Linear and Non-Linear Methods to Discriminate Cortical Parcels Based on Neurodynamics: Insights from sEEG Recordings
Abstract
1. Introduction
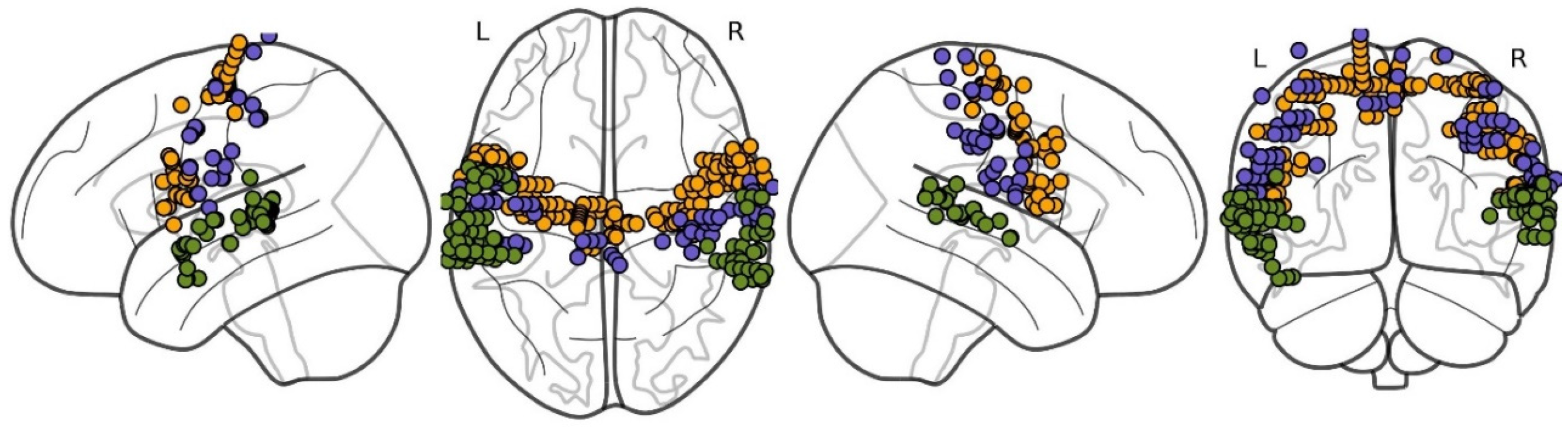
2. Materials and Methods
- precentral gyrus (PreCG): 141 channels (from 34 subjects);
- postcentral gyrus (PostCG): 64 channels (from 21 subjects);
- superior temporal gyrus (STG): 79 channels (from 26 subjects).
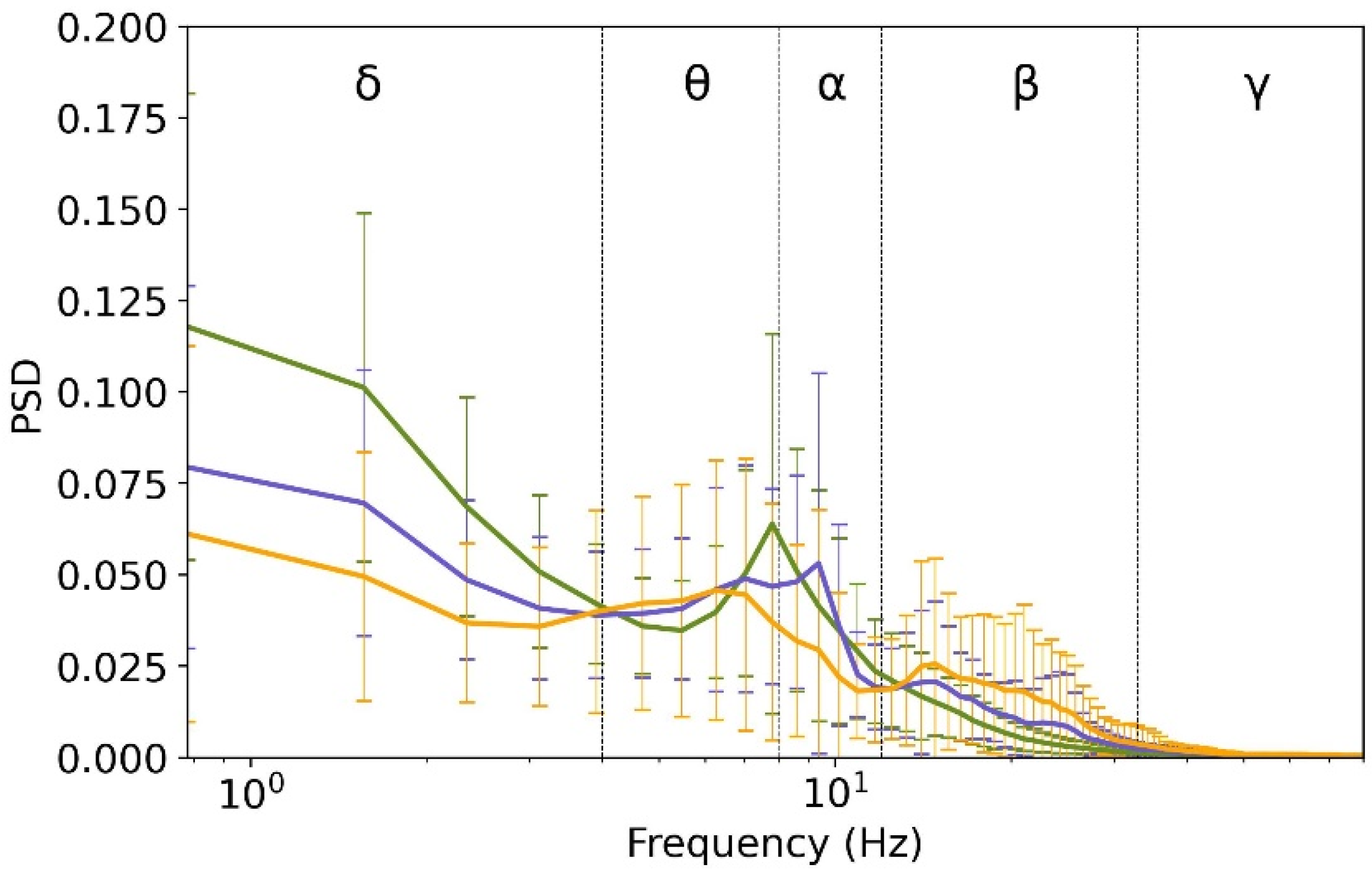
2.1. Spectral Analysis
2.2. Higuchi Fractal Dimension
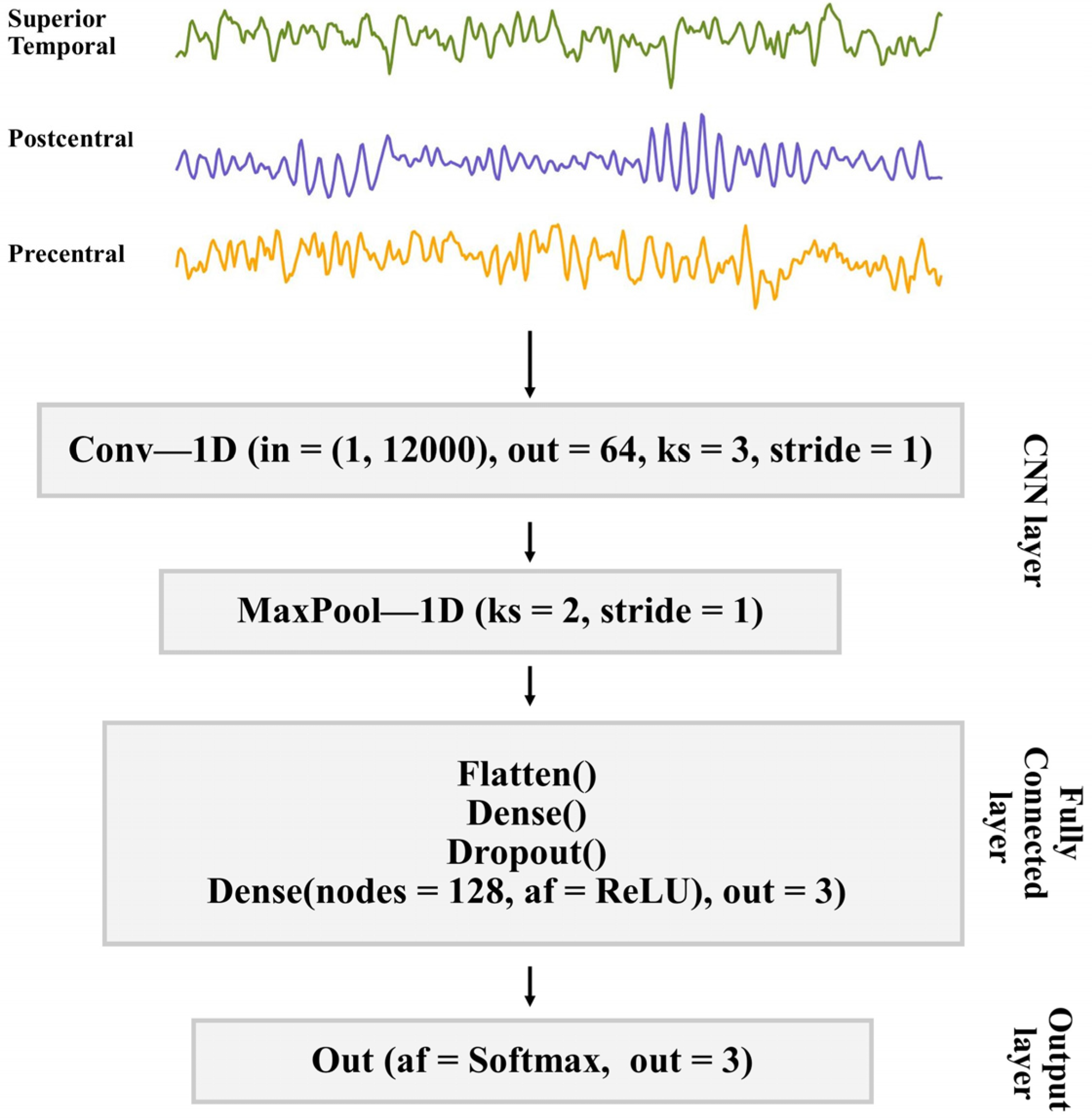
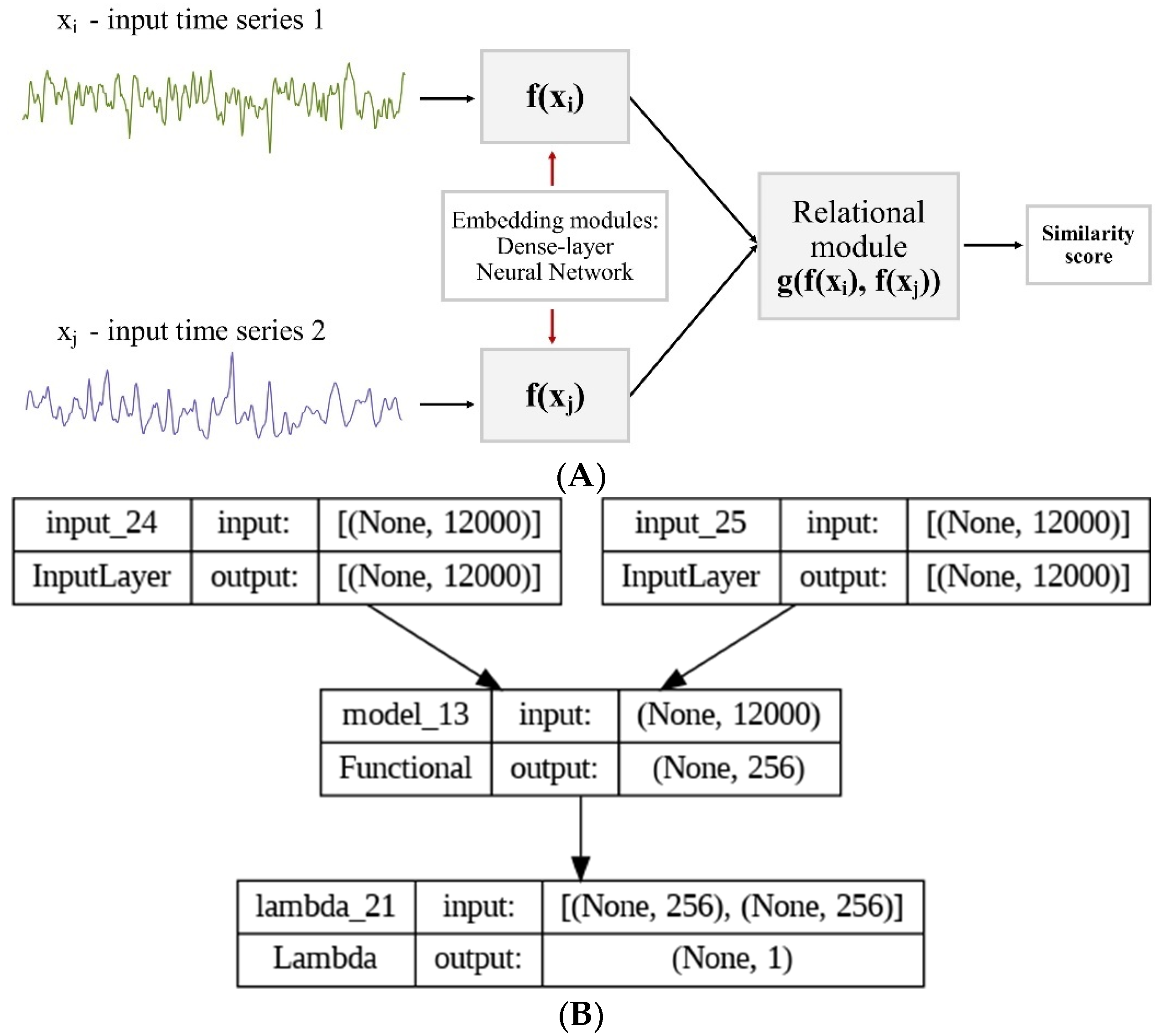
2.3. Deep Learning Classification
2.4. 1D-CNN Model
2.5. One-Shot Learning Model
2.6. Statistical Analysis
3. Results
3.1. Spectral Features in the Three Cortical Parcels
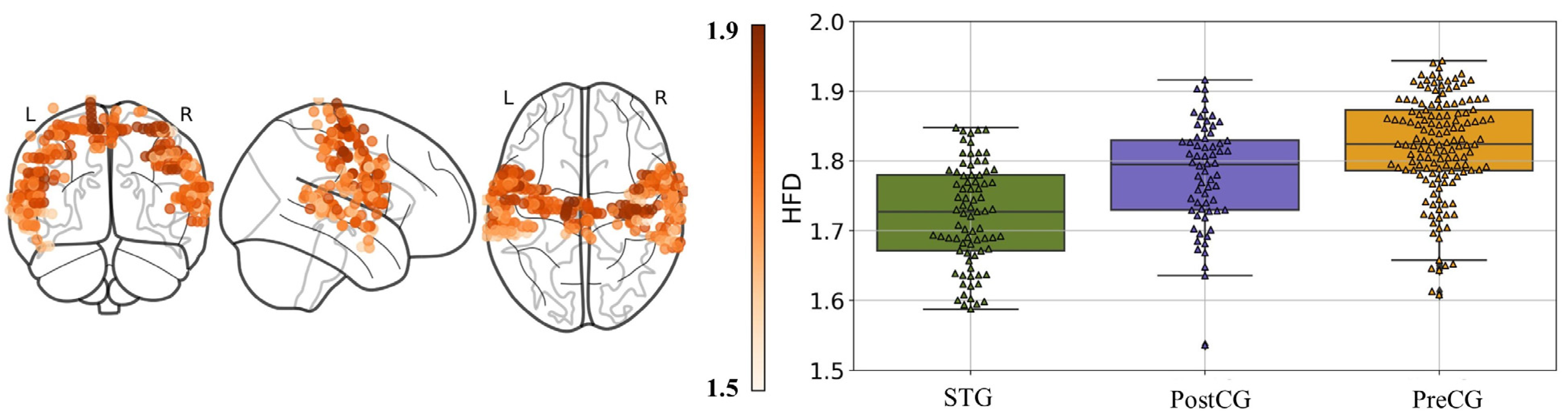
3.2. Higuchi Fractal Dimension in the Three Cortical Parcels
3.3. The Classification of the Three Cortical Parcels with 1D-CNN Model
3.4. Classification of the Three Cortical Parcels with One-Shot Learning Model
4. Discussion
Limitations of This Study and Future Developments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PreCG | Precentral gyrus |
| PostCG | Postcentral gyrus |
| STG | Superior temporal gyrus |
| sEEG | Stereotactical-intracranial electroencephalography |
| HFD | Higuchi Fractal Dimension |
| PSD | Power Spectral Density |
| 1D-CNN | One-dimensional convolutional neural network |
References
- Tecchio, F.; Bertoli, M.; Gianni, E.; L’Abbate, T.; Paulon, L.; Zappasodi, F. To Be Is To Become. Fractal Neurodynamics of the Body-Brain Control System. Front. Physiol. 2020, 11, 609768. [Google Scholar] [CrossRef]
- Li, J. Thoughts on neurophysiological signal analysis and classification. Brain Sci. Adv. 2020, 6, 210–223. [Google Scholar] [CrossRef]
- Moaveninejad, S.; D’Onofrio, V.; Tecchio, F.; Ferracuti, F.; Iarlori, S.; Monteriù, A.; Porcaro, C. Fractal Dimension as a discriminative feature for high accuracy classification in motor imagery EEG-based brain-computer interface. Comput. Methods Programs Biomed. 2024, 244, 107944. [Google Scholar] [CrossRef] [PubMed]
- Frauscher, B.; Von Ellenrieder, N.; Zelmann, R.; Doležalová, I.; Minotti, L.; Olivier, A.; Hall, J.; Hoffmann, D.; Nguyen, D.K.; Kahane, P.; et al. Atlas of the normal intracranial electroencephalogram: Neurophysiological awake activity in different cortical areas. Brain 2018, 141, 1130–1144. [Google Scholar] [CrossRef] [PubMed]
- Ruiz de Miras, J.; Soler, F.; Iglesias-Parro, S.; Ibáñez-Molina, A.J.; Casali, A.G.; Laureys, S.; Massimini, M.; Esteban, F.J.; Navas, J.; Langa, J.A. Fractal dimension analysis of states of consciousness and unconsciousness using transcranial magnetic stimulation. Comput. Methods Programs Biomed. 2019, 175, 129–137. [Google Scholar] [CrossRef]
- Yoder, K.J.; Brookshire, G.; Glatt, R.M.; Merrill, D.A.; Gerrol, S.; Quirk, C.; Lucero, C. Fractal Dimension Distributions of Resting-State Electroencephalography (EEG) Improve Detection of Dementia and Alzheimer’s Disease Compared to Traditional Fractal Analysis. Clin. Transl. Neurosci. 2024, 8, 27. [Google Scholar] [CrossRef]
- Armonaite, K.; Conti, L.; Olejarczyk, E.; Tecchio, F.; Roma, S.; Vergata, T. Insights on neural signal analysis with Higuchi fractal dimension. Commun. Appl. Ind. Math. 2024, 15, 17–27. [Google Scholar] [CrossRef]
- Kesić, S.; Spasić, S.Z. Application of Higuchi’s fractal dimension from basic to clinical neurophysiology: A review. Comput. Methods Programs Biomed. 2016, 133, 55–70. [Google Scholar] [CrossRef]
- Higuchi, T. Approach to an irregular time series on the basis of the fractal theory. Phys. D Nonlinear Phenom. 1988, 31, 277–283. [Google Scholar] [CrossRef]
- Olejarczyk, E.; Gotman, J.; Frauscher, B. Region-specific complexity of the intracranial EEG in the sleeping human brain. Sci. Rep. 2022, 12, 451. [Google Scholar] [CrossRef]
- Salimi, M.; Tabasi, F.; Abdolsamadi, M.; Dehghan, S.; Dehdar, K.; Nazari, M.; Javan, M.; Mirnajafi-Zadeh, J.; Raoufy, M.R. Disrupted connectivity in the olfactory bulb-entorhinal cortex-dorsal hippocampus circuit is associated with recognition memory deficit in Alzheimer’s disease model. Sci. Rep. 2022, 12, 4394. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Liu, C.; Xue, Y.; Chen, L. Changed firing activity of nigra dopaminergic neurons in Parkinson’s disease. Neurochem. Int. 2023, 162, 105465. [Google Scholar] [CrossRef]
- Nasca, C.; Barnhill, O.; DeAngelis, P.; Watson, K.; Lin, J.; Beasley, J.; Young, S.P.; Myoraku, A.; Dobbin, J.; Bigio, B.; et al. Multidimensional predictors of antidepressant responses: Integrating mitochondrial, genetic, metabolic and environmental factors with clinical outcomes. Neurobiol. Stress 2021, 15, 100407. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, L.M.; Négyessy, L.; Friedman, R.M.; Mishra, A.; Gore, J.C.; Roe, A.W. The Relationship of Anatomical and Functional Connectivity to Resting-State Connectivity in Primate Somatosensory Cortex. Neuron 2013, 78, 1116–1126. [Google Scholar] [CrossRef]
- Economo, M.N.; Viswanathan, S.; Tasic, B.; Bas, E.; Winnubst, J.; Menon, V.; Graybuck, L.T.; Nguyen, T.N.; Smith, K.A.; Yao, Z.; et al. Distinct descending motor cortex pathways and their roles in movement. Nature 2018, 563, 79–84. [Google Scholar] [CrossRef]
- Greicius, M.D.; Krasnow, B.; Reiss, A.L.; Menon, V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 253–258. [Google Scholar] [CrossRef]
- Raichle, M.E.; Macleod, A.M.; Snyder, A.Z.; Powers, W.J.; Gusnard, D.A.; Shulman, G.L. A default mode of brain function. Proc. Natl. Acad. Sci. USA 2001, 98, 676–682. [Google Scholar] [CrossRef]
- von Ellenrieder, N.; Gotman, J.; Zelmann, R.; Rogers, C.; Nguyen, D.K.; Kahane, P.; Dubeau, F.; Frauscher, B. How the Human Brain Sleeps: Direct Cortical Recordings of Normal Brain Activity. Ann. Neurol. 2020, 87, 289–301. [Google Scholar] [CrossRef]
- Solomon, M.O. PSD Computations Using Welch’s Method. Sandia Natl. Lab. 1991, 64, 23584. [Google Scholar]
- Spasić, S.; Kalauzi, A.; Ćulić, M.; Grbić, G.; Martać, L. Estimation of parameter kmax in fractal analysis of rat brain activity. Ann. N. Y. Acad. Sci. 2005, 1048, 427–429. [Google Scholar] [CrossRef]
- Paramanathan, P.; Uthayakumar, R. Application of fractal theory in analysis of human electroencephalographic signals. Comput. Biol. Med. 2008, 38, 372–378. [Google Scholar] [CrossRef]
- Smits, F.M.; Porcaro, C.; Cottone, C.; Cancelli, A.; Rossini, P.M.; Tecchio, F. Electroencephalographic Fractal Dimension in Healthy Ageing and Alzheimer’s Disease. PLoS ONE 2016, 11, e0149587. [Google Scholar] [CrossRef]
- Armonaite, K.; Nobili, L.; Paulon, L.; Balsi, M.; Conti, L.; Tecchio, F. Local neurodynamics as a signature of cortical areas: New insights from sleep. Cereb. Cortex 2022, 33, 3284–3292. [Google Scholar] [CrossRef] [PubMed]
- Armonaite, K.; Bertoli, M.; Paulon, L.; Gianni, E.; Balsi, M.; Conti, L.; Tecchio, F. Neuronal Electrical Ongoing Activity as Cortical Areas Signature: An Insight from MNI Intracerebral Recording Atlas. Cereb. Cortex 2021, 32, 2895–2906. [Google Scholar] [CrossRef]
- Liehr, L.; Massopust, P. On the mathematical validity of the Higuchi method. Phys. D Nonlinear Phenom. 2020, 402, 132265. [Google Scholar] [CrossRef]
- Ahmadlou, M.; Adeli, H.; Adeli, A. Fractality analysis of frontal brain in major depressive disorder. Int. J. Psychophysiol. 2012, 85, 206–211. [Google Scholar] [CrossRef]
- Phinyomark, A.; Larracy, R.; Scheme, E. Fractal Analysis of Human Gait Variability via Stride Interval Time Series. Front. Physiol. 2020, 11, 333. [Google Scholar] [CrossRef] [PubMed]
- Cottone, C.; Porcaro, C.; Cancelli, A.; Olejarczyk, E.; Salustri, C.; Tecchio, F. Neuronal electrical ongoing activity as a signature of cortical areas. Brain Struct. Funct. 2017, 222, 2115–2126. [Google Scholar] [CrossRef]
- Olejarczyk, E.; Zappasodi, F.; Ricci, L.; Pascarella, A.; Pellegrino, G.; Paulon, L.; Assenza, G.; Tecchio, F. Functional Source Separation-Identified Epileptic Network: Analysis Pipeline. Brain Sci. 2022, 12, 1179. [Google Scholar] [CrossRef]
- Yang, B.; Zhu, X.; Liu, Y.; Liu, H. A single-channel EEG based automatic sleep stage classification method leveraging deep one-dimensional convolutional neural network and hidden Markov model. Biomed. Signal Process. Control 2021, 68, 102581. [Google Scholar] [CrossRef]
- Cisotto, G.; Zanga, A.; Chlebus, J.; Zoppis, I.; Manzoni, S.; Markowska-Kaczmar, U. Comparison of Attention-based Deep Learning Models for EEG Classification. arXiv 2020, arXiv:2012.01074. [Google Scholar]
- Ullah, I.; Hussain, M.; Qazi, E.-U.-H.; Aboalsamh, H. An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Syst. Appl. 2018, 107, 61–71. [Google Scholar] [CrossRef]
- Truong, N.D.; Nguyen, A.D.; Kuhlmann, L.; Bonyadi, M.R.; Yang, J.; Ippolito, S.; Kavehei, O. Convolutional neural networks for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw. 2018, 105, 104–111. [Google Scholar] [CrossRef]
- Wu, X.; Kimura, A.; Iwana, B.K.; Uchida, S.; Kashino, K. Deep dynamic time warping: End-to-end local representation learning for online signature verification. In Proceedings of the 2019 International Conference on Document Analysis and Recognition (ICDAR), Sydney, Australia, 20–25 September 2019; pp. 1103–1110. [Google Scholar]
- Wang, X.; Sun, K.; Zhao, T.; Wang, W.; Gu, Q. Dynamic Speed Warping: Similarity-Based One-shot Learning for Device-free Gesture Signals. In Proceedings of the IEEE INFOCOM 2020-IEEE Conference on Computer Communications, Toronto, ON, Canada, 6–9 July 2020; pp. 556–565. [Google Scholar]
- Keitel, A.; Gross, J. Individual Human Brain Areas Can Be Identified from Their Characteristic Spectral Activation Fingerprints. PLoS Biol. 2016, 14, e1002498. [Google Scholar] [CrossRef] [PubMed]
- Barttfeld, P.; Uhrig, L.; Sitt, J.D.; Sigman, M.; Jarraya, B. Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl. Acad. Sci. USA 2015, 112, 887–892. [Google Scholar] [CrossRef]
- Fox, M.D.; Raichle, M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007, 8, 700–711. [Google Scholar] [CrossRef]
- He, B.J.; Zempel, J.M.; Snyder, A.Z.; Raichle, M.E. The temporal structures and functional significance of scale-free brain activity. Neuron 2010, 66, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Di Ieva, A.; Grizzi, F.; Jelinek, H.; Pellionisz, A.J.; Losa, G.A. Fractals in the neurosciences, part I: General principles and basic neurosciences. Neuroscientist 2014, 20, 403–417. [Google Scholar] [CrossRef]
- Muthukumaraswamy, S.D.; Liley, D.T. 1/F Electrophysiological Spectra in Resting and Drug-Induced States Can Be Explained By the Dynamics of Multiple Oscillatory Relaxation Processes. Neuroimage 2018, 179, 582–595. [Google Scholar] [CrossRef]
- Armonaite, K.; Conti, L.; Tecchio, F. Fractal Neurodynamics; Springer: Berlin/Heidelberg, Germany, 2024; Volume 36. [Google Scholar]
- VAROL ARISOY, M. Signature Verification Using Siamese Neural Network One-Shot Learning. Int. J. Eng. Innov. Res. 2021, 3, 248–260. [Google Scholar] [CrossRef]
- Weis, J.; Santra, A. One-shot learning for robust material classification using millimeter-wave radar system. IEEE Sensors Lett. 2018, 2, 2–5. [Google Scholar] [CrossRef]
- Burrello, A.; Schindler, K.; Benini, L.; Rahimi, A. One-shot Learning for iEEG Seizure Detection Using End-to-end Binary Operations: Local Binary Patterns with Hyperdimensional Computing. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; pp. 1–4. [Google Scholar]
- Jiang, R.; Zheng, X.; Sun, J.; Chen, L.; Xu, G.; Zhang, R. Classification for Alzheimer’s disease and frontotemporal dementia via resting-state electroencephalography-based coherence and convolutional neural network. Cogn. Neurodyn. 2025, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Ma, L.; Li, H. Efficient Neural Network Classification of Parkinson’s Disease and Schizophrenia Using Resting-State EEG Data. Brain Topogr. 2025, 38, 32. [Google Scholar] [CrossRef]
- Saha, S.; Baumert, M. Intra- and Inter-subject Variability in EEG-Based Sensorimotor Brain Computer Interface: A Review. Front. Comput. Neurosci. 2020, 13, 87. [Google Scholar] [CrossRef] [PubMed]

| Class | Precision (%) | Recall (%) | F1 Score (%) | |
|---|---|---|---|---|
| Train | PreCG | 89 | 100 | 94 |
| PostCG | 69 | 93 | 80 | |
| STG | 100 | 51 | 67 | |
| Test | PreG | 50 | 72 | 57 |
| PostG | 30 | 30 | 30 | |
| STG | 100 | 0 | 0 |
| Method | Precision (%) | Recall (%) | F1 Score (%) | Accuracy (%) | |
|---|---|---|---|---|---|
| Train | Macro avg | 86 | 81 | 80 | 85 |
| Weighted avg | 88 | 85 | 83 | ||
| Test | Macro avg | 58 | 34 | 29 | 50 |
| Weighted avg | 57 | 43 | 35 |
| Method | Precision (%) | Recall (%) | F1 Score (%) | Accuracy | |
|---|---|---|---|---|---|
| Train | Macro avg | 63 | 56 | 55 | 58 |
| Weighted avg | 63 | 56 | 55 | ||
| Test | Macro avg | 58 | 56 | 54 | 56 |
| Weighted avg | 58 | 56 | 54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armonaite, K.; Conti, L.; Laura, L.; Primavera, M.; Tecchio, F. Linear and Non-Linear Methods to Discriminate Cortical Parcels Based on Neurodynamics: Insights from sEEG Recordings. Fractal Fract. 2025, 9, 278. https://doi.org/10.3390/fractalfract9050278
Armonaite K, Conti L, Laura L, Primavera M, Tecchio F. Linear and Non-Linear Methods to Discriminate Cortical Parcels Based on Neurodynamics: Insights from sEEG Recordings. Fractal and Fractional. 2025; 9(5):278. https://doi.org/10.3390/fractalfract9050278
Chicago/Turabian StyleArmonaite, Karolina, Livio Conti, Luigi Laura, Michele Primavera, and Franca Tecchio. 2025. "Linear and Non-Linear Methods to Discriminate Cortical Parcels Based on Neurodynamics: Insights from sEEG Recordings" Fractal and Fractional 9, no. 5: 278. https://doi.org/10.3390/fractalfract9050278
APA StyleArmonaite, K., Conti, L., Laura, L., Primavera, M., & Tecchio, F. (2025). Linear and Non-Linear Methods to Discriminate Cortical Parcels Based on Neurodynamics: Insights from sEEG Recordings. Fractal and Fractional, 9(5), 278. https://doi.org/10.3390/fractalfract9050278







