Efficient Hybrid Parallel Scheme for Caputo Time-Fractional PDEs on Multicore Architectures
Abstract
1. Introduction
- Alleviate memory bottlenecks by distributing historical data across processors;
- Accelerate convergence through the concurrent evaluation of memory integrals and local operators;
- Enable adaptive domain decomposition for efficient modeling of complex anatomical geometries;
- Support real-time simulations for applications such as drug-response prediction, ECG signal reconstruction, and patient-specific medical imaging.
- A generalized parallel fractional framework capable of handling Caputo-type time derivatives and nonlinear spatial operators;
- A domain-decomposed architecture that enables concurrent computation of local solution components, thereby reducing overall simulation time;
- A symbolic–numeric hybrid strategy, in which the nonlinear systems arising from CTFPDE discretizations are solved using Newton-type methods;
- Comprehensive evaluation metrics, including convergence rate, CPU usage, residual error dynamics, memory efficiency, and biological interpretability.
- A fractional cardiac conduction system with nonlinear reaction–diffusion terms;
- A dynamical model of depression incorporating feedback mechanisms and long-term memory;
- A sub-diffusion drug delivery model in layered biological tissues.
- Problem Scope: We address CTFPDEs derived from biomedical models that exhibit spatial heterogeneity and memory effects, extending beyond the idealized benchmark problems commonly considered in the literature.
- Parallelization Strategy: The proposed schemes implement parfor-based parallelization in MATLAB, enabling efficient utilization of multi-core processors and yielding measurable reductions in computational time.
- Benchmarking and Validation: Comprehensive tests are conducted, encompassing comparisons with analytical solutions, performance benchmarks, and application-driven case studies, in order to confirm the accuracy and robustness of the proposed approach.
2. Construction and Analysis of the Next-Generation Computational Schemes
2.1. Construction of the Scheme
2.2. Theoretical Convergence Analysis
- is the Jacobian of at , assumed nonsingular;
- collects higher-order terms, with .
- is the Jacobian of evaluated at , assumed nonsingular;
- collects the higher-order terms, with .
3. Computational Efficiency and Numerical Outcomes
3.1. Implementation of Methodology, Convergence Enhancement, and Result Visualization
- -
- Iteration count;
- -
- Percentage convergence (P-Con);
- -
- Computational time (CPU seconds);
- -
- Memory usage (MB);
- -
- Percentage convergence under random initial values.
- -
- Criteria I: Element-wise scheme. Each solution component is updated independently in parallel:
- -
- Criteria II: Diagonalized scheme. The system is reformulated in matrix form and updated viawhere is a diagonal matrix that approximates a suitable operator to accelerate convergence.
- -
- Parallel implementation with MATLAB parfor. Both element-wise and diagonalized schemes were parallelized using MATLAB’s parfor construct, which distributes independent computations across multiple CPU cores. The main iteration loop was executed in parallel while ensuring data consistency and avoiding race conditions. Unlike OpenMP, MATLAB’s parfor replicates loop variables for each worker rather than automatically sharing memory, which may affect large-scale memory usage.This parallelization reduces computational time while preserving the accuracy of the serial version. To quantify performance, we measured the serial CPU time (Tseri), parallel CPU time (Tpara), and the speedup ratio, defined aswhere Tseri corresponds to execution on a single core without MATLAB parfor, and Tpara to execution on four cores with MATLAB parfor. Algorithm 1 and the flow chart in Figure 3 illustrate the complete implementation, including the computation of the COC and residual error for approximating the solution of (55). A higher speedup ratio indicates greater efficiency.
- -
- Initial Vector Sampling. For each numerical experiment, a single initial guess vector is drawn randomly from a feasible domain, with magnitude close to to improve the convergence rate. This unbiased initialization avoids selection bias and provides a fair evaluation of algorithmic robustness.
- -
- Selection Criterion. The iterative scheme is run on all sampled vectors, and the one yielding the highest accuracy is retained, measured bywhere . This high precision is achieved in MATLAB using the vpa function with digits = 64.
- -
- Stopping Criteria. The iteration is terminated once any of the following conditions is satisfied:
3.2. Applications in Biomedical Engineering
| Algorithm 1 Parallel VPA Weierstrass method for solving fractional PDEs |
|
| Algorithm 2 Parallel scheme for solving (55) using MATLAB parfor parallelization on multiple cores |
|
3.2.1. Drug Diffusion in Tissue with Nonlinear Reaction [49]
- Spatial domain: , discretized into N intervals with spacing .
- Time domain: , discretized into M intervals with spacing .
- Grid nodes: , and .
- Unknown: .
- ⊙ denotes element-wise operations;
- .
- Step size and tolerance were chosen to balance accuracy and convergence.
- Numerical results showed that the proposed approach preserved the model’s physical behavior over time and space.
- The interaction between fractional order, nonlinearity, and numerical discretization directly affected computational cost and convergence speed.
3.2.2. Brain Signal Propagation with Nonlinear Blood Flow Effects [51]
- : effective diffusion coefficient of the electrical signal (axonal/dendritic spread).
- : advective drift term (typically small; set unless modeling directed flow).
- : external source term.
- Spatial domain: , discretized into N intervals with spacing .
- Temporal domain: , discretized into M intervals with spacing .
- Grid nodes: , ; , .
- Unknowns: .
- ⊙ denotes element-wise operations;
- ;
- Step size and tolerance were chosen to balance accuracy and convergence in the presence of fractional dynamics and nonlinear flow effects.
- Numerical results showed that the proposed parallel scheme accurately reproduced signal propagation patterns while preserving key physiological features.
- Fractional order, nonlinear blood flow, and discretization parameters jointly influenced computational cost and convergence rate.
3.2.3. Fractional Heart Tissue Electrical Conduction with Nonlinear Reaction [53]
- Spatial domain: , discretized into N intervals with spacing .
- Time domain: , discretized into M intervals with spacing .
- Grid nodes: , ; , .
- Unknowns: .
- ⊙ denotes element-wise operations;
- Step size and tolerance were adjusted to balance precision and convergence in the presence of fractional dynamics and nonlinear ionic interactions.
- Numerical results confirmed that the proposed approach accurately reproduced the spatiotemporal propagation of electrical signals in cardiac tissue.
3.3. Comparative Discussion of Biomedical Examples
- Mathematical structure: Biomedical models with higher-order nonlinearities or significant fractional memory effects (e.g., cardiac conduction) tend to exhibit slower convergence. Nonetheless, the proposed methods ensure steady residual decay and maintain accuracy even under stiff nonlinear dynamics (see Figure 4, Figure 5, Figure 6, Figure 7, Figure 8 and Figure 9).
- Numerical parameters: Smaller step sizes and high-precision VPA computations reduce local truncation error and further improve accuracy. In addition, adaptive random initialization yields consistent results across all three biomedical applications.
- Efficiency: The proposed parallel schemes, particularly , reliably reduce CPU time and maximum error compared to classical fractional iterative methods. Performance gains become increasingly pronounced for larger problem sizes and denser fractional memory terms.
- Stability under perturbations: Convergence is preserved even when random initial vectors, parameter fluctuations, or noise in boundary conditions are introduced. This resilience is especially important for biomedical applications, where data uncertainty is inherent.
- Parallel scalability: Although MATLAB’s parfor differs from OpenMP in memory distribution, the results confirm effective parallel speedup on multi-core architectures. The scalability of the proposed approach makes it suitable for high-dimensional and long-time fractional simulations.
- Computational cost: The hierarchical formulation of the parallel schemes controls memory usage, delivering a favorable cost-to-accuracy ratio. Even for nonlinear biological PDEs with strong fractional effects, the proposed methods remain computationally competitive.
- Biomedical relevance: Each biomedical application represents a distinct physiological process—drug transport, neuronal dynamics, and cardiac tissue excitation. In all cases, the proposed methods produced reliable results, underscoring their translational potential in real-world biomedical modeling.
4. Conclusions
- Limitations. The present study has some limitations:
- Parallelization was implemented using MATLAB’s parfor construct rather than low-level OpenMP or GPU-based solutions, which may limit scalability for large-scale problems.
- Higher-dimensional cases may require additional stability and memory considerations; the numerical studies presented here were restricted to two-dimensional biological PDEs.
- Exact solutions were constructed for validation, whereas real biomedical data typically contain noise and parameter uncertainty, which were not considered in this work.
- Future work. Several directions are envisioned for extending this study:
- Implementing the techniques in hybrid CPU–GPU environments to improve scalability and computational speed.
- Extending the framework to multidimensional biomedical models, such as three-dimensional heart wave propagation.
- Incorporating uncertainty quantification to account for variability and noise in physiological data.
- Exploring adaptive step-size control and machine learning-assisted initialization to further improve reliability and convergence.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| – | Newly developed schemes |
| n | Iterations |
| CPU-time | Computational time in seconds |
| Computational local convergence order |
Appendix A. Symbolic Verification of Biomedical Models
Appendix A.1. Drug Diffusion in Tissue with Nonlinear Reaction

Appendix A.2. Brain Signal Propagation with Nonlinear Blood Flow Effects

Appendix A.3. Fractional Heart Tissue Electrical Conduction with Nonlinear Reaction

Appendix A.4. Remarks
References
- Rathore, A.S.; Mishra, S.; Nikita, S.; Priyanka, P. Bioprocess control: Current progress and future perspectives. Life 2021, 11, 557. [Google Scholar] [CrossRef]
- Su, W.H.; Chou, C.S.; Xiu, D. Deep learning of biological models from data: Applications to ODE models. Bull. Math. Biol. 2021, 83, 19. [Google Scholar] [CrossRef]
- Cruz, D.A.; Kemp, M.L. Hybrid computational modeling methods for systems biology. Prog. Biomed. Eng. 2021, 4, 012002. [Google Scholar] [CrossRef]
- Enderle, J.D.; Ropella, K.M.; Kelsa, D.M.; Hallowell, B. Ensuring that biomedical engineers are ready for the real world. IEEE Eng. Med. Biol. Mag. 2002, 21, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.M.; Wu, S.L. A parallel-in-time iterative algorithm for Volterra partial integro-differential problems with weakly singular kernel. J. Comput. Phys. 2020, 417, 109576. [Google Scholar] [CrossRef]
- Wen, J.; Tian, Y.E.; Skampardoni, I.; Yang, Z.; Cui, Y.; Anagnostakis, F.; Mamourian, E.; Zhao, B.; Toga, A.W.; Zalesky, A.; et al. The genetic architecture of biological age in nine human organ systems. Nat. Aging 2024, 4, 1290–1307. [Google Scholar] [CrossRef]
- Ghezal, A.; Al Ghafli, A.A.; Al Salman, H.J. Anomalous Drug Transport in Biological Tissues: A Caputo Fractional Approach with Non-Classical Boundary Modeling. Fractal Fract. 2025, 9, 508. [Google Scholar] [CrossRef]
- Sachse, F.B.; Moreno, A.P.; Seemann, G.; Abildskov, J.A. A model of electrical conduction in cardiac tissue including fibroblasts. Ann. Biomed. Eng. 2009, 37, 874–889. [Google Scholar] [CrossRef]
- Dai, X.; Wu, D.; Xu, K.; Ming, P.; Cao, S.; Yu, L. Viscoelastic Mechanics: From Pathology and Cell Fate to Tissue Regeneration Biomaterial Development. Acs Appl. Mater. Interfaces 2025, 17, 8751–8770. [Google Scholar] [CrossRef]
- Peng, C.; Guo, T.; Xie, C.; Bai, X.; Zhou, J.; Zhao, X.; He, E.; Xia, F. mBGT: Encoding brain signals with multimodal brain graph transformer. IEEE Trans. Consum. Electron. 2024, 71, 5812–5823. [Google Scholar] [CrossRef]
- Kaltenbacher, B.; Rundell, W. Inverse Problems for Fractional Partial Differential Equations; American Mathematical Society: Providence, RI, USA, 2023; Volume 230. [Google Scholar]
- Mubaraki, A.M.; Nuruddeen, R.I.; Gomez-Aguilar, J.F. Closed-form asymptotic solution for the transport of chlorine concentration in composite pipes. Phys. Scr. 2024, 99, 075201. [Google Scholar] [CrossRef]
- Lin, C.H.; Liu, C.H.; Chien, L.S.; Chang, S.C. Accelerating pattern matching using a novel parallel algorithm on GPUs. IEEE Trans. Comput. 2012, 62, 1906–1916. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Gu, X.M.; Ostermann, A. A preconditioning technique for an all-at-once system from Volterra subdiffusion equations with graded time steps. J. Sci. Comput. 2021, 88, 11. [Google Scholar] [CrossRef]
- He, J.H.; Anjum, N.; He, C.H.; Alsolami, A.A. Beyond laplace and fourier transforms: Challenges and future prospects. Therm. Sci. 2023, 27 Pt B, 5075–5089. [Google Scholar] [CrossRef]
- Haubold, H.J.; Mathai, A.M.; Saxena, R.K. Mittag-Leffler functions and their applications. J. Appl. Math. 2011, 2011, 298628. [Google Scholar] [CrossRef]
- Duffy, D.G. Green’s Functions with Applications; Chapman and Hall/CRC: Boca Raton, FL, USA, 2015. [Google Scholar]
- Hedin, L. New method for calculating the one-particle Green’s function with application to the electron-gas problem. Phys. Rev. 1965, 139, A796. [Google Scholar] [CrossRef]
- Rainer, B.; Kaltenbacher, B. Existence, uniqueness, and numerical solutions of the nonlinear periodic Westervelt equation. ESAIM Math. Model. Numer. Anal. 2025, 59, 2279–2304. [Google Scholar] [CrossRef]
- Kumar, M.; Umesh. Recent development of Adomian decomposition method for ordinary and partial differential equations. Int. J. Appl. Comput. Math. 2022, 8, 81. [Google Scholar] [CrossRef]
- Nadeem, M.; He, J.H.; Islam, A. The homotopy perturbation method for fractional differential equations: Part 1 Mohand transform. Int. J. Numer. Methods Heat Fluid Flow 2021, 31, 3490–3504. [Google Scholar] [CrossRef]
- Shihab, M.A.; Taha, W.M.; Hameed, R.A.; Jameel, A.; Ibrahim, S.M. Implementation of variational iteration method for various types of linear and nonlinear partial differential equations. Int. J. Electr. Comput. Eng. 2023, 13, 2131–2141. [Google Scholar] [CrossRef]
- Kamil Jassim, H.; Vahidi, J. A new technique of reduce differential transform method to solve local fractional PDEs in mathematical physics. Int. J. Nonlinear Anal. Appl. 2021, 12, 37–44. [Google Scholar]
- Li, C.; Zeng, F. Finite difference methods for fractional differential equations. Int. J. Bifurc. Chaos 2012, 22, 1230014. [Google Scholar] [CrossRef]
- Sacchetti, A.; Bachmann, B.; Löffel, K.; Künzi, U.M.; Paoli, B. Neural networks to solve partial differential equations: A comparison with finite elements. IEEE Access 2022, 10, 32271–32279. [Google Scholar] [CrossRef]
- Sheng, C.; Cao, D.; Shen, J. Efficient spectral methods for PDEs with spectral fractional Laplacian. J. Sci. Comput. 2021, 88, 4. [Google Scholar] [CrossRef]
- Rieder, A. A p-version of convolution quadrature in wave propagation. SIAM J. Numer. Anal. 2025, 63, 1729–1756. [Google Scholar] [CrossRef]
- Figueroa, A.; Jackiewicz, Z.; Löhner, R. Explicit two-step Runge-Kutta methods for computational fluid dynamics solvers. Int. J. Numer. Methods Fluids 2021, 93, 429–444. [Google Scholar] [CrossRef]
- Salem, M.G.; Abouelregal, A.E.; Elzayady, M.E.; Sedighi, H.M. Biomechanical response of skin tissue under ramp-type heating by incorporating a modified bioheat transfer model and the Atangana–Baleanu fractional operator. Acta Mech. 2024, 235, 5041–5060. [Google Scholar] [CrossRef]
- Li, J.M.; Wang, X.J.; He, R.S.; Chi, Z.X. An efficient fine-grained parallel genetic algorithm based on gpu-accelerated. In Proceedings of the 2007 IFIP International Conference on Network and Parallel Computing Workshops (NPC 2007), Dalian, China, 18–21 September 2007; pp. 855–862. [Google Scholar]
- Kelley, C.T. Solving Nonlinear Equations with Newton’s Method; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2003. [Google Scholar]
- Langtangen, H.P. Solving Nonlinear ODE and PDE Problems; Center for Biomedical Computing, Simula Research Laboratory and Department of Informatics, University of Oslo: Oslo, Norway, 2016. [Google Scholar]
- Nofal, T.A. Simple equation method for nonlinear partial differential equations and its applications. J. Egypt. Math. Soc. 2016, 24, 204–209. [Google Scholar] [CrossRef]
- Ramos, H.; Monteiro, M.T.T. A new approach based on the Newton’s method to solve systems of nonlinear equations. J. Comput. Appl. Math. 2017, 318, 3–13. [Google Scholar] [CrossRef]
- Noor, M.A.; Waseem, M. Some iterative methods for solving a system of nonlinear equations. Comput. Math. Appl. 2009, 57, 101–106. [Google Scholar] [CrossRef]
- Dehghan, M.; Shirilord, A. Three-step iterative methods for numerical solution of systems of nonlinear equations. Eng. Comput. 2022, 38, 1015–1028. [Google Scholar] [CrossRef]
- Darvishi, M.T.; Barati, A. Super cubic iterative methods to solve systems of nonlinear equations. Appl. Math. Comput. 2007, 188, 1678–1685. [Google Scholar] [CrossRef]
- Sharma, J.R.; Guha, R.K.; Sharma, R. An efficient fourth order weighted-Newton method for systems of nonlinear equations. Numer. Algorithms 2013, 62, 307–323. [Google Scholar] [CrossRef]
- Cordero, A.; Martínez, E.; Torregrosa, J.R. Iterative methods of order four and five for systems of nonlinear equations. J. Comput. Appl. Math. 2009, 231, 541–551. [Google Scholar] [CrossRef]
- Hueso, J.L.; Martínez, E.; Teruel, C. Convergence, efficiency and dynamics of new fourth and sixth order families of iterative methods for nonlinear systems. J. Comput. Appl. Math. 2015, 275, 412–420. [Google Scholar] [CrossRef]
- George, S.; Sadananda, R.; Padikkal, J.; Argyros, I.K. On the order of convergence of the Noor–Waseem method. Mathematics 2022, 10, 4544. [Google Scholar] [CrossRef]
- Solaiman, O.S.; Hashim, I. An iterative scheme of arbitrary odd order and its basins of attraction for nonlinear systems. Comput. Mater. Contin. Comput. 2021, 66, 1427–1444. [Google Scholar] [CrossRef]
- Bate, I.; Murugan, M.; George, S.; Senapati, K.; Argyros, I.K.; Regmi, S. On extending the applicability of iterative methods for solving systems of nonlinear equations. Axioms 2024, 13, 601. [Google Scholar] [CrossRef]
- Petković, M.; Carstensen, C.; Trajković, M. Weierstrass formula and zero-finding methods. Numer. Math. 1995, 69, 353–372. [Google Scholar] [CrossRef]
- Ehrlich, L.W. A modified Newton method for polynomials. Commun. ACM 1967, 10, 107–108. [Google Scholar] [CrossRef]
- Cordero, A.; Torregrosa, J.R.; Triguero-Navarro, P. Jacobian-Free Vectorial Iterative Scheme to Find Simple Several Solutions Simultaneously. Math. Methods Appl. Sci. 2025, 48, 5718–5730. [Google Scholar] [CrossRef]
- Petković, M. Computational efficiency of simultaneous methods. In Iterative Methods for Simultaneous Inclusion of Polynomial Zeros; Springer: Berlin/Heidelberg, Germany, 2006; pp. 221–249. [Google Scholar]
- Zhang, R.; Bai, H.; Zhao, F. L1-Finite Difference Method for Inverse Source Problem of Fractional Diffusion Equation. J. Phys. Conf. Ser. 2020, 1624, 032001. [Google Scholar] [CrossRef]
- King, C. Non-Linear Reaction-Diffusion of Oxygen in Biological Systems. Ph.D. Thesis, Washington State University, Pullman, WA, USA, 2023. [Google Scholar]
- Pujol, M.J.; Grimalt, P. A non-linear model of cerebral diffusion: Stability of finite differences method and resolution using the Adomian method. Int. J. Numer. Methods Heat Fluid Flow 2003, 13, 473–485. [Google Scholar] [CrossRef]
- Akay, M. (Ed.) Nonlinear Biomedical Signal Processing, Volume 2: Dynamic Analysis and Modeling; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 2. [Google Scholar]
- Toronov, V.; Myllylä, T.; Kiviniemi, V.; Tuchin, V.V. Dynamics of the brain: Mathematical models and non-invasive experimental studies. Eur. Phys. J. Spec. Top. 2013, 222, 2607–2622. [Google Scholar] [CrossRef]
- David, S.A.; Valentim, C.A.; Debbouche, A. Fractional modeling applied to the dynamics of the action potential in cardiac tissue. Fractal Fract. 2022, 6, 149. [Google Scholar] [CrossRef]
- Magin, R.L.; Ovadia, M. Modeling the cardiac tissue electrode interface using fractional calculus. J. Vib. Control 2008, 14, 1431–1442. [Google Scholar] [CrossRef]

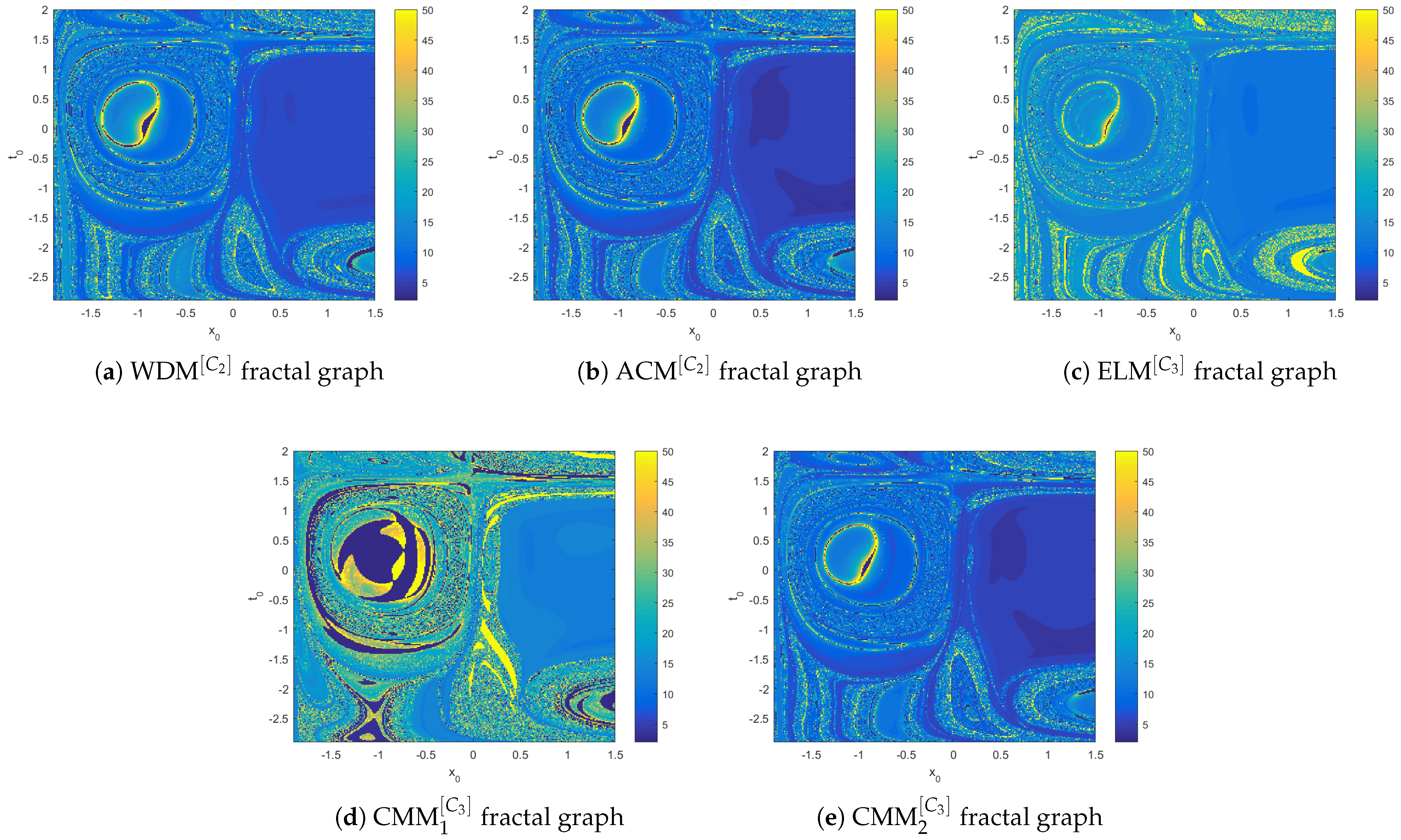


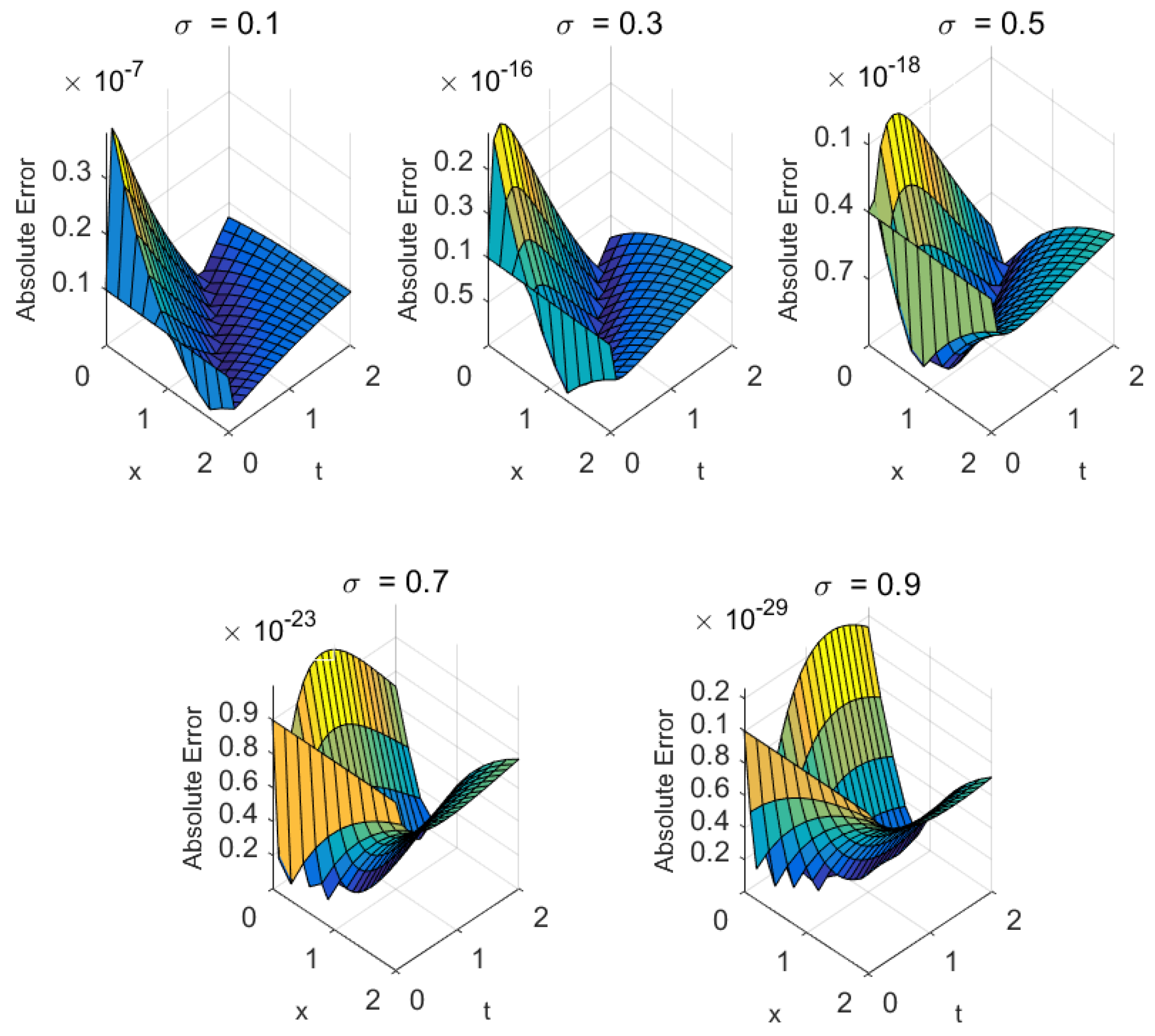

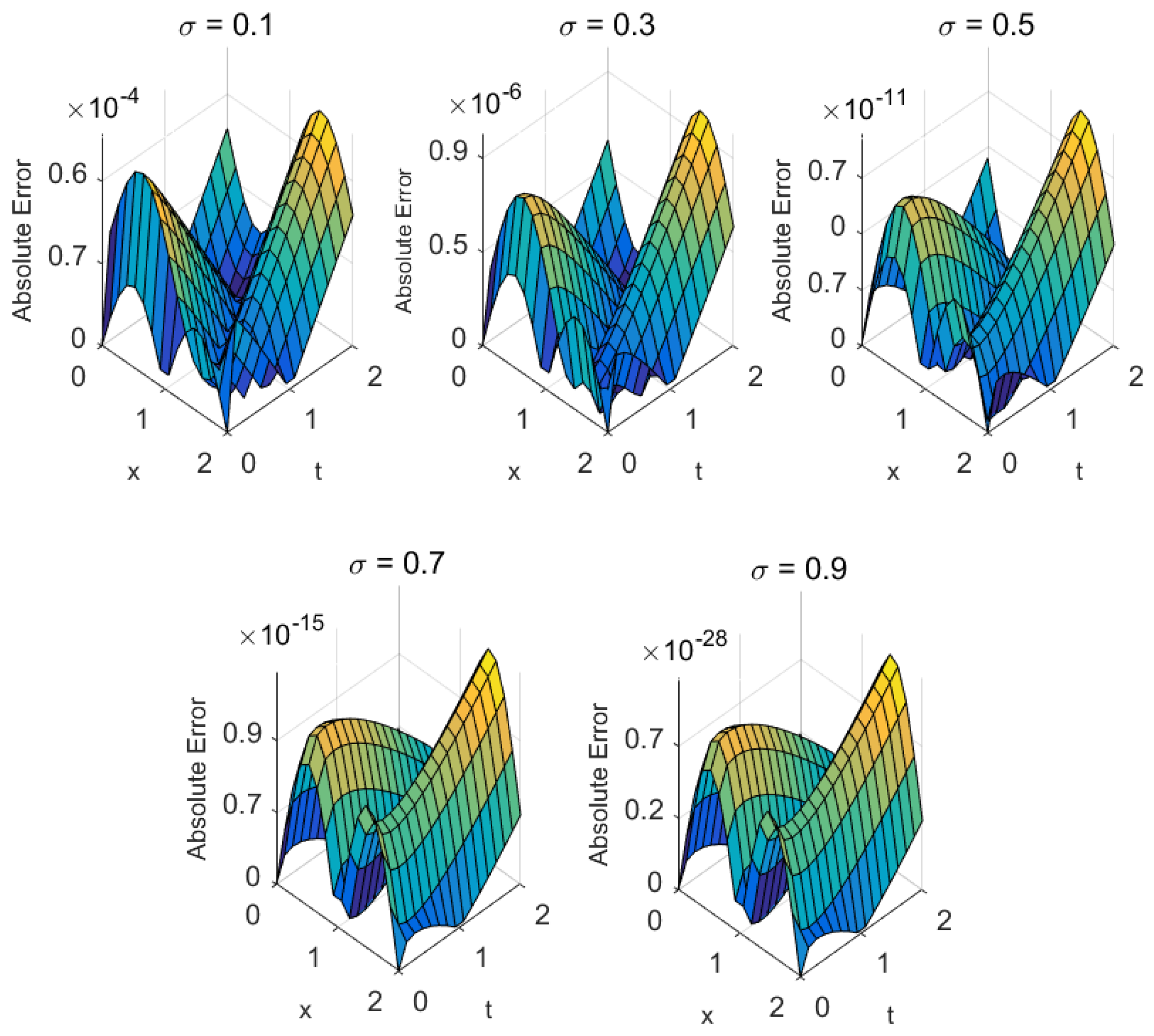

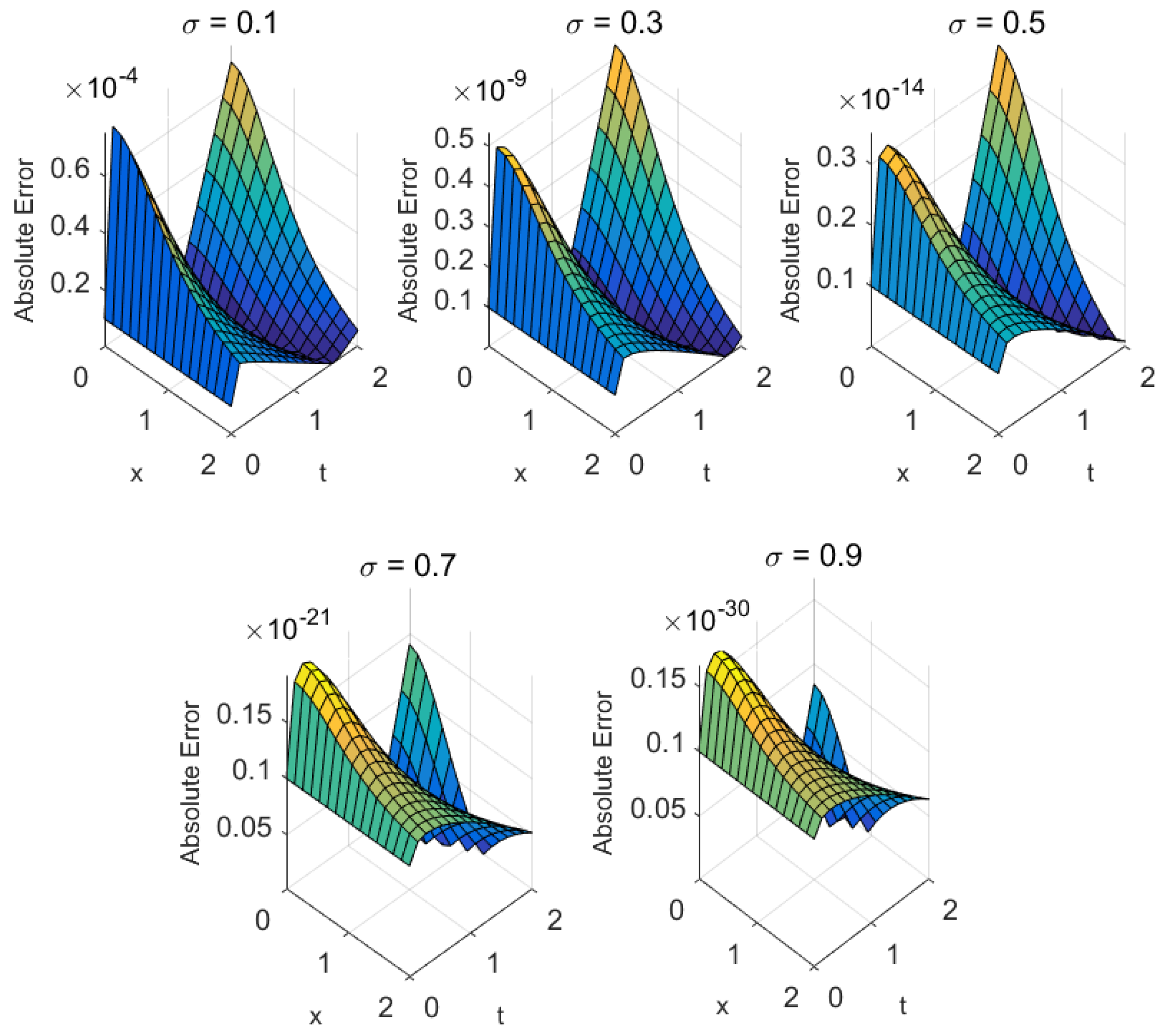
| Metric | ||||
|---|---|---|---|---|
| Additions/Subtractions | ||||
| Multiplications | ||||
| Efficiency |
| Method | n | Max-Error | Per-C | Ops [] | Memory (MB) | Elapsed Time (s) |
|---|---|---|---|---|---|---|
| 20 | 19 | 87.657 | 87.657 | |||
| 9 | 65 | 55.657 | 55.657 | |||
| 12 | 53 | 47.764 | 47.764 | |||
| 8 | 54 | 45.567 | 45.567 | |||
| 7 | 36 | 34.453 | 34.453 |
| Grid Points | -Norm | -Norm | CPU Time (s) |
|---|---|---|---|
| 30, 50 | 0.056 | ||
| 60, 90 | 0.120 | ||
| 120, 180 | 0.250 | ||
| 30, 50 | 0.060 | ||
| 60, 90 | 0.125 | ||
| 120, 180 | 0.260 | ||
| 30, 50 | 0.065 | ||
| 60, 90 | 0.130 | ||
| 120, 180 | 0.270 | ||
| 30, 50 | 0.070 | ||
| 60, 90 | 0.135 | ||
| 120, 180 | 0.280 | ||
| 30, 50 | 0.075 | ||
| 60, 90 | 0.145 | ||
| 120, 180 | 0.301 | ||
| Metric | |||||
|---|---|---|---|---|---|
| Criterion I | |||||
| -norm | |||||
| -norm | |||||
| Criterion II | |||||
| -norm | |||||
| -norm | |||||
| Metric | Iter. (n) | Max Error | Conv. (%) | Basic Ops | Memory (MB) | COC |
|---|---|---|---|---|---|---|
| 13 | 11.09 | 47 | 87.657 | 2.0014 | ||
| 13 | 19.76 | 51 | 55.657 | 2.0346 | ||
| 11 | 55.76 | 50 | 47.764 | 1.9993 | ||
| 9 | 63.54 | 54 | 45.567 | 3.1164 | ||
| 9 | 87.87 | 36 | 34.453 | 3.0087 |
| Application | Domain | Recorded Variables | Approx. Data Size |
|---|---|---|---|
| Bio-heat transfer |
| 0.03333 | 0.02000 | 0.98020 | 0.96722 | 0.95444 | 0.96724 |
| 0.73333 | 0.04000 | 0.47924 | 0.47237 | 0.46561 | 0.47240 |
| 0.46667 | 0.08000 | 0.62101 | 0.61653 | 0.61208 | 0.61655 |
| ⋮ (remaining entries omitted) | |||||
| 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | Mem U | |
|---|---|---|---|---|---|---|
| Using Criterion I | ||||||
| Using Criterion II | ||||||
| Metric | Average Iterations | COC | CPU Time (s) | Percentage Convergence | Memory Usage (MB) |
|---|---|---|---|---|---|
| Using Criterion I | |||||
| 17 | |||||
| 16 | |||||
| Using Criterion II | |||||
| 15 | |||||
| 16 | |||||
| Metric | (s) | (s) | Maximum Error | Percentage Convergence | Memory Usage (MB) | |
|---|---|---|---|---|---|---|
| Using Criterion I | ||||||
| 1.45 | 0.52 | 2.79 | 86.01% | 35.03 | ||
| 3.87 | 1.35 | 2.87 | 87.56% | 36.11 | ||
| Using Criterion II | ||||||
| 8.59 | 2.95 | 2.91 | 91.57% | 29.11 | ||
| 9.14 | 3.10 | 2.95 | 92.12% | 23.12 | ||
| Grid Points | -Norm | -Norm | C-Time |
|---|---|---|---|
| 30, 50 | 0.058 | ||
| 60, 90 | 0.115 | ||
| 120, 180 | 0.235 | ||
| 30, 50 | 0.065 | ||
| 60, 90 | 0.134 | ||
| 120, 180 | 0.260 | ||
| 30, 50 | 0.070 | ||
| 60, 90 | 0.135 | ||
| 120, 180 | 0.270 | ||
| 30, 50 | 0.075 | ||
| 60, 90 | 0.140 | ||
| 120, 180 | 0.283 | ||
| 30, 50 | 0.080 | ||
| 60, 90 | 0.146 | ||
| 120, 180 | 0.311 | ||
| Metric | |||||
|---|---|---|---|---|---|
| Using Criterion I | |||||
| -norm | |||||
| -norm | |||||
| Using Criterion II | |||||
| -norm | |||||
| -norm | |||||
| Metric | Iterations (n) | Max- Error | Percentage Convergence | Basic Ops | Memory Usage (MB) | COC |
|---|---|---|---|---|---|---|
| 23 | 35.09% | 47 | 76.147 | 2.00 | ||
| 21 | 41.96% | 51 | 67.347 | 2.03 | ||
| 16 | 65.13% | 50 | 53.704 | 2.01 | ||
| 11 | 77.04% | 54 | 49.500 | 3.12 | ||
| 10 | 93.98% | 36 | 44.413 | 3.00 |
| Application | Domain | Recorded Variables | Approx. Data Size |
|---|---|---|---|
| Bio-heat transfer |
| 0.03333 | 0.02000 | 2.1967 | 2.1976 | 2.1984 | 2.1980 |
| 0.73333 | 0.04000 | 2.8693 | 2.8712 | 2.8730 | 2.8721 |
| 0.46667 | 0.08000 | 2.6587 | 2.6599 | 2.6610 | 2.6605 |
| ⋮ (remaining entries omitted) | |||||
| 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | Mem U | |
|---|---|---|---|---|---|---|
| Using Criteria-I | ||||||
| Using Criteria-II | ||||||
| Metric | Average Iterations | COC | CPU Time (s) | Percentage Convergence | Memory Usage (MB) |
|---|---|---|---|---|---|
| Using Criterion I | |||||
| 17 | |||||
| 16 | |||||
| Using Criterion II | |||||
| 15 | |||||
| 16 | |||||
| Metric | (s) | (s) | Maximum Error | Percentage Convergence | Memory Usage (MB) | |
|---|---|---|---|---|---|---|
| Using Criterion I | ||||||
| Using Criterion II | ||||||
| Grid Points | -Norm | -Norm | CPU Time (s) |
|---|---|---|---|
| Metric | |||||
|---|---|---|---|---|---|
| Using Criterion I | |||||
| -norm | |||||
| -norm | |||||
| Using Criterion II | |||||
| -norm | |||||
| -norm | |||||
| Metric | Iterations (n) | Max- Error | Percentage Convergence | Basic Ops | Memory Usage (MB) | COC |
|---|---|---|---|---|---|---|
| 23 | 35.05% | 47 | 91.147 | 2.00 | ||
| 17 | 39.56% | 51 | 86.000 | 1.99 | ||
| 16 | 57.74% | 50 | 68.764 | 1.86 | ||
| 15 | 76.55% | 54 | 64.007 | 3.01 | ||
| 9 | 93.86% | 36 | 59.453 | 3.11 |
| Application | Domain | Recorded Variables | Data Size (Approx.) |
|---|---|---|---|
| Bio-heat transfer |
| 0.03333 | 0.02000 | 0.00000 | 0.00000 | 0.00000 | 0.01967 |
| 0.73333 | 0.04000 | 0.00000 | 0.01967 | 0.00000 | 0.02533 |
| 0.46667 | 0.08000 | 0.02533 | 0.00000 | 0.00000 | 0.06133 |
| ⋮ (remaining entries omitted) | |||||
| 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | Mem U | |
|---|---|---|---|---|---|---|
| Using Criterion I | ||||||
| Using Criterion II | ||||||
| Method | Average Iterations | COC | CPU Time (s) | Percentage Convergence | Memory Usage (MB) |
|---|---|---|---|---|---|
| Using Criterion I | |||||
| 23 | |||||
| 19 | |||||
| Using Criterion II | |||||
| 14 | |||||
| 8 | |||||
| Metric | (s) | (s) | Maximum Error | Percentage Convergence | Memory Usage (MB) | |
|---|---|---|---|---|---|---|
| Using Criterion I | ||||||
| Using Criterion II | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shams, M.; Carpentieri, B. Efficient Hybrid Parallel Scheme for Caputo Time-Fractional PDEs on Multicore Architectures. Fractal Fract. 2025, 9, 607. https://doi.org/10.3390/fractalfract9090607
Shams M, Carpentieri B. Efficient Hybrid Parallel Scheme for Caputo Time-Fractional PDEs on Multicore Architectures. Fractal and Fractional. 2025; 9(9):607. https://doi.org/10.3390/fractalfract9090607
Chicago/Turabian StyleShams, Mudassir, and Bruno Carpentieri. 2025. "Efficient Hybrid Parallel Scheme for Caputo Time-Fractional PDEs on Multicore Architectures" Fractal and Fractional 9, no. 9: 607. https://doi.org/10.3390/fractalfract9090607
APA StyleShams, M., & Carpentieri, B. (2025). Efficient Hybrid Parallel Scheme for Caputo Time-Fractional PDEs on Multicore Architectures. Fractal and Fractional, 9(9), 607. https://doi.org/10.3390/fractalfract9090607







