Abstract
Here, we propose a signal processing based approach for the extraction of the fetal heart rate (FHR) from Maternal Abdominal ECG (MAECG) in a non-invasive way. Datasets from a Physionet database has been used in this study for evaluating the performance of the proposed model that performs three major tasks; preprocessing of the MAECG signal, separation of Fetal QRS complexes from that of maternal and estimation of Fetal R peak positions. The MAECG signal is first preprocessed with improved multistep filtering techniques to detect the Maternal QRS (MQRS) complexes, which are dominant in the MAECG. A reference template is then reconstructed based on MQRS locations and removed from the preprocessed signal resulting in the raw FECG. This extracted FECG is further corrected and enhanced before obtaining the Fetal R peaks. The detection of FQRS and calculation of FHR has been compared against the reference Fetal Scalp ECG. Results indicate that the approach achieved good accuracy.
1. Introduction
Every single day, around 7000 sets of parents are unable to take their precious newborn home globally due to stillbirth. The major causes for stillbirth are the inability to continuously and non-invasively monitor the FHR continuously during pregnancy. A widely used method for FHR monitoring is Doppler Ultrasound scan. Other current electronic FHR monitoring systems include CTG, PCG, FMCG, SECG, most if not all of which are relatively expensive, requires technical expertise, may pose radioactive health hazards to the Fetus and can only be done very late in pregnancy [1]. This triggered the search for alternative FHR monitoring techniques during pregnancy, namely non-invasive FECG monitoring techniques [2,3]. Even though progress has been made in this sector, non-invasive FHR monitoring via digital signal processing is still a challenging [4] and evolving prospect.
We plan to address this issue and develop a model that is non-invasive, relatively inexpensive and easy to use. Towards this, we report a signal processing based model for the purpose mentioned above, that has been evaluated using the datasets from “Abdominal and Direct Fetal ECG Database” (ADFECGDB) [5]. The processing of this dataset is quite challenging because the data acquisition was done at the advanced stage (between 38 and 41 weeks of gestation) that caused many FQRS overlapping with MQRS complexes and plenty of motion artifacts generated by fetal and maternal movements. The accuracy achieved by this work (87%) is considerable bearing in mind the high amount of physiological noises available into the acquired signal.
2. Database Used
The ADFECGDB database contains data from 5 different women in labor, between 38 and 41 weeks of gestation. Each record had signals acquired from 4 different abdominal electrodes and a reference fetal scalp ECG measured directly from the fetal head. The sampling frequency of the data was 1000 Hz at a resolution of 16 bits.
3. Methodology
3.1. Preprocessing
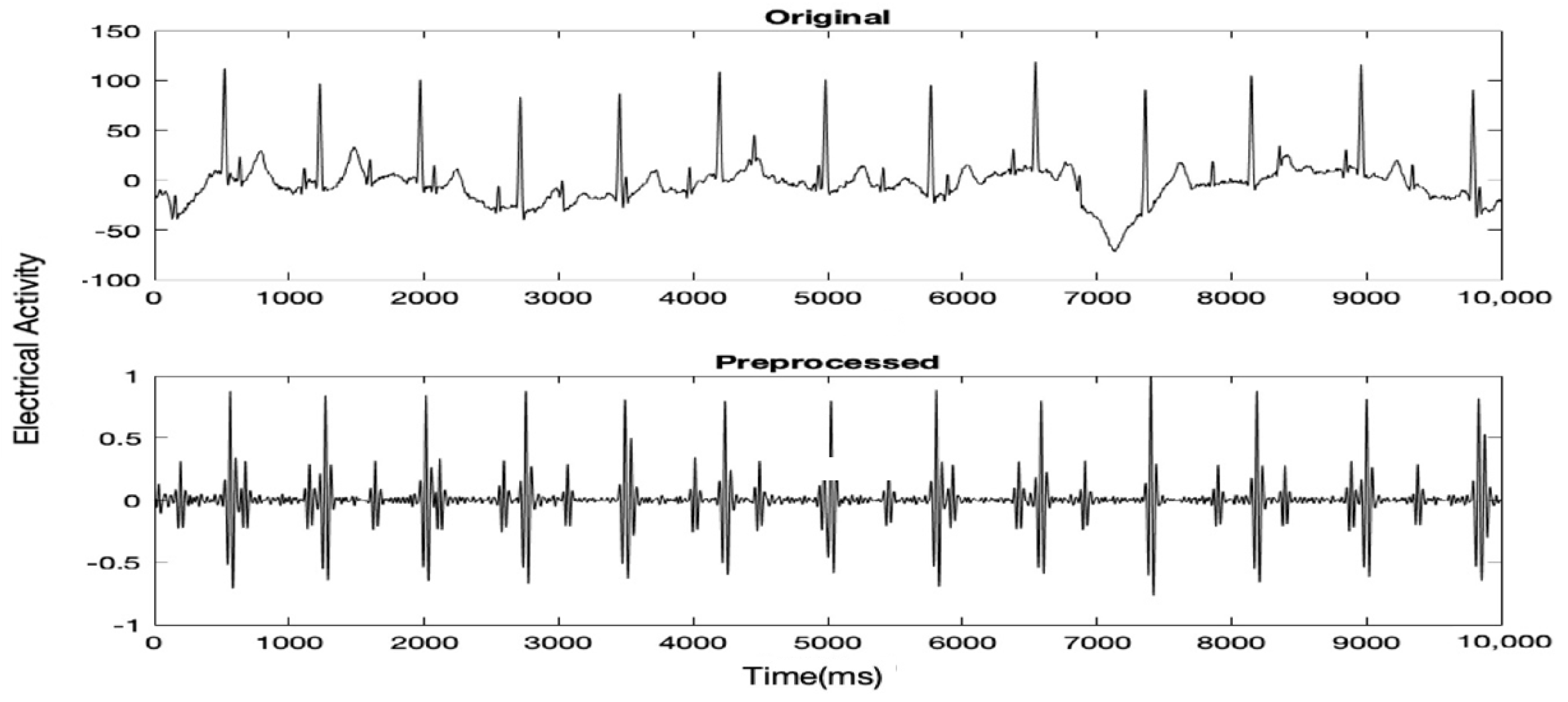
Initially, the raw MAECG signal was preprocessed to remove as much noise as possible while keeping the Fetal QRS (FQRS) and Maternal QRS (MQRS) complexes prominent. Typically, there are two types of noise in an ECG signal, high-frequency noise (40–60 Hz) and baseline drift or low- frequency noise. To remove high-frequency noise, an FIR low pass filter with Kaiser with a cutoff frequency of 100 Hz and ß = 0.5 (default parameter for sidelobe attenuation) was used. The order within the range of [10–100] which gave the highest SNR was chosen. For the removal of baseline drift, a Butterworth high pass filter with a cutoff frequency of 25 Hz and order of 4 was used. A further FIR low passes filter with Kaiser with a cutoff frequency of 40 Hz, order of 34 and ß = 0.5 was used to further remove any remaining high-frequency noise. Before feeding the signal to the filters, any DC drift was removed and the signal was normalized. Figure 1 shows 5 s of a MAECG signal before and after preprocessing.
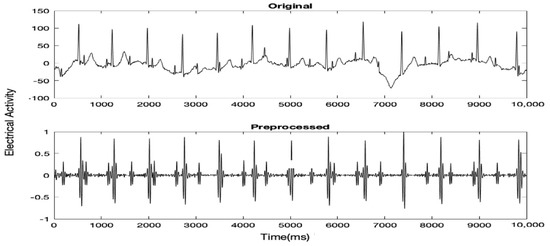
Figure 1.
5 s of a certain MAECG record before and after preprocessing.
3.2. Detection of MQRS Complexes
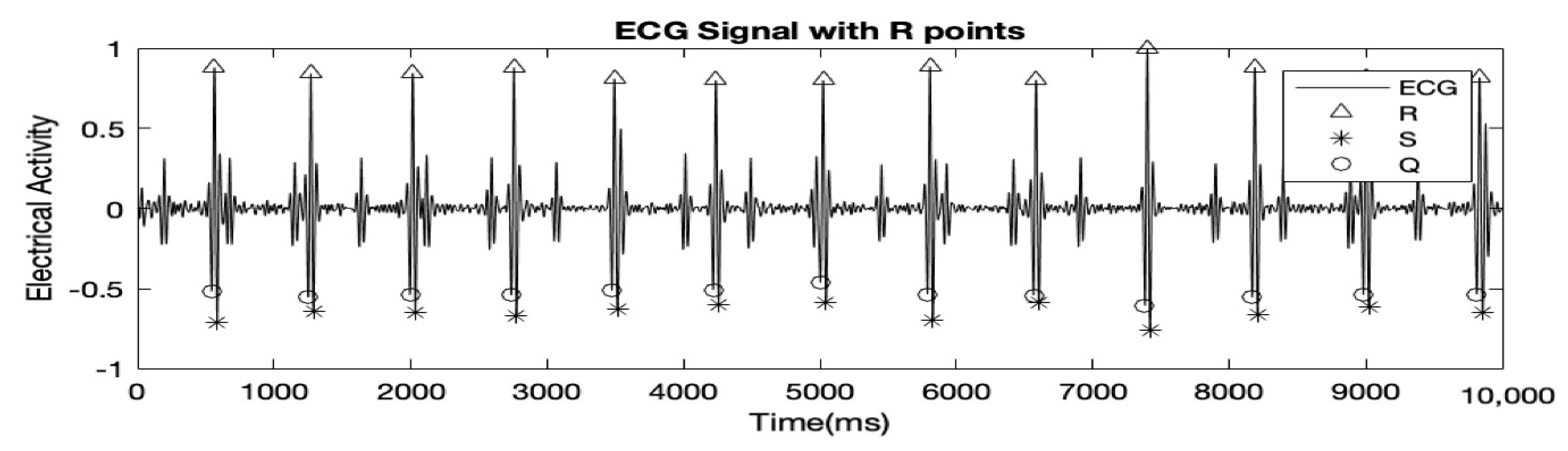
In this step, the MQRS complexes were detected using a modified version of Pan and Tompkins algorithm [6]. A derivative filter as implied in Pan and Tompkins algorithm was applied to the preprocessed MAECG signal to make the peaks more distinguished and prominent. The derived signal was increased by an exponent of 4 (as opposed to 2 in the original algorithm). A moving mean window of 120 points was then applied to make the resulting signal smoother. Signal values, which exceeded a threshold, were considered as peaks. The threshold was set as the product of the maximum peak value and the average signal value. The MQRS complexes were found using Equations (1)–(3).
Ri = max{f5(𝑡)}l<t<r
Qi = min{f5(𝑡)}l<t<R
Si = min{f5(𝑡)}R<t<r
Here, l and r are variables ‘left’ and ‘right’ which represent the time right before a peak and the time right after a peak respectively and Q, R and S hold the time locations of Q, R, and S peaks respectively. Figure 2 highlights the results after detection of MQRS.
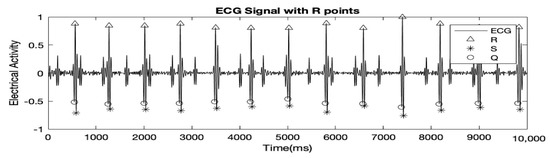
Figure 2.
10 s of the MAECG shown above with Q, R and S peaks detected.
3.3. FECG Extraction
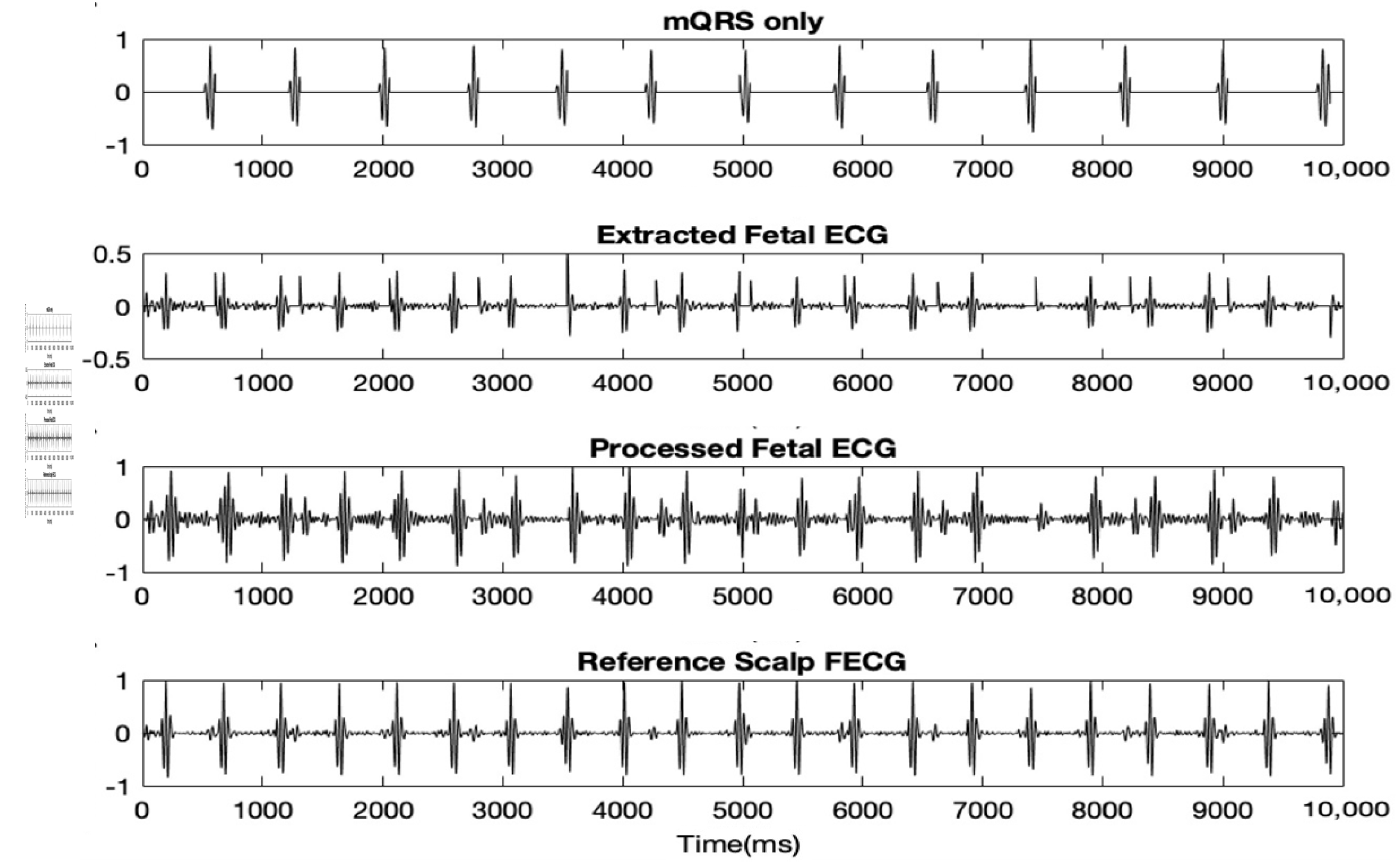
In this next step, the raw FECG was extracted, first by generating a clean MECG only and then deducting that clean signal from the preprocessed MAECG mixture. Equation (4) was used to generate the clean MECG signal, Z1 with mostly MQRS complexes only.
Case 1 is for zeroing everything in the preprocessed MAECG in between each MQRS complexes. Case 2 and 3 deal with zeroing everything from the start to first MQRS and last MQRS to the end of the signal respectively.
The clean MECG was then deducted from the preprocessed MAECG signal resulting in the raw FECG signal. This raw FECG signal was then again preprocessed the same way as the original raw MAECG.. Figure 3 shows the clean MECG, raw extracted FECG and preprocessed FECG. The accuracies of the results, when compared to the reference direct fetal scalp ECG are stated in Table 1
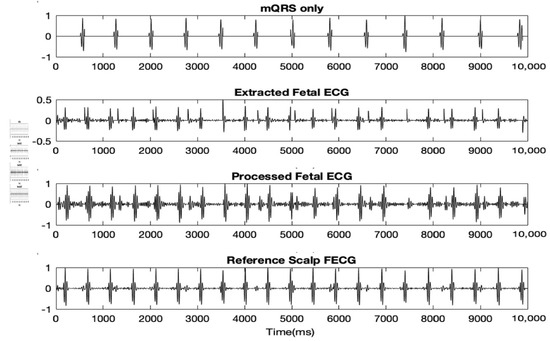
Figure 3.
10 s of clean MECG, extracted FECG and preprocessed FECG. Discussion.

Table 1.
Accuracy table of the results obtained when compared to the reference fetal scalp ECG.
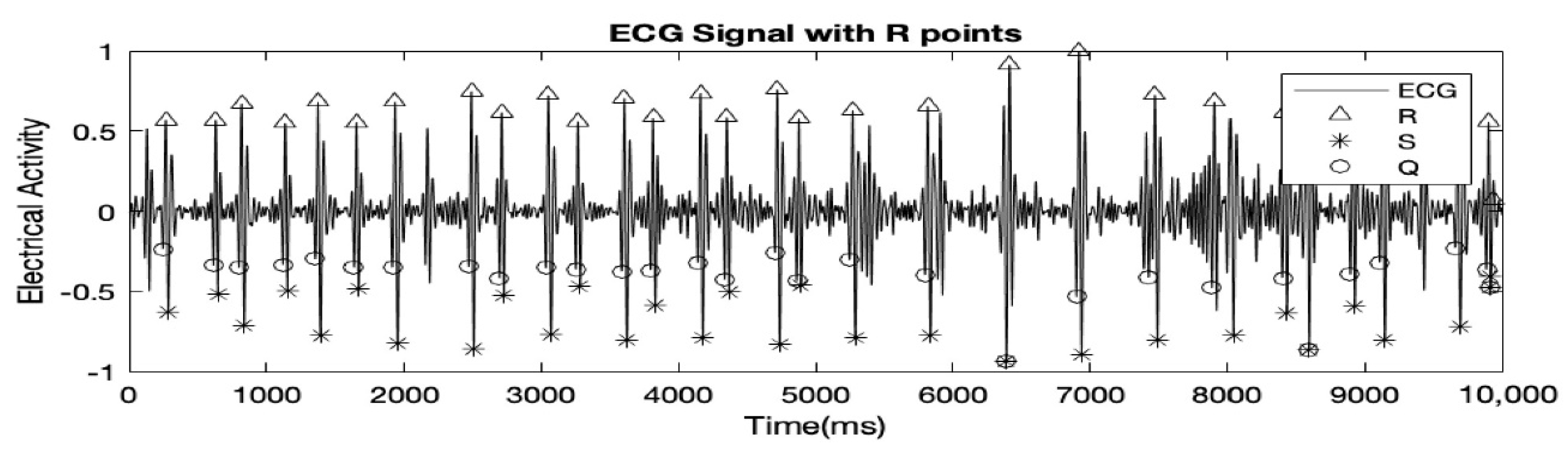
The record used in all the figures was ‘r04_edfm-4’. Table 1 shows that record ‘r01_edfm-1’ and ‘r04_edfm-4’ demonstrates high accuracies of over 95% because in both cases, there was only 1 False Negative (FN) peak and no False Positive (FP) peaks. Record ‘r04_edfm-1’ on the other hand showed a relatively lower accuracy of just over 81% because several FQRS complexes coincided with MQRS complexes resulting in a few of them being ultimately canceled out. As mentioned earlier, each of the records, R01, R04, R07, and R10 had 4 MAECG signals, but we excluded signal accuracies of some of the ECG signals altogether from the table. This is because, in the excluded signals, the FQRS complexes were as prominent and at times even more prominent than the MQRS complexes, leading to many FQRS complexes being falsely detected as MQRS complexes. The signals of the record R10 had high-frequency noise to the point where the MQRS and FQRS complexes were almost indistinguishable. This is demonstrated using Figure 4.
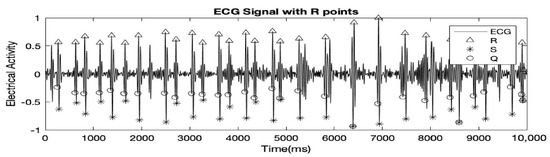
Figure 4.
FQRS complexes also detected as MQRS complexes wrongly.
References
- Behar, J.; Andreotti, F.; Zaunseder, S.; Oster, J.; Clifford, G.D. A practical guide to non-invasive foetal electrocardiogram extraction and analysis. Physiol. Meas. 2016, 37, R1. [Google Scholar] [CrossRef] [PubMed]
- Pieri, J.F.; Crowe, J.A.; Hayes-Gill, B.R.; Spencer, C.J.; Bhogal, K.; James, D.K. Compact long-term recorder for the transabdominal foetal and maternal electrocardiogram. Med. Biol. Eng. Comput. 2001, 39, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimy, M.I.; Ahmed, F.; Ali, M.A.M.; Zahedi, E. Real-Time Signal Processing for Fetal Heart Rate Monitoring. IEEE Trans. Biomed. Eng. 2003, 50, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.A.; Ibrahimy, M.I.; Reaz, M.B.I. Techniques of FECG signal analysis: Detection and processing for fetal monitoring. WIT Trans. Biomed. Health 2007, 12, 295–305. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).