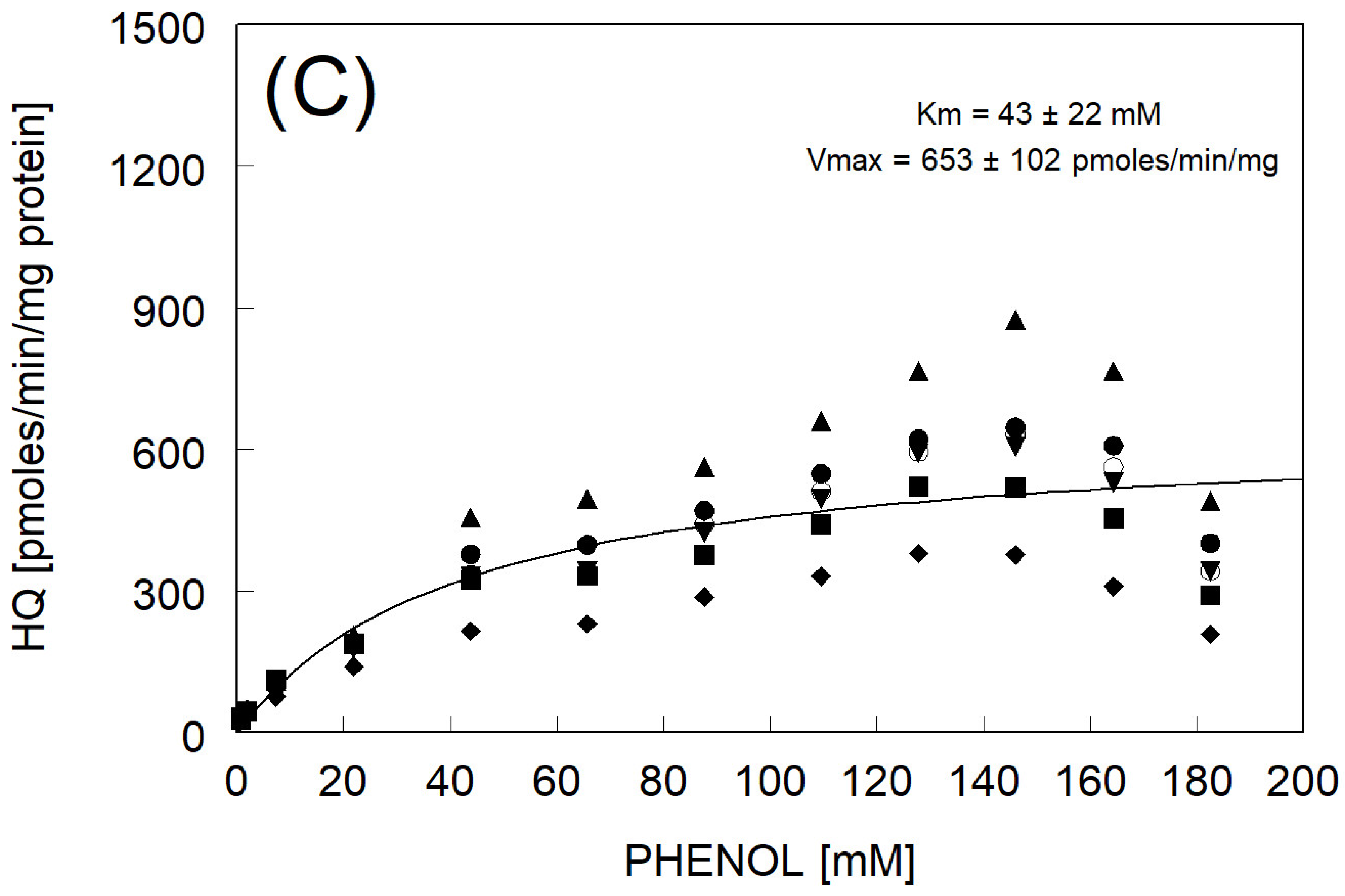
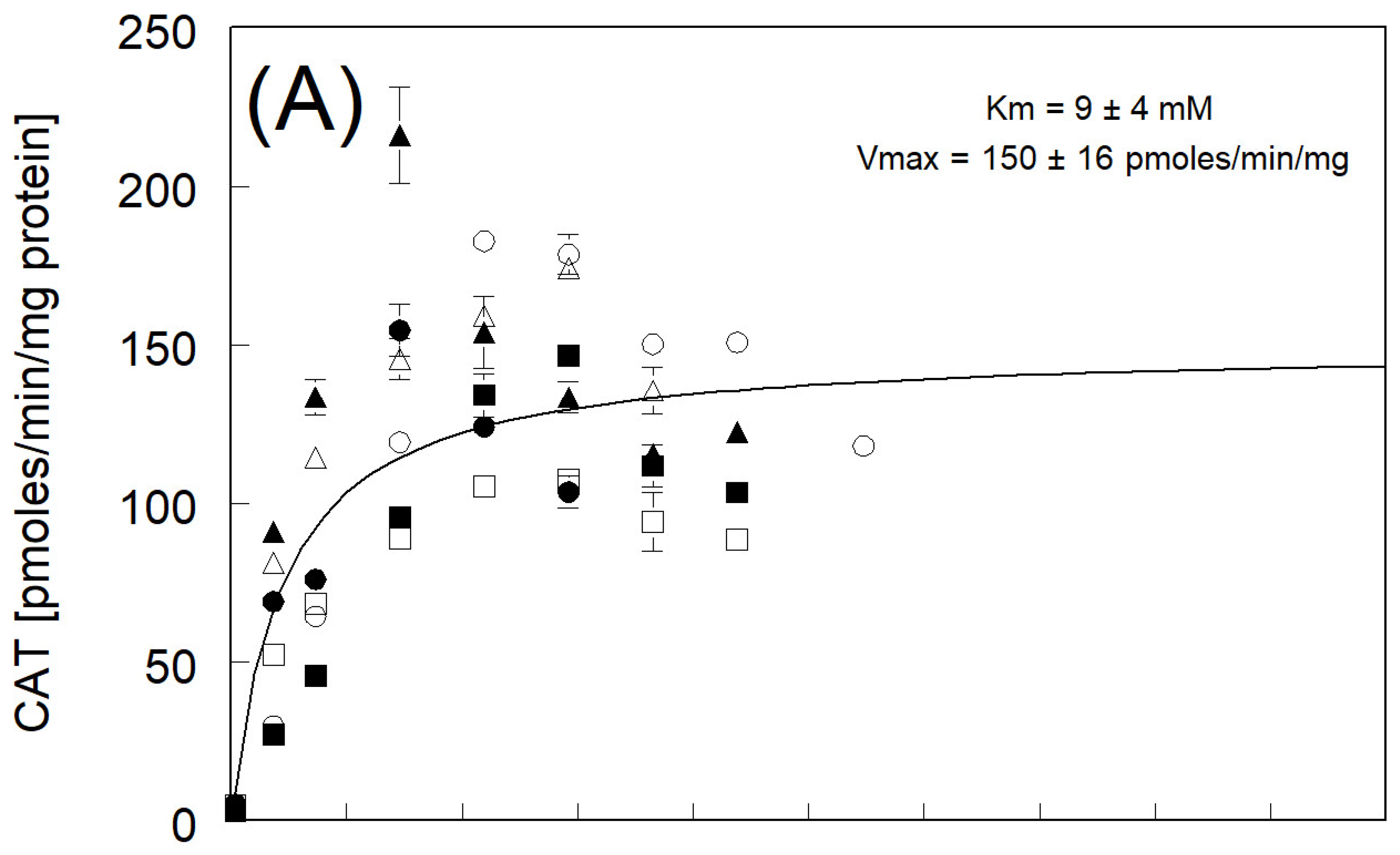
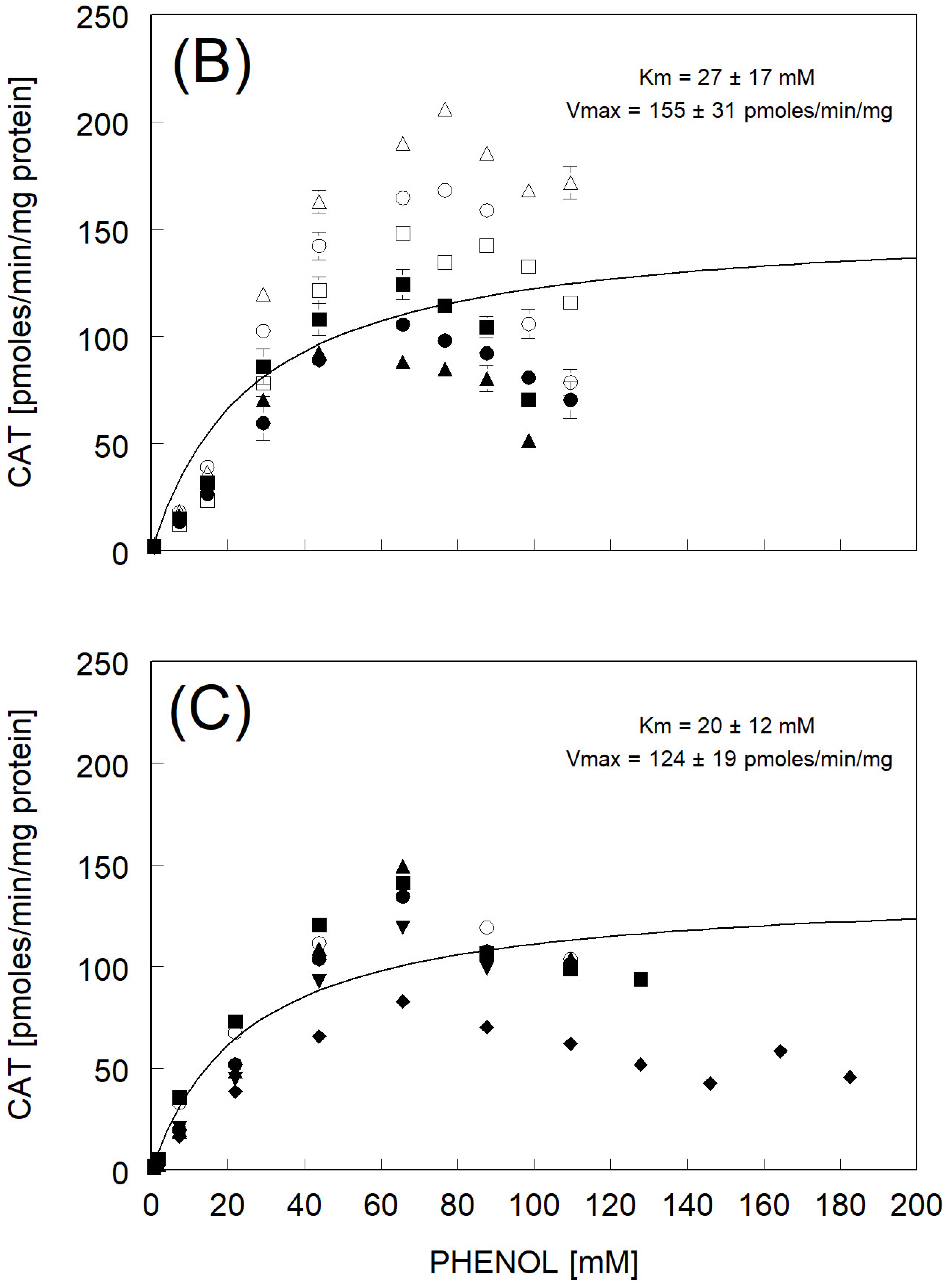
3.3. Catechol Formation
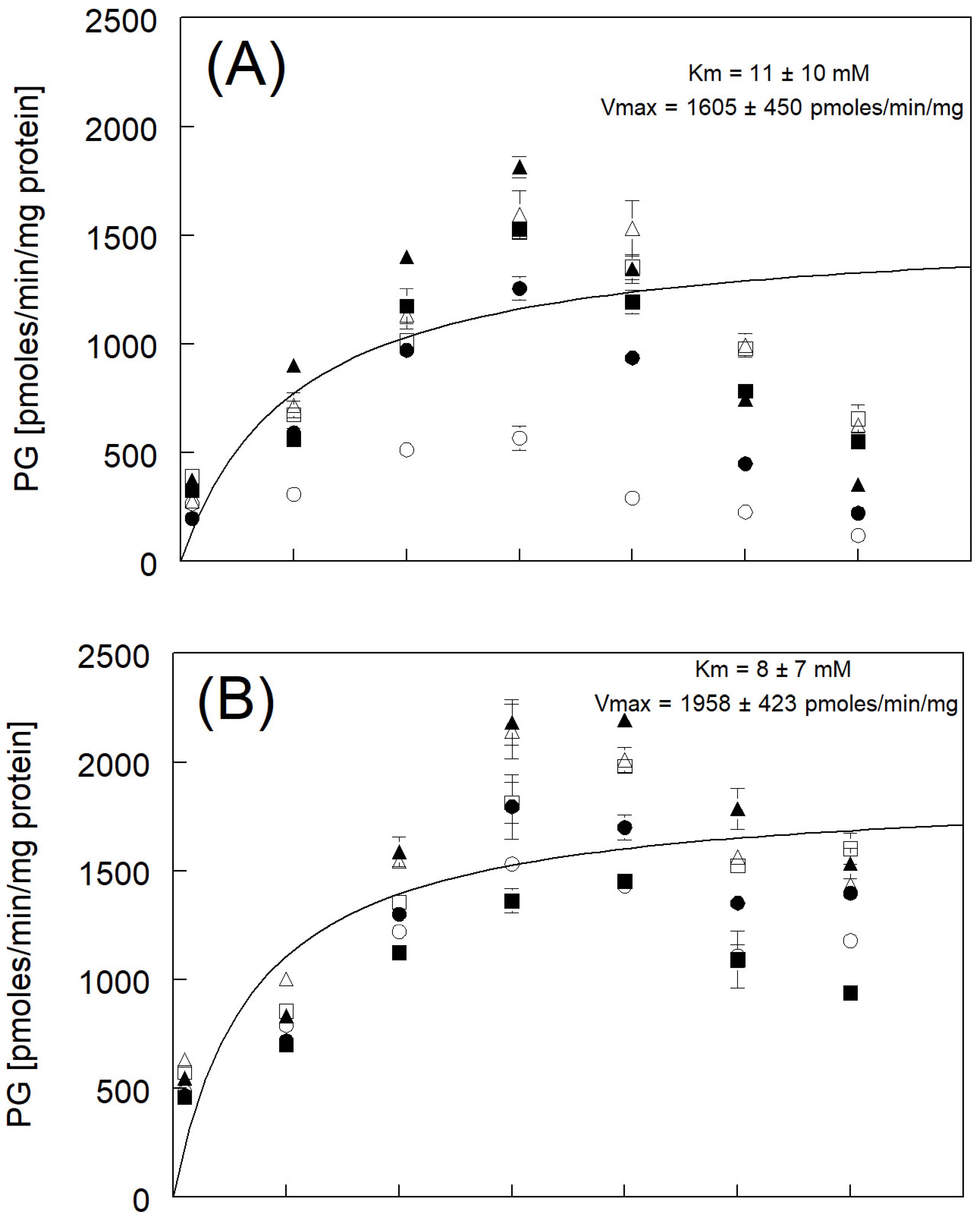
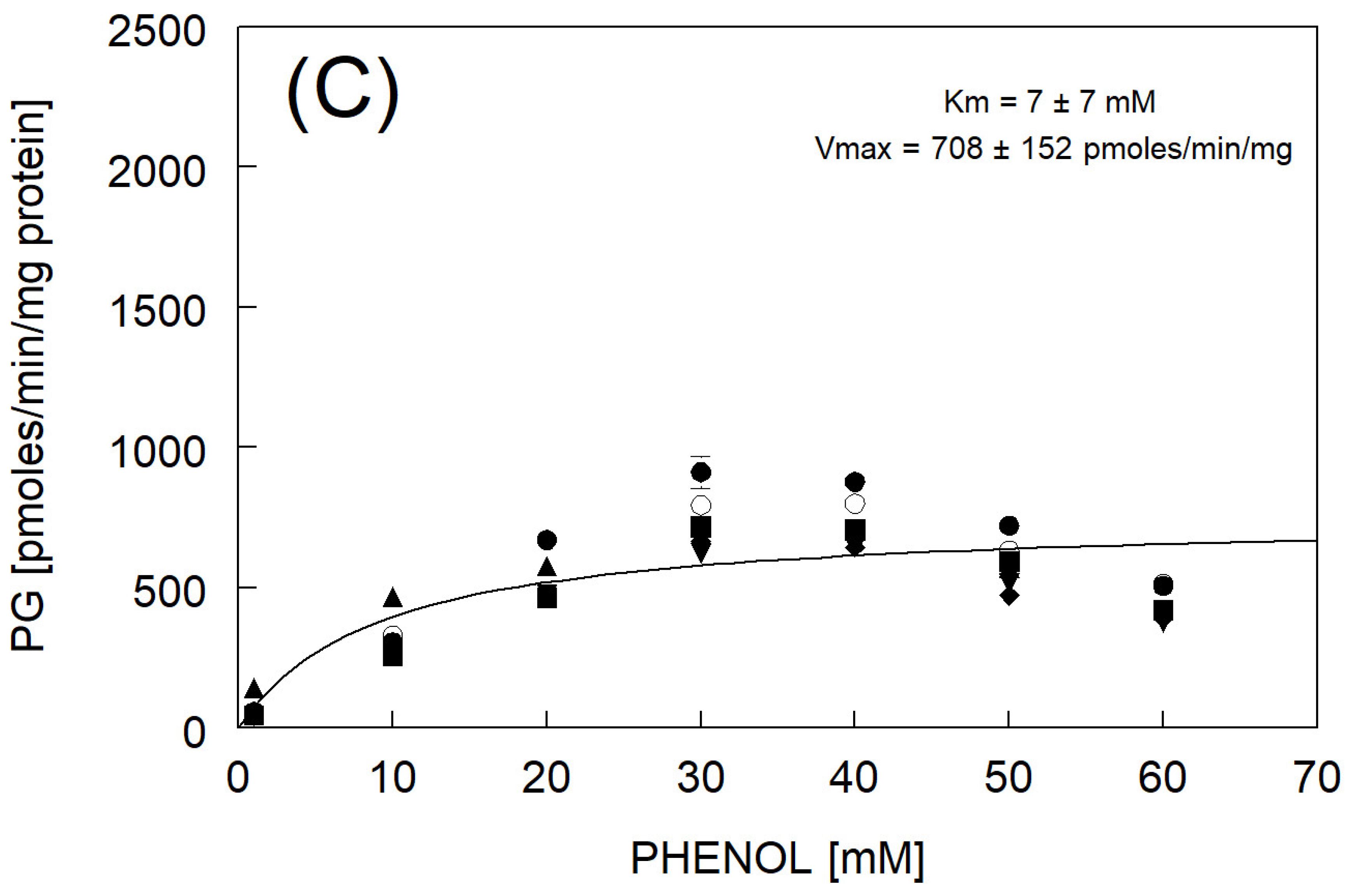
3.4. Phenylglucuronide Formation
4. Discussion
4.1. Oxidative Metabolism (Phase I)
4.2. Deactivation (Phase II)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sijm, D.; de Bruijn, J.; de Voogt, P.; de Wolf, W. Biotransformation in environmental risk assessment. In Proceedings of the SETAC-Europe Workshop, Noordwijkerhout, The Netherlands, 28 April–1 May 1996; SETAC-Europe: Brussels, Belgium, 1997; pp. 1–130. [Google Scholar]
- Nichols, J.W.; McKim, J.M.; Andersen, M.E.; Gargas, M.L.; Clewell, H.J., III; Erickson, R.J. A physiologically based toxicokinetic model for the uptake and disposition of waterborne organic chemicals in fish. Toxicol. Appl. Pharmacol. 1990, 106, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.W.; McKim, J.M.; Lien, G.J.; Hoffman, A.D.; Bertelsen, S.L. Physiologically based toxicokinetic modeling of three chlorinated ethanes in rainbow trout (Oncorhynchus mykiss). Toxicol. Appl. Pharmacol. 1991, 110, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.W.; McKim, J.M.; Lien, G.J.; Hoffman, A.D.; Bertelsen, S.L. A physiologically based toxicokinetic model for three waterborne chloroethanes in the channel catfish (Ictaturus punctatus). Aquat. Toxicol. 1993, 27, 83–112. [Google Scholar] [CrossRef]
- Lien, G.J.; Nichols, J.W.; McKim, J.M.; Gallinat, C.A. Modeling the accumulation of three waterborne chlorinated ethanes in fathead minnows (Pimephales promelas): A physiologically based approach. Environ. Toxicol. Chem. 1994, 13, 1195–1205. [Google Scholar] [CrossRef]
- Lien, G.J.; McKim, J.M.; Hoffman, A.D.; Jenson, C.T. A physiologically based toxicokinetic model for lake trout (Salvelinus namaycush). Aquat. Toxicol. 2001, 51, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Stadnicka, J.; Schirmer, K.; Ashauer, R. Predicting concentrations of organic chemicals in fish by using toxicokinetic models. Environ. Sci. Technol. 2012, 46, 3273–3280. [Google Scholar] [CrossRef]
- Brinkman, M.; Schlechtriem, C.; Reininghaus, M.; Eichbaum, K.; Buchinger, S.; Reifferscheid, G.; Hollert, H.; Preuss, T.G. Cross-species extrapolation of uptake and disposition of neutral organic chemicals in fish using a multispecies physiologically-based toxicokinetic model framework. Environ. Sci. Technol. 2016, 50, 1914–1923. [Google Scholar] [CrossRef]
- Golosovskaia, E.; Orn, S.; Ahrens, L.; Chelcea, I.; Andersson, P.L. Studying mixture effects on uptake and tissue distribution of PFAS in zebrafish (Danio rerio) using physiologically based kinetic (PBK) modelling. Sci. Total Environ. 2024, 912, 168738. [Google Scholar] [CrossRef]
- Nichols, J.W.; Schultz, I.R.; Fitzsimmons, P.N.I. A review of methods, and strategies for incorporating intrinsic clearance estimates into chemical kinetic models. Aquat. Toxicol. 2006, 78, 74–90. [Google Scholar] [CrossRef]
- Nichols, J.W.; Fitzsimmons, P.N.; Burkhard, L.P. In vitro-in vivo extrapolation of quantitative hepatic biotransformation data for fish. II. Modeled effects on chemical bioaccumulation. Environ. Toxicol. Chem. 2007, 26, 1304–1319. [Google Scholar] [CrossRef]
- Nichols, J.W.; Fitzsimmons, P.N.; Hoffman, A.D.; Wong, K. In vitro-in vivo extrapolation of hepatic biotransformation data for fish. III. An in-depth case study with pyrene. Environ. Toxicol. Chem. 2023, 42, 1501–1515. [Google Scholar] [CrossRef] [PubMed]
- Pery, A.R.R.; Devillers, J.; Brochot, C.; Mombelli, E.; Palluel, O.; Piccini, B.; Brion, F.; Beaudouin, R. A physiologically based toxicokinetic model for the zebrafish Danio rerio. Environ. Sci. Technol. 2014, 48, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Grech, A.; Tebby, C.; Brochot, C.; Bois, F.Y.; Bado-Nilles, A.; Dorne, J.L.; Quignot, N.; Beaudouin, R. Generic physiologically-based toxicokinetic modelling for fish: Integration of environmental factors and species variability. Sci. Total Environ. 2019, 651, 516–531. [Google Scholar] [CrossRef] [PubMed]
- Chelcea, I.; Orn, S.; Hamers, T.; Koekkoek, J.; Legradi, J.; Vogs, C.; Andersson, P.L. Physiologically based toxicokinetic modeling of bisphenols in zebrafish (Danio rerio) accounting for variations in metabolic rates, brain distribution, and liver accumulation. Environ. Sci. Technol. 2022, 56, 10216–10228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Yao, L.; Zhang, Y.Y.; Chen, C.E.; Zhao, J.L.; Ying, G.G. Enhanced prediction of internal concentrations of phenolic endocrine disrupting chemicals and their metabolites in fish by a physiologically based toxicokinetic incorporating metabolism (PBTK-MT) model. Environ. Pollut. 2022, 314, 120290. [Google Scholar] [CrossRef] [PubMed]
- Mit, C.; Bado-Nilles, A.; Daniele, G.; Giroud, B.; Vulliet, E.; Beaudouin, R. The toxicokinetics of bisphenol A and its metabolites in fish elucidated by a PBTK model. Aquat. Toxicol. 2022, 247, 106174. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimmons, P.N.; Lien, G.J.; Nichols, J.W. A compilation of in vitro rate and affinity values for xenobiotic biotransformation in fish, measured under physiological conditions. Comp. Biochem. Physiol. Part C 2007, 145, 485–506. [Google Scholar] [CrossRef] [PubMed]
- Sikka, H.C.; Rutkowski, J.P.; Kandaswami, C. Comparative metabolism of benzo[a]pyrene by liver microsomes from brown bullhead and carp. Aquat. Toxicol. 1990, 16, 101–112. [Google Scholar] [CrossRef]
- Yuan, Z.-X.; Kumar, S.; Sikka, H.C. Comparative metabolism of benzo[a]pyrene by liver microsomes of channel catfish and brown bullhead. Environ. Toxicol. Chem. 1997, 16, 835–836. [Google Scholar] [CrossRef]
- Yuan, Z.-X.; Honey, S.A.; Kumar, S.; Sikka, H.C. Comparative metabolism of dibenzo[a,l]pyrene by liver microsomes from rainbow trout and rats. Aquat. Toxicol. 1999, 45, 1–8. [Google Scholar] [CrossRef]
- Clarke, D.J.; George, S.G.; Burchell, B. Glucuronidation in fish. Aquat. Toxicol. 1991, 20, 35–56. [Google Scholar] [CrossRef]
- Lech, J.J.; Stratham, C.N. Role of glucuronide formation in the selective toxicity of 3-fluoromethyl-4-nitrophenol (TFM) for the sea lamprey and rainbow trout. Toxicol. Appl. Pharmacol. 1975, 31, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Koivusaari, U.; Forlin, L. Xenobiotic biotransformation in the rainbow trout liver and kidney during starvation. Comp. Biochem. Physiol. 1985, 82C, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Gregus, Z.; Watkins, J.B.; Thompson, T.N.; Harvey, M.J.; Rozman, K.; Klassen, C.D. Hepatic Phase I and Phase II biotransformations in quail and trout: Comparison to other species commonly used in toxicity testing. Toxicol. Appl. Pharmacol. 1983, 67, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Sivarajah, K.; Franklin, S.; Williams, W.P. The effects of polychlorinated biphenyls on plasma steroid levels and hepatic microsomal enzymes in fish. J. Fish Biol. 1978, 13, 401–409. [Google Scholar] [CrossRef]
- Lech, J.J. Isolation and identification of 3-fluoromethyl-4-nitrophenol glucuronide from bile of rainbow trout exposed to 3-fluoromethyl-4-nitrophenol. Toxicol. Appl. Pharmacol. 1973, 24, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Lech, J.J. Glucuronide formation in rainbow trout—Effect of salicylamide on the acute toxicity, conjugation, and excretion of trifluoromethyl-4-nitrophenol. J. Biochem. Pharmacol. 1974, 23, 2403–2410. [Google Scholar] [CrossRef] [PubMed]
- Hanninen, O.; Lindstrom-Seppa, P.; Koivusaari, U.; Vaisanen, M.; Julkunen, A.; Juvonen, R. Glucuronidation and glucosidation reactions in aquatic species from boreal regions. Biochem. Soc. Trans. 1984, 12, 13–17. [Google Scholar] [CrossRef]
- Pathiratne, A.; George, S. Comparison of xenobiotic metabolizing enzymes of tilapia with those of other fish species and interspecies relationships between gene families. Mar. Environ. Res. 1996, 42, 293–296. [Google Scholar] [CrossRef]
- Forlin, L.; Lemaire, P.; Livingstone, D.R. Comparative studies of hepatic xenobiotic metabolizing and antioxidant enzymes in different fish species. Mar. Environ. Res. 1995, 39, 201–204. [Google Scholar] [CrossRef]
- Perdu-Durand, E.F.; Cravedi, J.P. Characterization of xenobiotic metabolizing enzymes in sturgeon (acipenser baeri). Comp. Biochem. Physiol. 1989, 93B, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Goksoyr, A.; Andersson, T.; Hansson, T.; Klungsoyr, J.; Zhang, Y.; Forlin, L. Species characteristics of the hepatic xenobiotic and steroid biotransformation systems of two teleost fish, atlantic cod (Gadus morhua) and rainbow trout (Salmo gairdneri). Toxicol. Appl. Pharmacol. 1987, 89, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom-Seppa, P.; Koivusaari, U.; Hanninen, O. Extrahepatic metabolism in north European freshwater fish. Comp. Biochem. Physiol. 1981, 69C, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Davi, M.L.; Gnudi, F. Phenolic compounds in surface water. Water Res. 1999, 33, 3213–3219. [Google Scholar] [CrossRef]
- Ramos, R.L.; Moreira, V.R.; Amaral, M.C.S. Phenolic compounds in water: Review of occurrence, risk, and retention by membrane technology. J. Environ. Manag. 2024, 351, 119772. [Google Scholar] [CrossRef] [PubMed]
- Phipps, G.L.; Holcombe, G.W.; Fiandt, J.T. Acute toxicity of phenol and substituted phenols to the fathead minnow. Bull. Environ. Contam. Toxicol. 1981, 26, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, S.P.; Henry, T.R.; Niemi, G.J.; Carlson, R.W.; Snarski, V.M. Use of respiratory-cardiovascular responses of rainbow trout (Salmo gairdneri) in identifying acute toxicity syndromes in fish: Part 3. Polar narcotics. Environ. Toxicol. Chem. 1989, 8, 247–261. [Google Scholar]
- Nagel, R.; Urich, K. Quinol sulphate, a new conjugate of phenol in goldfish. Xenobiotica 1983, 13, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Nagel, R. Species differences, influence of dose and application on biotransformation of phenol in fish. Xenobiotica 1983, 13, 101–106. [Google Scholar] [CrossRef]
- Layiwola, P.J.; Linnecar, D.C.F. The biotransformation of [14C]phenol in some fresh water fish. Xenobiotica 1981, 11, 167–171. [Google Scholar] [CrossRef]
- McKim, J.M., Jr.; McKim, J.M., Sr.; Naumann, S.; Hammermeister, D.E.; Hoffman, A.D.; Klaassen, C.D. In vivo microdialysis sampling of phenol and phenyl glucuronide in the blood of unanesthetized rainbow trout: Implications for toxicokinetic studies. Fundam. Appl. Toxicol. 1993, 20, 190–198. [Google Scholar] [CrossRef]
- McKim, J.M.; Kolanczyk, R.C.; Lien, G.J.; Hoffman, A.D. Dynamics of renal excretion of phenol and major metabolites in the rainbow trout (Oncorhynchus mykiss). Aquat. Toxicol. 1999, 45, 265–277. [Google Scholar] [CrossRef]
- Kolanczyk, R.; Schmieder, P. Rate and capacity of hepatic microsomal ring hydroxylation of phenol to hydroquinone and catechol in rainbow trout (Oncorhynchus mykiss). Toxicology 2002, 176, 77–90. [Google Scholar] [CrossRef]
- Kolanczyk, R.; Schmieder, P.; Bradbury, S.; Spizzo, T. Biotransformation of 4-methoxyphenol in rainbow trout (Oncorhynchus mykiss) hepatic microsomes. Aquat. Toxicol. 1999, 45, 47–61. [Google Scholar] [CrossRef]
- Forlin, L.; Andersson, T. Storage conditions of rainbow trout liver cytochrome P-450 and conjugating enzymes. Comp. Biochem. Physiol. 1985, 80B, 569–572. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Estabrook, R.; Peterson, W.J.; Baron, J.; Hildebrandt, A.G. The spectrophotometric measurement of turbid suspensions of cytochromes associated with drug metabolism. Methods Pharmacol. 1972, 2, 303–350. [Google Scholar]
- Pohl, R.J.; Fouts, J.R. A rapid method for assaying the metabolism of 7-ethoxyresorufin by microsomal subcellular fractions. Anal. Biochem. 1980, 107, 150–155. [Google Scholar] [CrossRef]
- McKim, J.M.; Nichols, J.W. Use of physiologically based toxicokinetic models in a mechanistic approach to aquatic toxicology. In Aquatic Toxicology: Molecular, Biochemical, and Cellular Perspectives; Malins, D.C., Ostrander, G.K., Eds.; Lewis: Boca Raton, FL, USA, 1994. [Google Scholar]
- Sahawata, T.; Neal, R.A. Biotransformation of phenol to hydroquinone and catechol by rat liver microsomes. Mol. Pharmacol. 1983, 23, 453–460. [Google Scholar]
- Schlosser, P.M.; Bond, J.A.; Medinsky, M.A. Benzene and phenol metabolism by mouse and rat liver microsomes. Carcinogenesis 1993, 14, 2477–2486. [Google Scholar] [CrossRef]
- Schlosser, M.J.; Shurina, R.D.; Kalf, G.F. Prostaglandin H synthase catalyzed oxidation of hydroquinone to sulfhydryl-binding and DNA-damaging metabolite. Chem. Res. Toxicol. 1990, 3, 333–339. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J. Molecular mechanisms of quinone cytotoxicity-review. Chem.-Biol. Interact. 1991, 80, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Holcombe, G.W.; Phipps, G.L.; Sulaiman, A.H.; Hoffman, A.D. Simultaneous multiple species testing: Acute toxicity of 13 chemicals to 12 diverse freshwater amphibian, fish, and invertebrate families. Arch. Environ. Contam. Toxicol. 1987, 16, 697–710. [Google Scholar] [CrossRef] [PubMed]
- DeGraeve, G.M.; Geiger, D.L.; Meyer, J.S.; Bergman, H.L. Acute and embryo-larval toxicity of phenolic compounds to aquatic biota. Arch. Environ. Contam. Toxicol. 1980, 9, 557–568. [Google Scholar] [CrossRef] [PubMed]
| Species | Sex | Fish Weight (g) | Liver Weight (g) | HSI (%) | Microsomal Protein (mg/g Liver) | P450 Protein (nmol/mg Microsomal Protein) | EROD (pmol/min/mg Microsomal Protein) | |
|---|---|---|---|---|---|---|---|---|
| fish #1 | Rainbow | male | 913 | 10.3 | 1.1 | 16.5 | 0.50 ** | 18.2 |
| fish #2 | Rainbow | male | 863 | 9.1 | 1.1 | 12.7 | 0.52 ** | 32.7 |
| fish #3 | Rainbow | male | 502 | 5.6 | 1.1 | 17.5 | 0.54 ** | 2.9 |
| fish #4 | Rainbow | female | 861 | 18.9 | 2.2 | 18.0 | 0.17 ** | 2.8 |
| fish #5 | Rainbow | female | 823 | 9.9 | 1.2 | 18.3 | 0.35 ** | 7.6 |
| fish #6 | Rainbow | female | 561 | 10.2 | 1.8 | 19.2 | 0.22 ** | 1.4 |
| avg ± std err | 754 ± 72 | 10.7 ± 1.8 | 1.4 ± 0.5 | 17.0 ± 0.9 * | 0.38 ± 0.07 | 10.9 ± 5.0 | ||
| fish #1 | Brook | male | 1376 | 19.3 | 1.4 | 8.4 | 0.57 *** | 6.8 |
| fish #2 | Brook | male | 1093 | 14.2 | 1.3 | 7.9 | 0.58 *** | 5.2 |
| fish #3 | Brook | male | 987 | 11.7 | 1.2 | 11.0 | 0.61 *** | 9.6 |
| fish #4 | Brook | female | 1282 | 29.2 | 2.3 | 8.0 | 0.47 *** | 3.0 |
| fish #5 | Brook | female | 974 | 18.4 | 1.9 | 9.8 | 0.38 *** | 2.4 |
| fish #6 | Brook | female | 779 | 12.8 | 1.6 | 9.5 | 0.47 *** | 6.3 |
| avg ± std err | 1082 ± 89 | 17.6 ± 2.6 | 1.6 ± 0.4 | 9.1 ± 0.5 * | 0.51 ± 0.04 | 5.6 ± 1.1 | ||
| fish #1 | Lake | male | 954 | 10.3 | 1.1 | 11.1 | 0.49 | 11.3 |
| fish #2 | Lake | male | 1140 | 15.0 | 1.3 | 13.3 | 0.41 | 5.7 |
| fish #3 | Lake | male | 1209 | 13.9 | 1.1 | 11.1 | 0.40 | 5.3 |
| fish #4 | Lake | male | 1227 | 20.9 | 1.7 | 8.0 | 0.38 | 8.3 |
| fish #5 | Lake | male | 1592 | 18.8 | 1.2 | 12.0 | 0.48 | 3.8 |
| fish #6 | Lake | female | 893 | 10.4 | 1.2 | 9.4 | 0.58 | 6.6 |
| avg ± std err | 1169 ± 101 | 14.9 ± 1.8 | 1.3 ± 0.2 | 10.8 ± 0.8 * | 0.46 ± 0.03 | 6.8 ± 1.1 |
| RAINBOW TROUT | HYDROQUINONE | ||
|---|---|---|---|
| SEX | Km | Vmax | |
| fish #1 | male | 15 ± 4 | 525 ± 35 |
| fish #2 | male | 22 ± 14 | 1013 ± 193 |
| fish #3 | male | 13 ± 3 | 405 ± 20 |
| fish #4 | female | 17 ± 9 | 1383 ± 191 |
| fish #5 | female | 11 ± 6 | 527 ± 64 |
| fish #6 | female | 15 ± 7 | 608 ± 72 |
| avg of individual Km and Vmax ± std err (N = 6) | 15 ± 2 ** | 744 ± 154 | |
| Km and Vmax fitted to avg rate | 16 ± 7 | 742 ± 89 | |
| BROOK TROUT | HYDROQUINONE | ||
| SEX | Km | Vmax | |
| fish #1 | male | 54 ± 34 | 1038 ± 283 |
| fish #2 | male | 38 ± 29 | 997 ± 278 |
| fish #3 | male | 18 ± 16 | 956 ± 234 |
| fish #4 | female | 45 ± 39 | 1654 ± 574 |
| fish #5 | female | 243 ± 112 | 2277 ± 782 |
| fish #6 | female | 232 ± 106 | 2800 ± 942 |
| avg of individual Km and Vmax ± std err (N = 6) | 105 ± 42 | 1620 ± 316 | |
| avg of individual Km and Vmax ± std err (N = 4) * | 38 ± 8 ** | 1161 ± 165 | |
| Km and Vmax fitted to avg rate | 42 ± 23 | 1099 ± 233 | |
| LAKE TROUT | HYDROQUINONE | ||
| SEX | Km | Vmax | |
| fish #1 | male | 46 ± 21 | 680 ± 119 |
| fish #2 | male | 30 ± 15 | 526 ± 68 |
| fish #3 | male | 55 ± 29 | 966 ± 176 |
| fish #4 | male | 45 ± 24 | 656 ± 109 |
| fish #5 | male | 31 ± 16 | 381 ± 53 |
| fish #6 | female | 46 ± 25 | 680 ± 119 |
| avg of individual Km and Vmax ± std err (N = 6) | 42 ± 4 ** | 657 ± 81 | |
| Km & Vmax fitted to avg rate | 43 ± 22 | 653 ± 102 | |
| RAINBOW TROUT | CATECHOL | ||
|---|---|---|---|
| SEX | Km | Vmax | |
| fish #1 | male | 8 ± 6 | 146 ± 32 |
| fish #2 | male | 24 ± 16 | 166 ± 38 |
| fish #3 | male | 6 ± 6 | 187 ± 39 |
| fish #4 | female | 18 ± 12 | 190 ± 35 |
| fish #5 | female | 8 ± 3 | 111 ± 7 |
| fish #6 | female | 8 ± 3 | 180 ± 15 |
| avg of individual Km and Vmax ± std err N = 6) | 12 ± 3 ** | 163 ± 12 | |
| Km and Vmax fitted to avg rate | 9 ± 4 | 150 ± 16 | |
| BROOK TROUT | CATECHOL | ||
| SEX | Km | Vmax | |
| fish #1 | male | 28 ± 16 | 118 ± 22 |
| fish #2 | male | 25 ± 18 | 137 ± 32 |
| fish #3 | male | 17 ± 13 | 97 ± 19 |
| fish #4 | female | 23 ± 20 | 171 ± 43 |
| fish #5 | female | 45 ± 25 | 205 ± 45 |
| fish #6 | female | 42 ± 20 | 273 ± 51 |
| avg of individual Km and Vmax ± std err (N = 6) | 30 ± 5 | 167 ± 26 | |
| avg of individual Km and Vmax ± std err (N = 4) * | 23 ± 2 ** | 131 ± 16 | |
| Km and Vmax fitted to avg rate | 27 ± 17 | 155 ± 31 | |
| LAKE TROUT | CATECHOL | ||
| SEX | Km | Vmax | |
| fish #1 | male | 31 ± 18 | 154 ± 32 |
| fish #2 | male | 16 ± 10 | 140 ± 21 |
| fish #3 | male | 25 ± 18 | 143 ± 30 |
| fish #4 | male | 32 ± 18 | 142 ± 28 |
| fish #5 | male | 36 ± 15 | 113 ± 20 |
| fish #6 | female | 10 ± 7 | 112 ± 13 |
| avg of individual Km and Vmax ± std err (N = 6) | 25 ± 4 ** | 134 ± 7 | |
| Km and Vmax fitted to avg rate | 20 ± 12 | 124 ± 19 | |
| RAINBOW TROUT | PHENYLGLUCURONIDE | ||
|---|---|---|---|
| SEX | Km | Vmax | |
| fish #1 | male | 10 ± 9 | 1383 ± 369 |
| fish #2 | male | 15 ± 16 | 1935 ± 773 |
| fish #3 | male | 7 ± 7 | 1872 ± 446 |
| fish #4 | female | 1 ± 1 | 438 ± 80 |
| fish #5 | female | 17 ± 15 | 2076 ± 742 |
| fish #6 | female | 8 ± 8 | 1591 ± 399 |
| avg of individual Km and Vmax ± std err (N = 6) | 10 ± 3 | 1549 ± 245 * | |
| Km and Vmax fitted to avg rate | 11 ± 10 | 1605 ± 450 | |
| BROOK TROUT | PHENYLGLUCURONIDE | ||
| SEX | Km | Vmax | |
| fish #1 | male | 7 ± 6 | 1771 ± 315 |
| fish #2 | male | 5 ± 5 | 1431 ± 250 |
| fish #3 | male | 8 ± 7 | 2204 ± 449 |
| fish #4 | female | 9 ± 7 | 1811 ± 425 |
| fish #5 | female | 7 ± 6 | 1968 ± 322 |
| fish #6 | female | 6 ± 6 | 2108 ± 416 |
| avg of individual Km and Vmax ± std err (N = 6) | 7 ± 1 | 1882 ± 113 * | |
| Km and Vmax fitted to avg rate | 8 ± 7 | 1958 ± 423 | |
| LAKE TROUT | PHENYLGLUCURONIDE | ||
| SEX | Km | Vmax | |
| fish #1 | male | 9 ± 10 | 887 ± 236 |
| fish #2 | male | 10 ± 11 | 727 ± 193 |
| fish #3 | male | 5 ± 2 | 726 ± 70 |
| fish #4 | male | 8 ± 8 | 627 ± 142 |
| fish #5 | male | 11 ± 9 | 738 ± 173 |
| fish #6 | female | 11 ± 10 | 815 ± 201 |
| avg of individual Km and Vmax ± std err (N = 6) | 9 ± 1 | 753 ± 36 * | |
| Km and Vmax fitted to avg rate | 7 ± 7 | 708 ± 152 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolanczyk, R.C.; Solem, L.E.; Schmieder, P.K.; McKim, J.M., III. A Comparative Study of Phase I and II Hepatic Microsomal Biotransformation of Phenol in Three Species of Salmonidae: Hydroquinone, Catechol, and Phenylglucuronide Formation. Fishes 2024, 9, 284. https://doi.org/10.3390/fishes9070284
Kolanczyk RC, Solem LE, Schmieder PK, McKim JM III. A Comparative Study of Phase I and II Hepatic Microsomal Biotransformation of Phenol in Three Species of Salmonidae: Hydroquinone, Catechol, and Phenylglucuronide Formation. Fishes. 2024; 9(7):284. https://doi.org/10.3390/fishes9070284
Chicago/Turabian StyleKolanczyk, Richard C., Laura E. Solem, Patricia K. Schmieder, and James M. McKim, III. 2024. "A Comparative Study of Phase I and II Hepatic Microsomal Biotransformation of Phenol in Three Species of Salmonidae: Hydroquinone, Catechol, and Phenylglucuronide Formation" Fishes 9, no. 7: 284. https://doi.org/10.3390/fishes9070284
APA StyleKolanczyk, R. C., Solem, L. E., Schmieder, P. K., & McKim, J. M., III. (2024). A Comparative Study of Phase I and II Hepatic Microsomal Biotransformation of Phenol in Three Species of Salmonidae: Hydroquinone, Catechol, and Phenylglucuronide Formation. Fishes, 9(7), 284. https://doi.org/10.3390/fishes9070284





