A Review of Non-Linear Optical Imaging Techniques for Cancer Detection
Abstract
1. Introduction
2. Biochemical Imaging
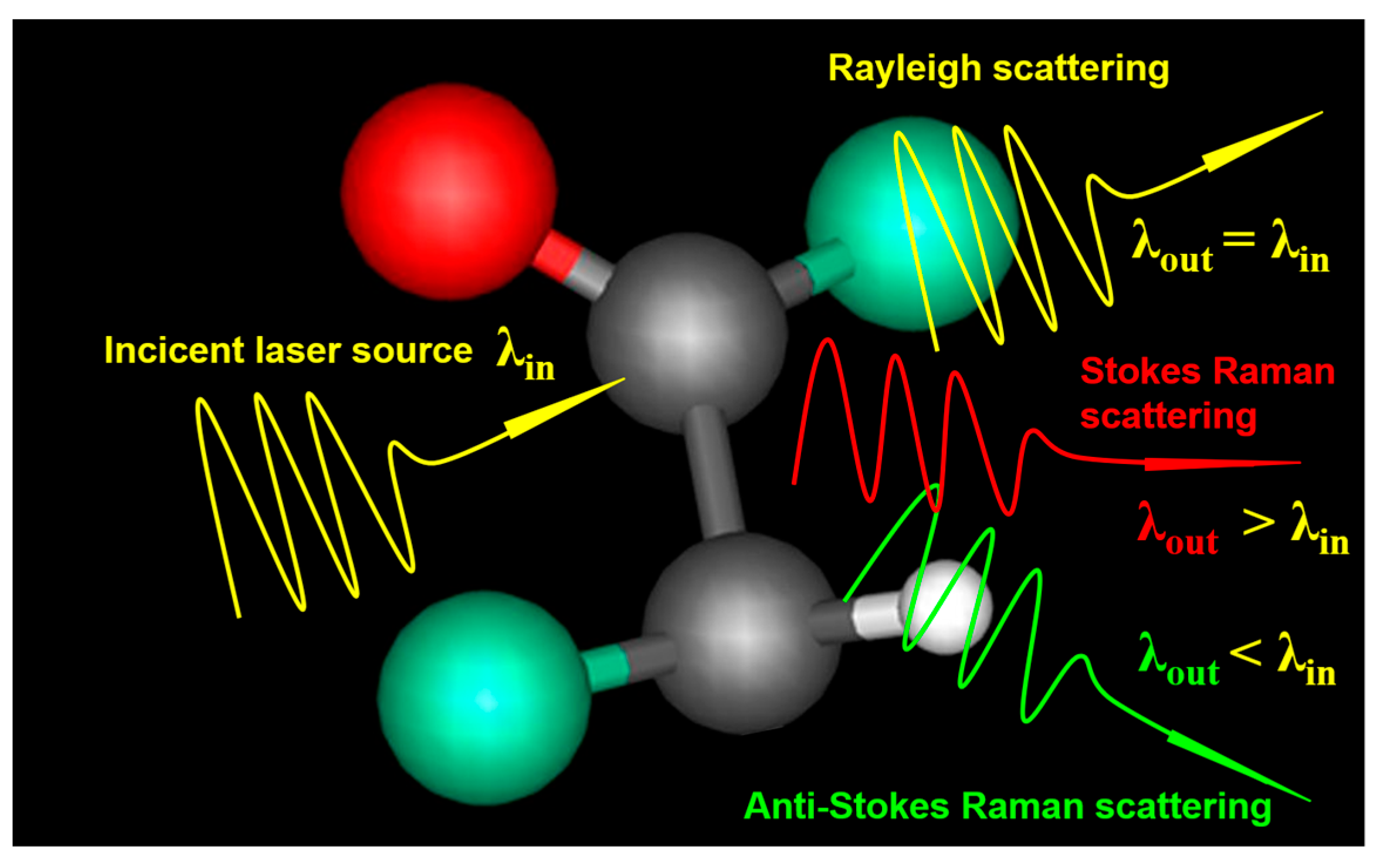
2.1. Spontaneous Raman Scattering
2.1.1. Principle
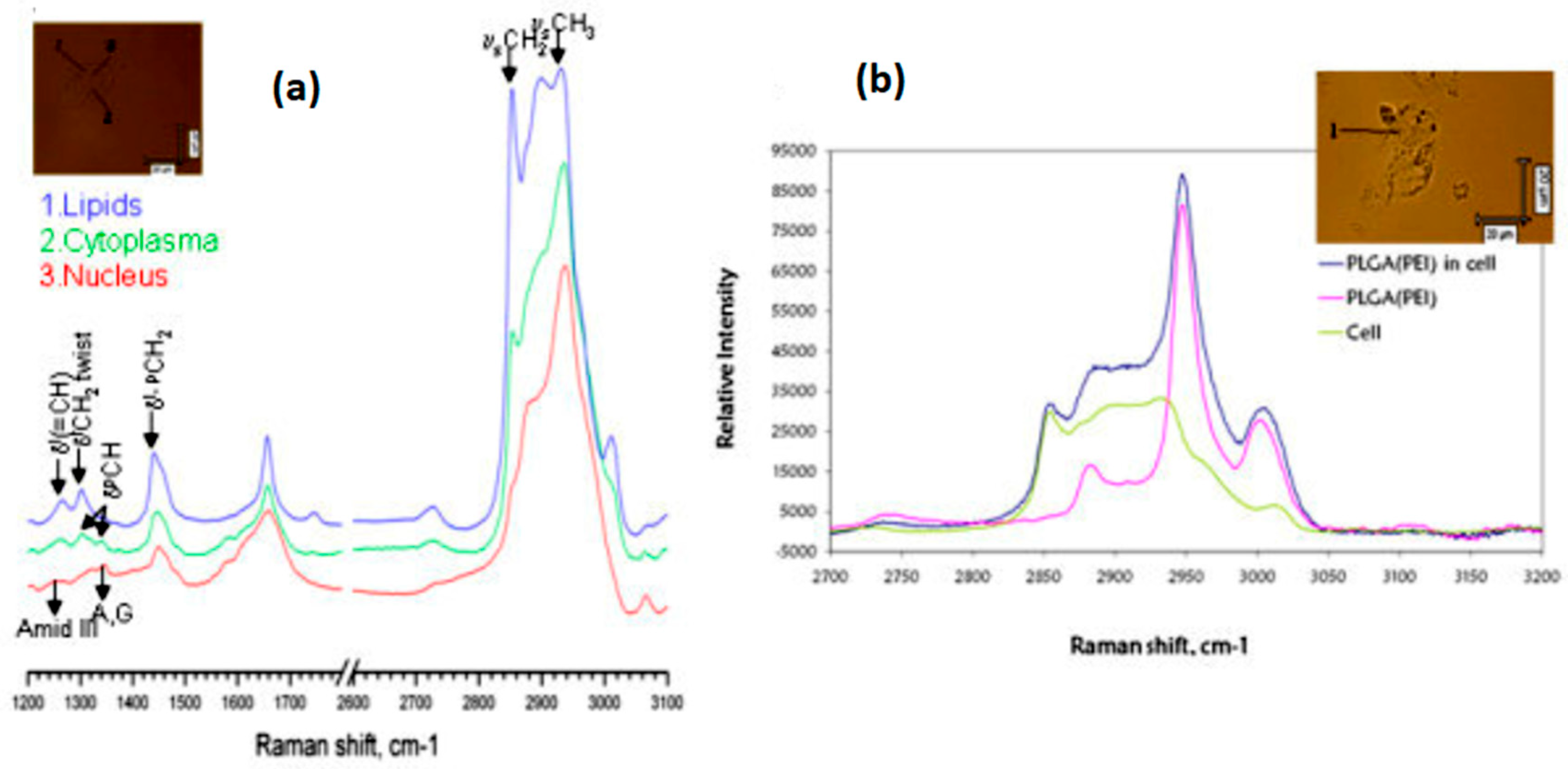
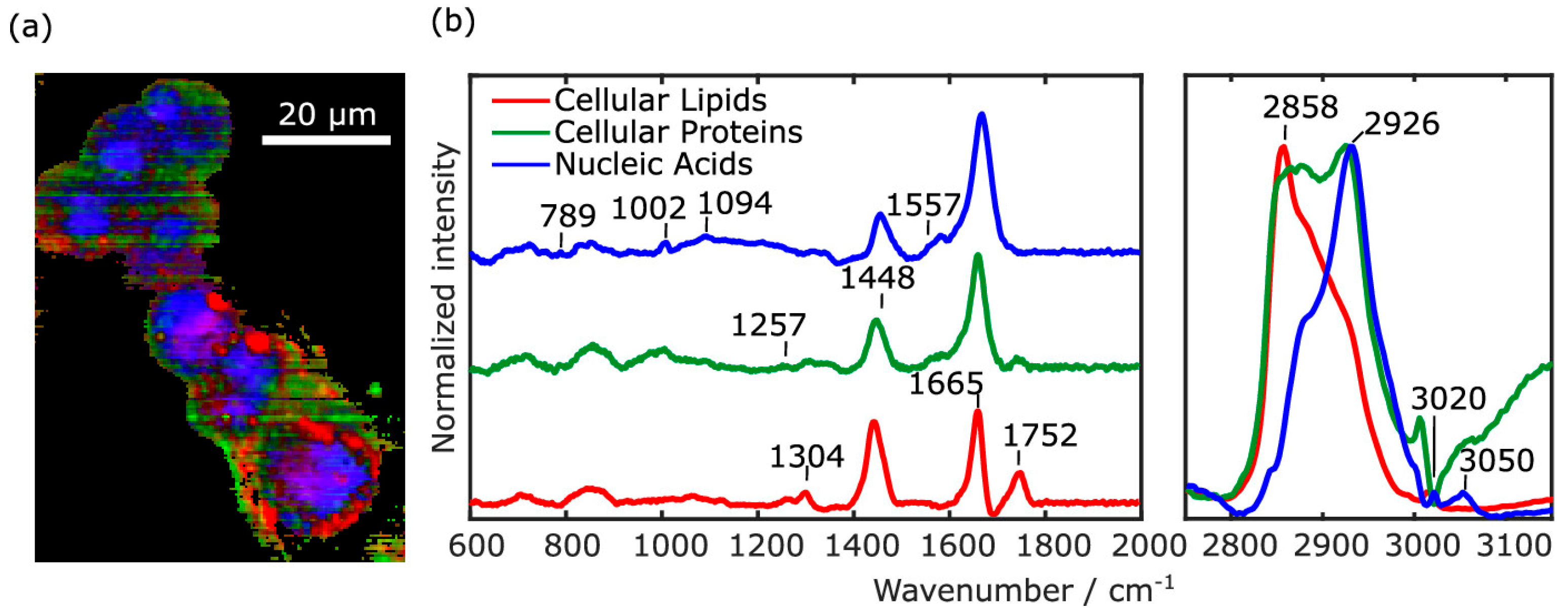
2.1.2. Applications in Cancer Detection
2.2. Stimulated Raman Scattering
2.2.1. Principle
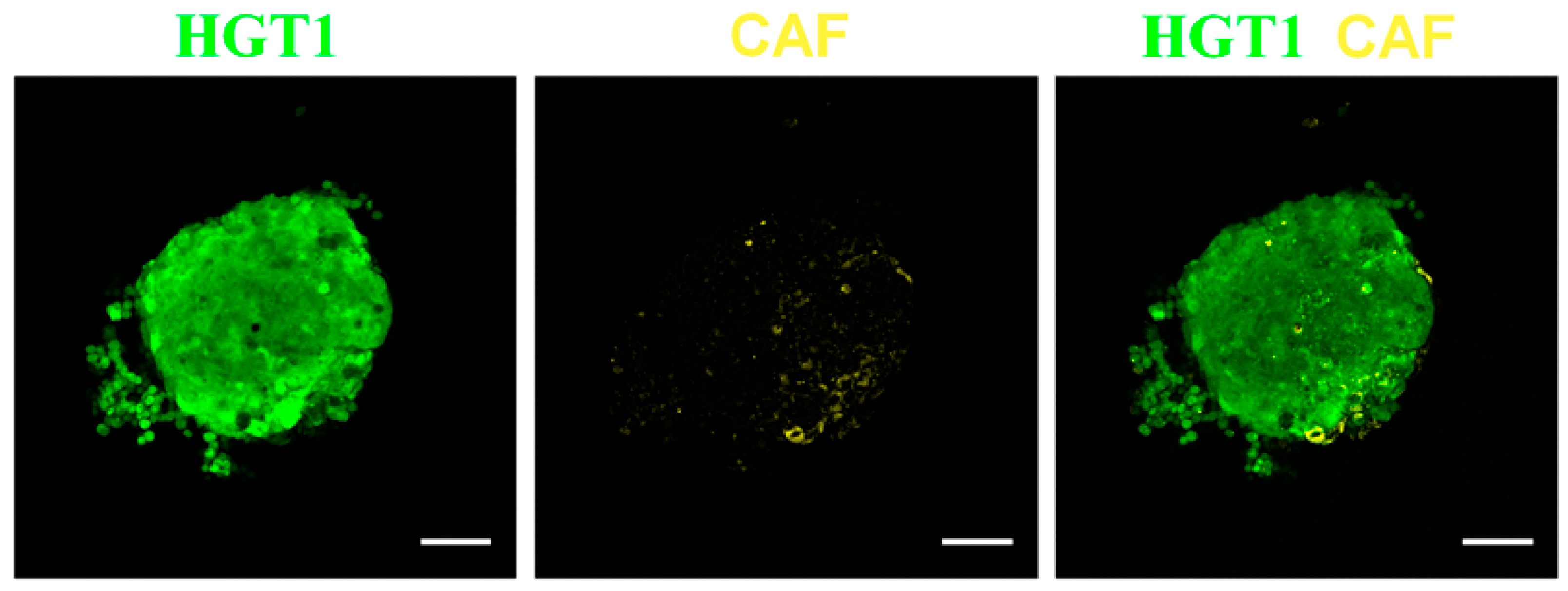
2.2.2. Applications in Cancer Detection
2.3. Coherent Anti-Stokes Raman Scattering
2.3.1. Principle
2.3.2. Applications in Cancer Detection
3. Metabolic Imaging
3.1. Two-Photon Excitation Fluorescence Microscopy
3.1.1. Principle
3.1.2. Applications in Cancer Detection
3.2. Transient Absorption Microscopy
3.2.1. Principle
3.2.2. Applications in Cancer Detection
4. Structural Imaging
4.1. Second Harmonic Generation
4.1.1. Principle
4.1.2. Applications in Cancer Detection
4.2. Third Harmonic Generation
4.2.1. Principle
4.2.2. Applications in Cancer Detection
5. Discussion
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gospodarowicz, M.; O’Sullivan, B. Prognostic factors in cancer. Semin. Surg. Oncol. 2003, 21, 13–18. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman Techniques: Fundamentals and Frontiers. Nanoscale Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Larson, A. Multiphoton microscopy. Nat. Photon. 2011, 5, 1. [Google Scholar] [CrossRef]
- Schenke-Layland, K. Non-invasive multiphoton imaging of extracellular matrix structures. J. Biophotonics 2008, 1, 451–462. [Google Scholar] [CrossRef]
- Hu, F.; Shi, L.; Min, W. Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nat. Methods 2019, 16, 830–842. [Google Scholar] [CrossRef]
- Van Steenbergen, V.; Boesmans, W.; Li, Z.; de Coene, Y.; Vints, K.; Baatsen, P.; Dewachter, I.; Ameloot, M.; Clays, K.; Berghe, P.V. Molecular understanding of label-free second harmonic imaging of microtubules. Nat. Commun. 2019, 10, 3530. [Google Scholar] [CrossRef]
- Szeto, H.H.; Schiller, P.W.; Zhao, K.; Luo, G. Fluorescent dyes alter intracellular targeting and function of cell-penetrating tetrapeptides. FASEB J. 2004, 19, 118–120. [Google Scholar] [CrossRef]
- Jhan, J.-W.; Chang, W.-T.; Chen, H.-C.; Lee, Y.T.; Wu, M.F.; Chen, C.H.; Liau, I. Integrated multiple multi-photon imaging and Raman spectroscopy for charac-terizing structure-constituent correlation of tissues. Opt. Express 2008, 16, 16431–16441. [Google Scholar] [CrossRef]
- Quansah, E.; Shaik, T.A.; Çevik, E.; Wang, X.; Höppener, C.; Meyer-Zedler, T.; Krafft, C. Investigating biochemical and structural changes of glycated collagen using mul-timodal multiphoton imaging, Raman spectroscopy, and atomic force microscopy. Anal. Bioanal. Chem. 2023, 415, 6257–6267. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, T.H.; Kokanyan, N.; Fontana, M.D. Use of Stokes and anti-Stokes Raman scattering for new applications. J. Raman Spectrosc. 2018, 50, 418–424. [Google Scholar] [CrossRef]
- Griffiths, D.J. Introduction to Electrodynamics, 3rd ed.; Pearson Education Dorling Kindersley: Glenview, IL, USA, 2007; ISBN 81-7758-293-3. [Google Scholar]
- Qi, Y.F.; Liu, Y.H.; Liu, D.M. Research progress on application of Raman spectroscopy in tumor diagnosis. Laser Optoelectron. Prog. 2020, 57, 220001. [Google Scholar]
- Ilchenko, O.; Pilhun, Y.; Kutsyk, A. Towards Raman imaging of centimeter scale tissue areas for real-time opto-molecular vis-ualization of tissue boundaries for clinical applications. Light Sci. Appl. 2022, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Auner, G.W.; Koya, S.K.; Huang, C.; Broadbent, B.; Trexler, M.; Auner, Z.; Elias, A.; Curtin Mehne, K.; Brusatori, M.A. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018, 37, 691–717. [Google Scholar] [CrossRef]
- Hollon, T.; Orringer, D.A. Label-free brain tumor imaging using Raman-based methods. J. Neuro-Oncol. 2021, 151, 393–402. [Google Scholar] [CrossRef]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer by using Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 12371–12376. [Google Scholar] [CrossRef]
- Hanna, K.; Krzoska, E.; Shaaban, A.M.; Muirhead, D.; Abu-Eid, R.; Speirs, V. Raman spectroscopy: Current applications in breast cancer diagnosis, challenges and future prospects. Br. J. Cancer 2021, 126, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.P.; Barroso, E.M.; Bakker Schut, T.C.; Caspers, P.J.; van Lanschot, C.G.F.; Choi, D.-H.; van der Kamp, M.F.; Smits, R.W.H.; van Doorn, R.; Verdijk, R.M.; et al. Raman spectroscopy for cancer detection and cancer surgery guidance: Translation to the clinics. Analyst 2017, 142, 3025–3047. [Google Scholar] [CrossRef]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of tumors during tissue-conserving surgery with integrated autofluo-rescence and Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar]
- Xu, J.; Yu, T.; Zois, C.E.; Cheng, J.-X.; Tang, Y.; Harris, A.L.; Huang, W.E. Unveiling Cancer Metabolism through Spontaneous and Coherent Raman Spectroscopy and Stable Isotope Probing. Cancers 2021, 13, 1718. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Rojas, E.; Estrela-Lopis, I.; Donath, E.; Moya, S.E. Spontaneous confocal Raman microscopy--a tool to study the uptake of nanopar-ticles and carbon nanotubes into cells. Nanoscale Res. Lett. 2011, 6, 429. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, P.; Kovalev, A.; Volkmer, A. Vibrational imaging based on stimulated Raman scattering microscopy. New J. Phys. 2009, 11, 033026. [Google Scholar] [CrossRef]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-free biomedical imaging with high sensitivity by stimulated Raman scat-tering microscopy. Science 2008, 322, 1857–1861. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, X.; Chi, M.; Wu, Y.; Zhang, M.; Gao, S. Spontaneous-stimulated Raman co-localization dual-modal analysis approach for efficient identification of tumor cells. Talanta 2024, 277, 126297. [Google Scholar] [CrossRef]
- Ni, H.; Dessai, C.P.; Lin, H.; Wang, W.; Chen, S.; Yuan, Y.; Ge, X.; Ao, J.; Vild, N.; Cheng, J.-X. High-content stimulated Raman histology of human breast cancer. Theranostics 2024, 14, 1361–1370. [Google Scholar] [CrossRef]
- Ji, M.; Lewis, S.; Camelo-Piragua, S.; Ramkissoon, S.H.; Snuderl, M.; Venneti, S.; Fisher-Hubbard, A.; Garrard, M.; Fu, D.; Wang, A.C.; et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci. Transl. Med. 2015, 7, 309ra163. [Google Scholar] [CrossRef]
- Saar, B.G.; Freudiger, C.W.; Reichman, J.; Stanley, C.M.; Holtom, G.R.; Xie, X.S. Video-Rate Molecular imaging in vivo with stimulated raman scattering. Science 2010, 330, 1368–1370. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, D.; von Spreckelsen, N.; Mawrin, C.; Ion-Margineanu, A.; Fürtjes, G.; Jünger, S.T.; Khalid, F.; Freudiger, C.W.; Timmer, M.; Ruge, M.I.; et al. Novel rapid intraoperative qualitative tumor detection by a residual con-volutional neural network using label-free stimulated Raman scattering microscopy. Acta Neuropathol. Commun. 2022, 10, 109. [Google Scholar] [CrossRef]
- Evans, C.L.; Xie, X.S. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909. [Google Scholar] [CrossRef]
- Zumbusch, A.; Holtom, G.R.; Xie, X.S. Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Phys. Rev. Lett. 1999, 82, 4142–4145. [Google Scholar] [CrossRef]
- Gachet, D.; Billard, F.; Rigneault, H. Background-free coherent anti-Stokes Raman spectroscopy near transverse interfaces: A vectorial study. J. Opt. Soc. Am. B 2008, 25, 1655–1666. [Google Scholar] [CrossRef]
- Heuke, S.; Rigneault, H. Coherent Stokes Raman scattering microscopy (CSRS). Nat. Commun. 2023, 14, 3337. [Google Scholar] [CrossRef] [PubMed]
- Vito, G.M.; Bifone, A.; Piazza, V. Rotating-polarization CARS microscopy: Combining chemical and molecular orientation sen-sitivity. Opt. Express 2012, 20, 29369–29377. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Yi, R.; Liu, L.; Qu, J. Coherent Anti-Stokes Raman Scattering Microscopy and Its Applications. Front. Phys. 2020, 8, 598420. [Google Scholar] [CrossRef]
- McCullagh, E.A.; Poleg, S.; Stich, D.; Moldovan, R.; Klug, A. Coherent Anti-Stokes Raman Spectroscopy (CARS) Application for Imaging Myelination in Brain Slices. J. Vis. Exp. 2022, 185, e64013. [Google Scholar] [CrossRef]
- Le, T.T.; Huff, T.B.; Cheng, J.-X. Coherent anti-Stokes Raman scattering imaging of lipids in cancer metastasis. BMC Cancer 2009, 9, 42. [Google Scholar] [CrossRef]
- Mitra, R.; Chao, O.; Urasaki, Y.; Goodman, O.B.; Le, T.T. Detection of lipid-rich prostate circulating tumour cells with coherent anti-stokes raman scattering microscopy. BMC Cancer 2012, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhao, W.; Yue, S. Coherent Raman Scattering Microscopy in Oncology Pharmacokinetic Research. Front. Pharmacol. 2021, 12, 630167. [Google Scholar] [CrossRef]
- Vernuccio, F.; Vanna, R.; Ceconello, C.; Bresci, A.; Manetti, F.; Sorrentino, S.; Ghislanzoni, S.; Lambertucci, F.; Motiño, O.; Martins, I.; et al. Full-Spectrum CARS Microscopy of Cells and Tissues with Ultrashort White-Light Continuum Pulses. J. Phys. Chem. B 2023, 127, 4733–4745. [Google Scholar] [CrossRef]
- Williams, R.M.; Zipfel, W.R.; Webb, W.W. Multiphoton microscopy in biological research. Curr. Opin. Chem. Biol. 2001, 5, 603–608. [Google Scholar] [CrossRef]
- Stringari, C.; Abdeladim, L.; Malkinson, G.; Mahou, P.; Solinas, X.; Lamarre, I.; Brizion, S.; Galey, J.-B.; Supatto, W.; Legouis, R.; et al. Multicolor two-photon imaging of endogenous fluorophores in living tissues by wavelength mixing. Sci. Rep. 2017, 7, 3792. [Google Scholar] [CrossRef] [PubMed]
- Vergen, J.; Hecht, C.; Zholudeva, L.V.; Marquardt, M.M.; Hallworth, R.; Nichols, M.G. Metabolic imaging using two-photon excited NADH intensity and fluorescence lifetime imaging. Microsc. Microanal. 2012, 18, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Centonze, V.E.; White, J.G. Multiphoton excitation provides optical sections from deeper within scattering specimens than Confocal imaging. Biophys. J. 1998, 75, 2015–2024. [Google Scholar] [CrossRef]
- Denk, W.; Strickler, J.H.; Webb, W.W. Two-Photon Laser Scanning Fluorescence Microscopy. Science 1990, 248, 73–76. [Google Scholar] [CrossRef]
- Helmchen, F.; Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2005, 2, 932–940. [Google Scholar] [CrossRef]
- Denk, W.; Delaney, K.; Gelperin, A.; Kleinfeld, D.; Strowbridge, B.; Tank, D.; Yuste, R. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. J. Neurosci. Methods 1994, 54, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Bousso, P.; Bhakta, N.R.; Lewis, R.S.; Robey, E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon mi-croscopy. Science 2002, 296, 1876–1880. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, S.; Shi, M.; Fan, J.; Xie, J.; Xiao, G.; Yu, L.; Wu, J.; Dai, Q. Long-term intravital subcellular imaging with confocal scanning light-field microscopy. Nat. Biotechnol. 2024, 38802562. [Google Scholar] [CrossRef]
- Jun, L.I.U. Two-photon microscopy in pre-clinical and clinical cancer research. Front. Optoelectron. 2015, 8, 141–151. [Google Scholar]
- Alzeeb, G.; Dubreuil, M.; Arzur, D.; Rivet, S.; Corcos, L.; Le Grand, Y.; Le Jossic-Corcos, C. Gastric cancer multicellular spheroid analysis by two-photon microscopy. Biomed. Opt. Express 2022, 13, 3120–3130. [Google Scholar] [CrossRef] [PubMed]
- Noh, C.-K.; Lim, C.S.; Lee, G.H.; Cho, M.K.; Lee, H.W.; Roh, J.; Kim, Y.B.; Lee, E.; Park, B.; Kim, H.M.; et al. A Diagnostic Method for Gastric Cancer Using Two-Photon Microscopy with Enzyme-Selective Fluorescent Probes: A Pilot Study. Front. Oncol. 2021, 11, 634219. [Google Scholar] [CrossRef]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-associated fibroblasts: From basic science to anticancer therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef]
- Franchi-Mendes, T.; Lopes, N.; Brito, C. Heterotypic Tumor Spheroids in Agitation-Based Cultures: A Scaffold-Free Cell Model That Sustains Long-Term Survival of Endothelial Cells. Front. Bioeng. Biotechnol. 2021; 9, 649949. [Google Scholar]
- Li, L.; Hong, S.; Kang, D.; Huang, X.; Zhang, S.; Zhan, Z.; Zhou, Y.; Chen, J. Two-photon imaging reveals histopathological changes in gastric tumor microenvironment induced by neoadjuvant treatment. Biomed. Opt. Express 2023, 14, 5085–5096. [Google Scholar] [CrossRef]
- Hong, S.T.; Kim, T.H.; Choi, J.W.; Park, S.J.; Kwon, S.A.; Paik, K.C.; Han, M.S.; Kim, E.S.; Chun, H.J.; Heo, J.N.; et al. Two-Photon Probes for pH: Detection of Human Colon Cancer using Two-Photon Microscopy. Anal Chem. 2017, 89, 9830–9835. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, S.; Knap, B.; Przystupski, D.; Saczko, J.; Kędzierska, E.; Knap-Czop, K.; Kotlińska, J.; Michel, O.; Kotowski, K.; Kulbacka, J. Photodynamic therapy-mechanisms, photosensitizers and combinations. Biomed. Pharmacother. 2018, 106, 1098–1107. [Google Scholar] [CrossRef]
- Benninger, R.K.; Piston, D.W. Two-photon excitation microscopy for the study of living cells and tissues. Curr. Protoc. Cell Biol. 2013, 59, 4.11.1–4.11.24. [Google Scholar] [CrossRef]
- Juvekar, V.; Lee, D.J.; Park, T.G.; Samanta, R.; Kasar, P.; Kim, C.; Rotermund, F.; Kim, H.M. Two-photon excitation photosensitizers for photodynamic therapy: From small-molecules to nano-complex systems. Coord. Chem. Rev. 2024, 506, 215711. [Google Scholar] [CrossRef]
- Jung, H.S.; Kim, H.J. Definitive surgery and intraoperative photodynamic therapy for locally advanced non-small cell lung cancer: A case report. World J. Surg. Oncol. 2022, 20, 265. [Google Scholar] [CrossRef]
- Rodrigues, J.A.; Correia, J.H. Photodynamic Therapy for Colorectal Cancer: An Update and a Look to the Future. Int. J. Mol. Sci. 2023, 24, 12204. [Google Scholar] [CrossRef]
- Xu, C.; Law, S.K.; Leung, A.W.N. Comparison of the Differences between Two-Photon Excitation, Upconversion, and Con-ventional Photodynamic Therapy on Cancers in In Vitro and In Vivo Studies. Pharmaceuticals 2024, 17, 663. [Google Scholar] [CrossRef] [PubMed]
- Starkey, J.R.; Rebane, A.K.; Drobizhev, M.A.; Meng, F.; Gong, A.; Elliott, A.; McInnerney, K.; Spangler, C.W. New Two-Photon Activated Photodynamic Therapy Sensitizers Induce Xenograft Tumor Regressions after Near-IR Laser Treatment through the Body of the Host Mouse. Clin. Cancer Res. 2008, 14, 6564–6573. [Google Scholar] [CrossRef] [PubMed]
- Bower, A.J.; Li, J.; Chaney, E.J.; Marjanovic, M.; Spillman, D.R.; Boppart, S.A. High-speed imaging of transient metabolic dynamics using two-photon fluorescence lifetime imaging microscopy. Optica 2018, 5, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Ranawat, H.; Pal, S.; Mazumder, N. Recent trends in two-photon auto-fluorescence lifetime imaging (2P-FLIM) and its biomedical applications. Biomed. Eng. Lett. 2019, 9, 293–310. [Google Scholar] [CrossRef]
- Liang, W.; Chen, D.; Guan, H.; Park, H.C.; Li, K.; Li, A.; Li, M.J.; Gannot, I.; Li, X. Label-Free Metabolic Imaging In Vivo by Two-Photon Fluorescence Lifetime Endomi-croscopy. ACS Photonics 2022, 9, 4017–4029. [Google Scholar] [CrossRef]
- Karrobi, K.; Tank, A.; Fuzail, M.A.; Kalidoss, M.; Tilbury, K.; Zaman, M.; Ferruzzi, J.; Roblyer, D. Fluorescence Lifetime Imaging Microscopy (FLIM) reveals spatial-metabolic changes in 3D breast cancer spheroids. Sci. Rep. 2023, 13, 3624. [Google Scholar] [CrossRef]
- Sundström, V. Femtobiology. Annu. Rev. Phys. Chem. 2008, 59, 53–77. [Google Scholar] [CrossRef]
- Berera, R.; van Grondelle, R.; Kennis, J.T.M. Ultrafast transient absorption spectroscopy: Principles and application to photosynthetic systems. Photosynth. Res. 2009, 101, 105–118. [Google Scholar] [CrossRef]
- Zhu, Y.; Cheng, J.-X. Transient absorption microscopy: Technological innovations and applications in materials science and life science. J. Chem. Phys. 2020, 152, 020901. [Google Scholar] [CrossRef]
- Wang, E.; Whitcomb, L.A.; Chicco, A.J.; Wilson, J.W. Transient absorption spectroscopy and imaging of redox in muscle mitochondria. Biomed. Opt. Express 2022, 13, 2103–2116. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Wang, Z.; Shao, M.; Zhang, C.; Chen, X.; Sun, J.; Kwok, R.T.K.; Lam, J.W.Y.; Tang, B.Z. Study of transient absorption spectroscopy of a D–π–A structure aggregation-induced emission luminogen and its photodynamic therapy application. J. Mater. Chem. B 2024, 12, 8349–8356. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Luo, Z.; Liu, X.; Huang, Z. Unveiling the Spatiotemporal and Dose Responses within a Single Live Cancer Cell to Pho-toswitchable Upconversion Nanoparticle Therapeutics Using Hybrid Hyperspectral Stimulated Raman Scattering and Transient Absorption Microscopy. Anal. Chem. 2024, 96, 6148–6157. [Google Scholar] [CrossRef]
- Tanzer, M.L. Current concepts of extracellular matrix. J. Orthop. Sci. 2006, 11, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Rozario, T.; DeSimone, D.W. The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol. 2010, 341, 126–140. [Google Scholar] [CrossRef]
- James, D.S.; Campagnola, P.J. Recent Advancements in Optical Harmonic Generation Microscopy: Applications and Perspectives. BME Front. 2021, 2021, 3973857. [Google Scholar] [CrossRef]
- LaComb, R.; Nadiarnykh, O.; Townsend, S.S.; Campagnola, P.J. Phase Matching Considerations in Second Harmonic Generation from Tissues: Effects on Emission Directionality, Conversion Efficiency and Observed Morphology. Opt. Commun. 2007, 281, 1823–1832. [Google Scholar] [CrossRef]
- Boyd, R.W. Nonlinear Optics; Academic Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Williams, R.M.; Zipfel, W.R.; Webb, W.W. Interpreting second-harmonic generation images of collagen I fibrils. Biophys. J. 2005, 88, 1377–1386. [Google Scholar] [CrossRef] [PubMed]
- Weigelin, B.; Bakker, G.-J.; Friedl, P. Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci. 2016, 129, 245–255. [Google Scholar] [CrossRef]
- Farrar, M.J.; Wise, F.W.; Fetcho, J.R.; Schaffer, C.B. In Vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys. J. 2011, 100, 1362–1371. [Google Scholar] [CrossRef]
- Popova, N.V.; Jücker, M. The Functional Role of Extracellular Matrix Proteins in Cancer. Cancers 2022, 14, 238. [Google Scholar] [CrossRef]
- Kular, J.K.; Basu, S.; Sharma, R.I. The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J. Tissue Eng. 2014, 5, 2041731414557112. [Google Scholar] [CrossRef]
- Aghigh, A.; Bancelin, S.; Rivard, M.; Pinsard, M.; Ibrahim, H.; Légaré, F. Second harmonic generation microscopy: A powerful tool for bio-imaging. Biophys. Rev. 2023, 15, 43–70. [Google Scholar] [CrossRef] [PubMed]
- Mohler, W.; Millard, A.C.; Campagnola, P.J. Second harmonic generation imaging of endogenous structural proteins. Methods 2003, 29, 97–109. [Google Scholar] [CrossRef]
- Jiang, J.; Yuste, R. Second-Harmonic generation imaging of membrane potential with photon counting. Microsc. Microanal. 2008, 14, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.J.; Morrison, J.L.; Botting, K.J.; Kee, T.W. Second-harmonic generation and two-photon-excited autofluorescence microscopy of cardiomyocytes: Quantification of cell volume and myosin filaments. J. Biomed. Opt. 2008, 13, 064018. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jin, Y.; Liu, H.; Wang, F.; Deng, X. The forward and backward second-harmonic generation from crystallized collagen fibre with tightly focused linearly polarized beams. J. Opt. 2012, 14, 055301. [Google Scholar] [CrossRef]
- Keikhosravi, A.; Bredfeldt, J.S.; Sagar, A.K.; Eliceiri, K.W. Second-harmonic generation imaging of cancer. Methods Cell Biol. 2014, 123, 531–546. [Google Scholar]
- Mierke, C.T. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Rep Prog Phys. 2019, 82, 064602. [Google Scholar] [CrossRef]
- Song, K.; Yu, Z.; Zu, X.; Li, G.; Hu, Z.; Xue, Y. Collagen Remodeling along Cancer Progression Providing a Novel Opportunity for Cancer Diagnosis and Treatment. Int. J. Mol. Sci. 2022, 23, 10509. [Google Scholar] [CrossRef]
- Desa, D.E.; Wu, W.; Brown, R.M.; Brown, E.B., 4th; Hill, R.L.; Turner, B.M.; Brown, E.B., 3rd. Second-Harmonic Generation Imaging Reveals Changes in Breast Tumor Collagen Induced by Neoadjuvant Chemotherapy. Cancers 2022, 14, 857. [Google Scholar] [CrossRef]
- Venning, F.A.; Wullkopf, L.; Erler, J.T. Targeting ECM Disrupts Cancer Progression. Front. Oncol. 2015, 5, 224. [Google Scholar] [CrossRef] [PubMed]
- Nejim, Z.; Navarro, L.; Morin, C.; Badel, P. Quantitative analysis of second harmonic generated images of collagen fibers: A review. Res. Biomed. Eng. 2022, 39, 273–295. [Google Scholar] [CrossRef]
- Ávila, F.J.; Bueno, J.M. Analysis and quantification of collagen organization with the structure tensor in second harmonic microscopy images of ocular tissues. Appl. Opt. 2015, 54, 9848–9854. [Google Scholar] [CrossRef] [PubMed]
- Bueno, J.M.; Ávila, F.J.; Hristu, R.; Stanciu, S.G.; Eftimie, L.; Stanciu, G.A. Objective analysis of collagen organization in thyroid nodule capsules using second harmonic generation microscopy images and the Hough transform. Appl. Opt. 2020, 59, 6925. [Google Scholar] [CrossRef]
- Alizadeh, M.; Ghotbi, M.; Loza-Alvarez, P.; Merino, D. Comparison of Different Polarization Sensitive Second Harmonic Generation Imaging Techniques. Methods Protoc. 2019, 2, 49. [Google Scholar] [CrossRef]
- Yu, W.; Li, X.; Wang, B.; Li, Y.; Shen, B.; Qu, J.; Liu, L. Nonlinear polarization tensor measurement with a vectorial complex field in second-harmonic-generation microscopy. Phys. Rev. A 2023, 107, 013505. [Google Scholar] [CrossRef]
- Golaraei, A.; Kontenis, L.; Cisek, R.; Tokarz, D.; Done, S.J.; Wilson, B.C.; Barzda, V. Changes of collagen ultrastructure in breast cancer tissue determined by second-harmonic generation double Stokes-Mueller polarimetric microscopy. Biomed. Opt. Express 2016, 7, 4054–4068. [Google Scholar] [CrossRef]
- Tokarz, D.; Cisek, R.; Joseph, A.; Asa, S.L.; Wilson, B.C.; Barzda, V. Characterization of pathological thyroid tissue using polarization-sensitive second harmonic generation microscopy. Mod. Pathol. 2020, 100, 1280–1287. [Google Scholar] [CrossRef]
- Hompland, T.; Erikson, A.; Lindgren, M.; Lindmo, T.; Davies, C.d.L. Second-Harmonic Generation in Collagen as a Potential Cancer Diagnostic Parameter. J. Biomed. Opt. 2008, 13, 054050. [Google Scholar] [CrossRef]
- Rouède, D.; Schaub, E.; Bellanger, J.-J.; Ezan, F.; Scimeca, J.-C.; Baffet, G.; Tiaho, F. Determination of Extracellular Matrix Collagen Fibril Architectures and Pathological Remodeling by Polarization Dependent Second Harmonic Microscopy. Sci. Rep. 2017, 7, 12197. [Google Scholar] [CrossRef]
- Ambekar, R.; Lau, T.-Y.; Walsh, M.; Bhargava, R.; Toussaint, K.C. Quantifying Collagen Structure in Breast Biopsies Using Second-Harmonic Generation Imaging. Biomed. Opt. Express 2012, 3, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Pan, S.; Zheng, W.; Huang, Z. Polarization-Resolved Second-Harmonic Generation Imaging for Liver Fibrosis Assessment without Labeling. Appl. Phys. Lett. 2013, 103, 173701. [Google Scholar] [CrossRef]
- Watanabe, T.; Thayil, A.; Jesacher, A.; Grieve, K.; Debarre, D.; Wilson, T.; Booth, M.; Srinivas, S. Characterisation of the dynamic behaviour of lipid droplets in the early mouse embryo using adaptive harmonic generation microscopy. BMC Cell Biol. 2010, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Müller; Squier; Wilson; Brakenhoff 3D microscopy of transparent objects using third-harmonic generation. J. Microsc. 1998, 191, 266–274. [CrossRef] [PubMed]
- Chu, S.W.; Chen, S.Y.; Chern, G.W.; Tsai, T.H.; Chen, Y.C.; Lin, B.L.; Sun, C.K. Studies of chi(2)/chi(3) tensors in submicron-scaled bio-tissues by polarization harmonics optical microscopy. Biophys. J. 2004, 86, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Gavgiotaki, E.; Filippidis, G.; Tsafas, V.; Bovasianos, S.; Kenanakis, G.; Georgoulias, V.; Tzardi, M.; Agelaki, S.; Athanassakis, I. Third Harmonic Generation microscopy distinguishes malignant cell grade in human breast tissue biopsies. Sci. Rep. 2020, 10, 11055. [Google Scholar] [CrossRef]
- Gavgiotaki, E.; Filippidis, G.; Markomanolaki, H.; Kenanakis, G.; Agelaki, S.; Georgoulias, V.; Athanassakis, I. Distinction between breast cancer cell subtypes using third harmonic generation microscopy. J. Biophotonics 2016, 10, 1152–1162. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Ghader, A.; Asgari, H.; Keshavarz, M.; Tazehmahalleh, F.E.; Ara, M.H.M.; Malekzadeh, M.; Ghaznavi, H.; Shakeri-Zadeh, A. The capability of nonlinear optical characteristics as a predictor for cellular uptake of nanoparticles and cell damage. Photodiagn. Photodyn. Ther. 2019, 27, 442–448. [Google Scholar] [CrossRef]
- Combes, G.F.; Vučković, A.-M.; Bakulić, M.P.; Antoine, R.; Bonačić-Koutecky, V.; Trajković, K. Nanotechnology in Tumor Biomarker Detection: The Potential of Liganded Nanoclusters as Nonlinear Optical Contrast Agents for Molecular Diagnostics of Cancer. Cancers 2021, 13, 4206. [Google Scholar] [CrossRef]
- Sun, L.; Liu, H.; Ye, Y.; Lei, Y.; Islam, R.; Tan, S.; Tong, R.; Miao, Y.-B.; Cai, L. Smart nanoparticles for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 418. [Google Scholar] [CrossRef]
- Aneesa, V.M.; Safna, K.P.; Hussan, S.; Lekshmi, S.; Babu, T.D.; Muraleedharan, K. Analysis of non-linear optical properties of phytochemical photosensitizers in cancer photodynamic therapy by quantum computational. Results Chem. 2024, 8, 101580. [Google Scholar] [CrossRef]
- Hoque, E.; Biswas, M.K.; Hossain, M.; Bhattacharja, S.; Hasan, K.M.E.; Sharafuddin, S.M.; Das, S.K.; Haque, Y. Nonlinear optical phase shift in blood plasmas for neoplasia diagnosis. Opt. Express 2023, 31, 23056–23065. [Google Scholar] [CrossRef] [PubMed]
- Ghader, A.; Behruzi, M.; Ara, M.H.M.; Ghaznavi, H.; Ardakani, A.A. Nonlinear optical response of cancer cells following conventional and nano-technology based treatment strategies: Results of chemo-, thermo- and radiation therapies. Photodiagn. Photodyn. Ther. 2021, 37, 102686. [Google Scholar] [CrossRef] [PubMed]

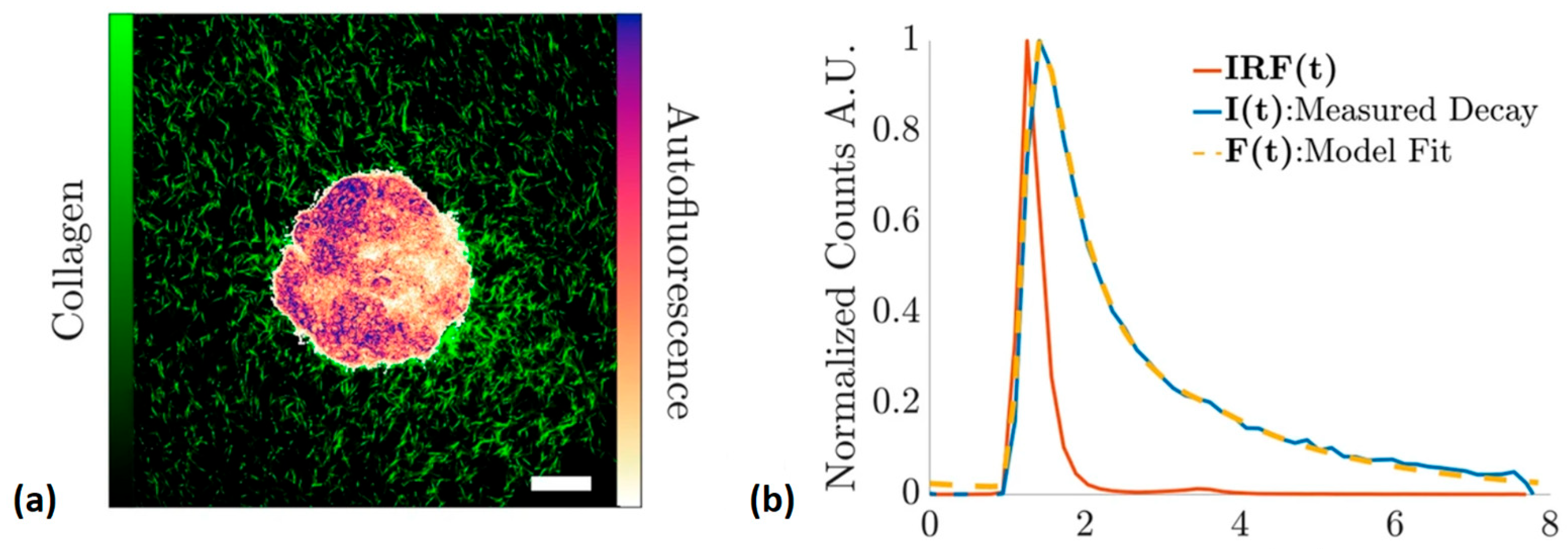
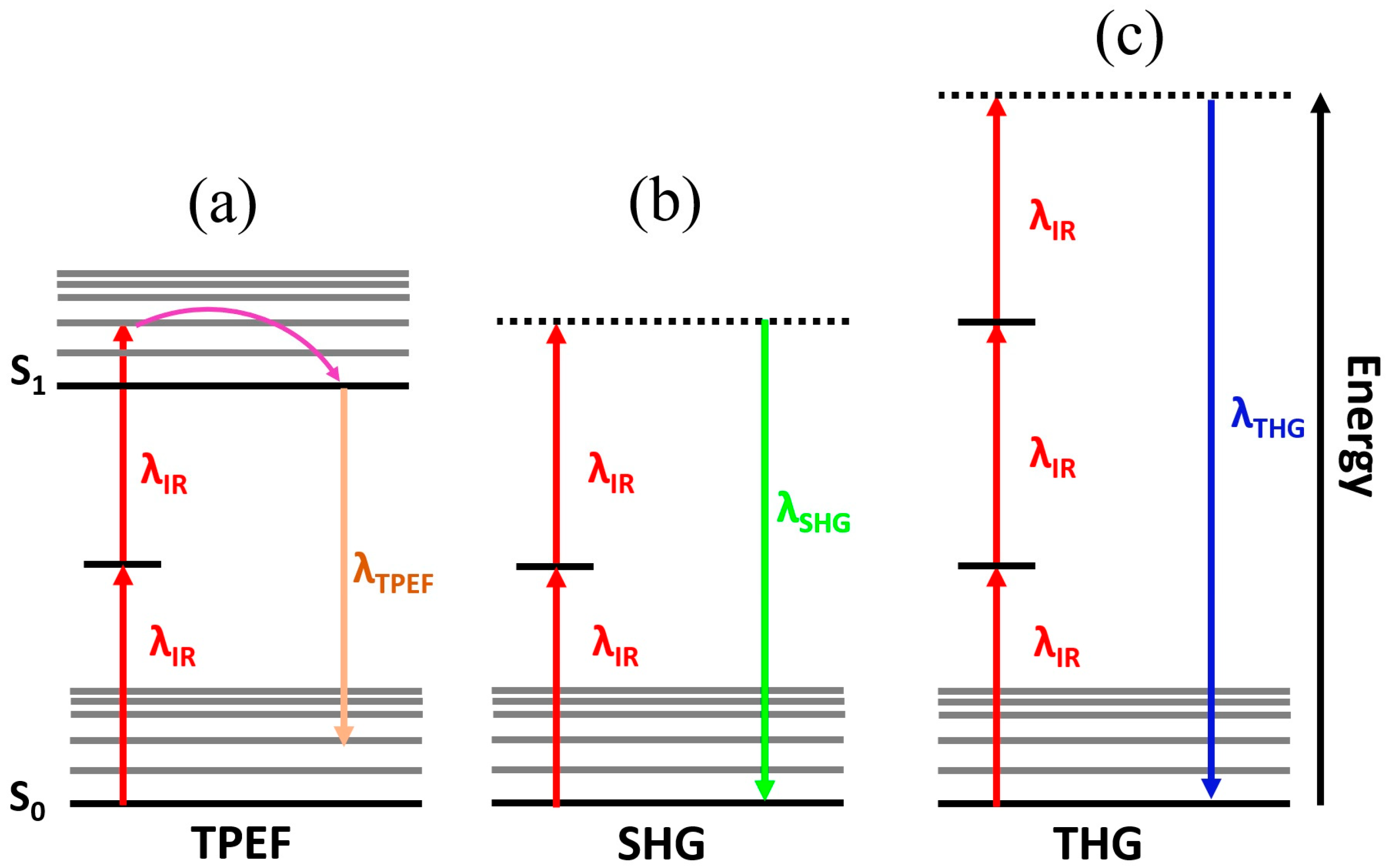
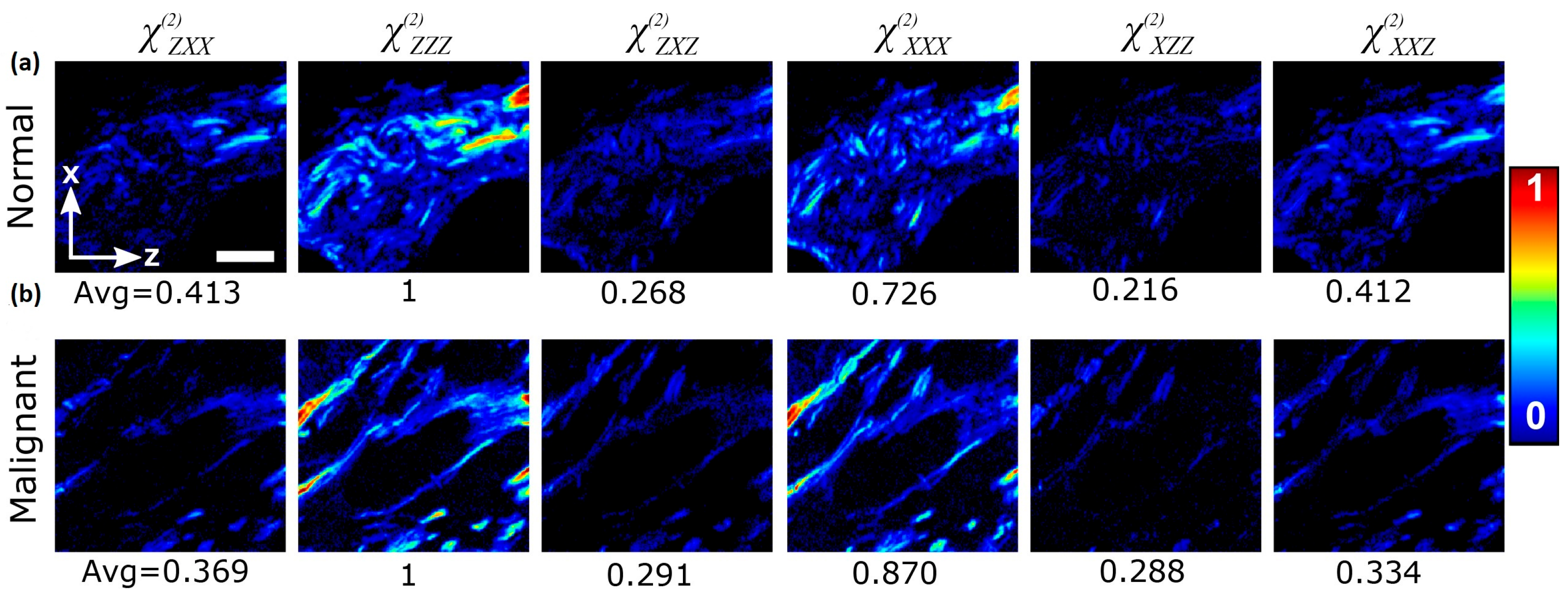

| Technique | Principle | Contrast Mechanism | Applications | Key Features |
|---|---|---|---|---|
| RS | Inelastic scattering of light, where energy is transferred to molecular vibrations. | Chemical composition based on vibrational modes. | Chemical imaging, material science, cancer diagnostics. |
|
| SRS | Enhanced Raman signal by stimulating Raman-active modes with pump and probe lasers. | Chemical composition based on vibrational modes | Fast imaging of live cells, tissue imaging. |
|
| CARS | Non-linear process where anti-Stokes shifted light is generated, enhancing Raman signal. | Chemical composition based on vibrational modes | Label-free imaging, lipid mapping, biomedical imaging. |
|
| TPEF | Non-linear excitation where two photons are absorbed simultaneously to excite a fluorophore. | Fluorescence emission from excited states | Deep tissue imaging, neuroscience, cellular imaging. |
|
| FLIM | Measures the decay time of fluorescence after two-photon excitation. | Temporal decay of fluorescence signal, independent of intensity. | Cellular metabolism, protein interactions, cancer diagnostics. |
|
| TAM | Measures the absorption of a probe pulse following excitation by a pump pulse. | Differential absorption signal revealing electronic and vibrational dynamics. | Study of ultrafast dynamics, excited state lifetimes, and charge transfer in materials and biological systems. |
|
| SHG | Non-linear optical process where two photons combine to form a single photon with twice the energy. | Structural information from non-centrosymmetric molecules (e.g., collagen). | Imaging of collagen, muscle fibers, and other non-centrosymmetric structures. |
|
| THG | Non-linear optical process where three photons combine to form a single photon with three times the energy. | Interface-sensitive imaging of boundaries between different refractive indices. | Imaging of interfaces, cellular structures, and developmental biology. |
|
| Limitation | Recommendation |
|---|---|
| High-costs | Cost reduction strategies, including the development of simplified designs based on cost-effective materials, open-sources manufacturing processes. |
| Technological equipment | Development of robust, affordable, and user-friendly systems and data analysis software. Advancements in miniaturizing equipment. |
| Tissue penetration | Enhance tissue penetration depth with the implementation of adaptive optics and the combination with multimodal approaches. |
| Data Analysis | Incorporating artificial intelligence for automatic image processing, pattern recognition, and data analysis to enhance the differentiation of cancerous tissues. |
| In vivo trials | More in vivo studies and clinical trials are needed to demonstrate the efficacy and reliability of non-linear optical techniques in real-clinic cancer detection, to ensure broad applicability. |
| Multi-modal imaging | Combining the contrast mechanisms of the reviewed techniques with other imaging techniques such as PET or MRI could offer a more comprehensive diagnostic approach. |
| Standardization protocols | It is critical to establish standardized protocols for the use of non-linear optical techniques in cancer detection. This includes guidelines for image acquisition, data analysis, and interpretation of results. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ávila, F.J. A Review of Non-Linear Optical Imaging Techniques for Cancer Detection. Optics 2024, 5, 416-433. https://doi.org/10.3390/opt5040031
Ávila FJ. A Review of Non-Linear Optical Imaging Techniques for Cancer Detection. Optics. 2024; 5(4):416-433. https://doi.org/10.3390/opt5040031
Chicago/Turabian StyleÁvila, Francisco J. 2024. "A Review of Non-Linear Optical Imaging Techniques for Cancer Detection" Optics 5, no. 4: 416-433. https://doi.org/10.3390/opt5040031
APA StyleÁvila, F. J. (2024). A Review of Non-Linear Optical Imaging Techniques for Cancer Detection. Optics, 5(4), 416-433. https://doi.org/10.3390/opt5040031






