Mutants with Enhanced Cellobiose-Fermenting Ability from Thermotolerant Kluyveromyces marxianus DMKU 3-1042, Which Are Beneficial for Fermentation with Cellulosic Biomass
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
2.2. Mutagenesis and Screening
2.3. Fermentation Tests and Analysis of Fermentation Parameters
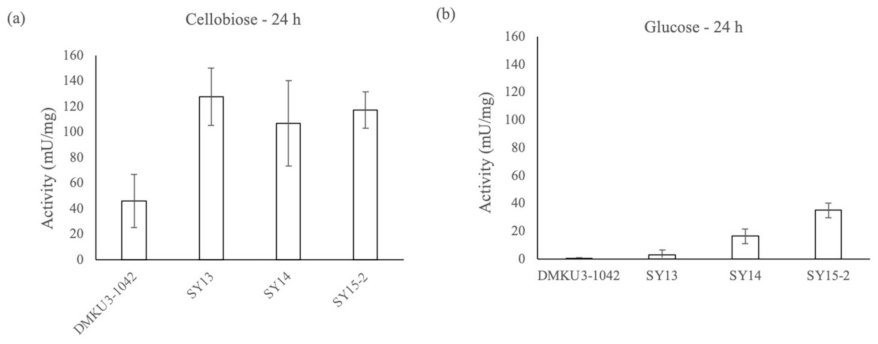
2.4. ß-Glucosidase Assay
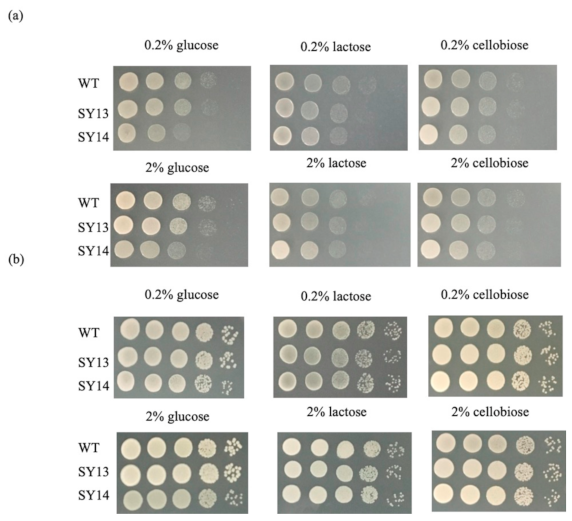
2.5. Growth Tests of Mutants on YAY Agar Plates Containing Glucose, Lactose or Cellobiose
2.6. Preparation of Genomic DNA, Genomic Sequencing and Determination of Mutations
2.7. Insertion of Transporter Genes into the Genome of a Mutant Strain, SY14
3. Results
3.1. Mutagenesis of K. marxianus DMKU3-1042 and Screening of Strains with High Cellobiose-Fermenting Activity
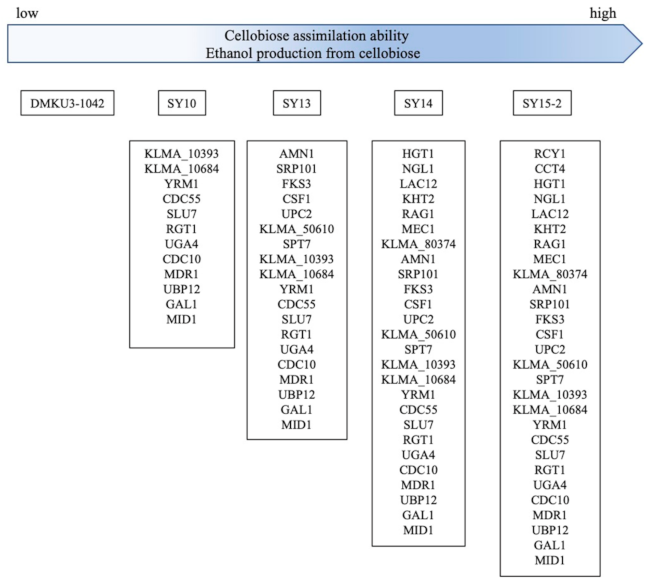
3.2. Mutation Points of Mutant Strains
3.3. Glucose-Fermenting Abilities of Mutant Strains
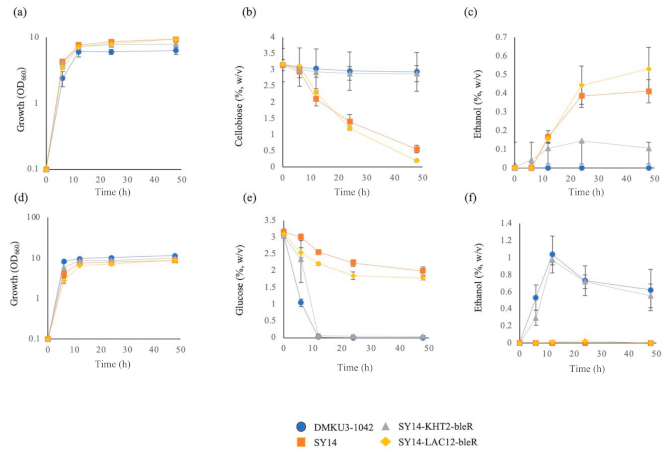
3.4. Introduction of KHT2 and LAC2 Genes into SY14
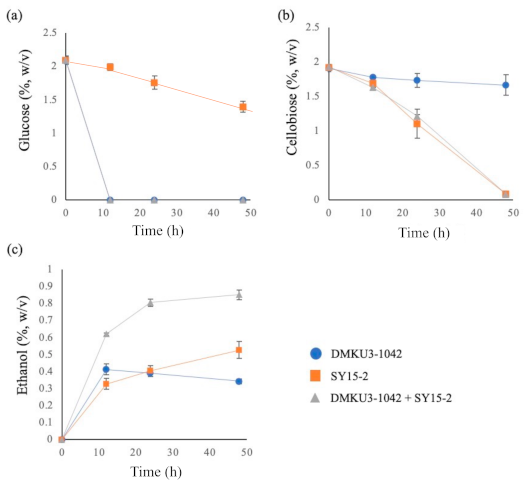
3.5. Co-Culture of SY15-2 and the Parental Strain in a Medium Supplemented with Cellobiose and Glucose
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bayer, E.A.; Chanzy, H.; Lamed, R.; Shoham, Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Klarskov, K.; Piens, K.; Ståhlberg, J.; Høj, P.B.; Beeumen, J.V.; Claeyssens, M. Cellobiohydrolase I from Trichoderma reesei: Identification of an active-site nucleophile and additional information on sequence including the glycosylation pattern of the core protein. Carbohydr. Res. 1997, 304, 143–154. [Google Scholar] [CrossRef]
- Xu, Q.; Singh, A.; Himmel, M.E. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr. Opin. Biotechnol. 2009, 20, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Christian, P.K. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J. Biotechnol. 2013, 163, 133–142. [Google Scholar]
- Saloheimo, M.; Kuja-Panula, J.; Ylösmäki, E.; Ward, M.; Penttilä, M. Enzymatic properties and intracellular localization of the novel Trichoderma reesei beta-glucosidase BGLII (cel1A). Appl. Environ. Microbiol. 2002, 68, 4546–4553. [Google Scholar] [CrossRef]
- Herpoël-Gimbert, I.; Margeot, A.; Dolla, A.; Jan, G.; Mollé, D.; Lignon, S.; Mathis, H.; Sigoillot, J.-C.; Monot, F.; Asther, M. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels. 2008, 23, 18. [Google Scholar] [CrossRef]
- Berlin, A.; Maximenko, V.; Gilkes, N.; Saddler, J. Optimization of enzyme complexes for lignocellulose hydrolysis. Biotechnol. Bioeng. 2007, 97, 287–296. [Google Scholar] [CrossRef]
- Baba, Y.; Sumitani, J.-I.; Tani, S.; Kawaguchi, T. Characterization of Aspergillus aculeatus β-glucosidase 1 accelerating cellulose hydrolysis with Trichoderma cellulase system. AMB Express. 2015, 24, 3. [Google Scholar] [CrossRef]
- Dekker, R.F. Kinetic, inhibition, and stability properties of a commercial beta-D-glucosidase (cellobiase) preparation from aspergillus niger and its suitability in the hydrolysis of lignocellulose. Biotechnol. Bioeng. 1986, 28, 1438–1442. [Google Scholar] [CrossRef]
- Limtong, S.; Sringiew, C.; Yongmanitchai, W. Production of fuel ethanol at high temperature from sugar cane juice by a newly isolated Kluyveromyces marxianus. Bioresour. Technol. 2007, 98, 3367–3374. [Google Scholar] [CrossRef] [PubMed]
- Goshima, T.; Tsuji, M.; Inoue, H.; Yano, S.; Hoshino, T.; Matsushika, A. Bioethanol production from Lignocellulosic biomass by a novel Kluyveromyces marxianus strain. Biosci. Biotechnol. Biochem. 2013, 77, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Nitiyon, S.; Keo-Oudone, C.; Murata, M.; Lertwattanasakul, N.; Limtong, S.; Kosaka, T.; Yamada, M. Efficient conversion of xylose to ethanol by stress-tolerant Kluyveromyces marxianus BUNL-21. Springerplus 2016, 27, 185. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Beniwal, A.; Kokkiligadda, A.; Vij, S. Evolutionary adaptation of Kluyveromyces marxianus strain for efficient conversion of whey lactose to bioethanol. Process. Biochem. 2017, 62, 69–79. [Google Scholar] [CrossRef]
- Kosaka, T.; Nakajima, Y.; Ishii, A.; Yamashita, M.; Yoshida, S.; Murata, M.; Kato, K.; Shiromaru, Y.; Kato, S.; Kanasaki, Y.; et al. Capacity for survival in global warming: Adaptation of mesophiles to the temperature upper limit. PLoS ONE 2019, 14, e0215614. [Google Scholar]
- Rodrussamee, N.; Lertwattanasakul, N.; Hirata, K.; Suprayogi; Limtong, S.; Kosaka, T.; Yamada, M. Growth and ethanol fermentation ability on hexose and pentose sugars and glucose effect under various conditions in thermotolerant yeast Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 2011, 90, 1573–1586. [Google Scholar] [CrossRef]
- Nurcholis, M.; Murata, M.; Limtong, S.; Kosaka, T.; Yamada, M. MIG1 as a positive regulator for the histidine biosynthesis pathway and as a global regulator in thermotolerant yeast Kluyveromyces marxianus. Sci. Rep. 2019, 9, 9926. [Google Scholar] [CrossRef]
- Lertwattanasakul, N.; Kosaka, T.; Hosoyama, A.; Suzuki, Y.; Rodrussamee, N.; Matsutani, M.; Murata, M.; Fujimoto, N.; Suprayogi; Tsuchikane, K.; et al. Genetic basis of the highly efficient yeast Kluyveromyces marxianus: Complete genome sequence and transcriptome analyses. Biotechnol. Biofuels. 2015, 18, 47. [Google Scholar] [CrossRef]
- Tingle, M.A.; Halvorson, H.O. A comparison of β-glucanase and β-glucosidase in Saccharomyces lactis. Biochim. Biophys. Acta 1971, 250, 165–171. [Google Scholar] [CrossRef]
- Rubio-Texeira, M.; Arévalo-Rodríguez, M.; Lequerica, J.; Lequerica, L.; Polaina, J. Lactose utilization by Saccharomyces cerevisiae strains expressing Kluyveromyces lactis LAC genes. J. Biotechnol. 2001, 84, 97–106. [Google Scholar] [CrossRef]
- Sadie, C.J.; Rose, S.H.; Den Haan, R.; Van Zyl, W.H. Co-expression of a cellobiose phosphorylase and lactose permease enables intracellular cellobiose utilisation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2011, 90, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Varela, J.A.; Puricelli, M.; Ortiz-Merino, R.A.; Giacomobono, R.; Braun-Galleani, S.; Wolfe, K.H.; Morrissey, J.P. Origin of Lactose Fermentation in Kluyveromyces lactis by Interspecies Transfer of a Neo-functionalized Gene Cluster during Domestication. Curr. Biol. 2019, 29, 4284–4290. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Singhal, S.; Mathur, T.; Upadhyay, D.J.; Rattan, A. Antifungal potential of disulfiram. Nippon Ishinkin Gakkai Zasshi 2007, 48, 109–113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nguyen, M.T.; Lertwattanasakul, N.; Rodrussamee, N.; Limtong, S.; Kosaka, T.; Yamada, M. A Kluyveromyces marxianus 2-deoxyglucose-resistant mutant with enhanced activity of xylose utilization. Int. Microbiol. 2015, 18, 235–244. [Google Scholar]
- Orikasa, Y.; Mikumo, D.; Ohwada, T. A 2-Deoxyglucose-Resistant Mutant of Saccharomyces cerevisiae Shows Enhanced Maltose Fermentative Ability by the Activation of MAL Genes. Foods 2018, 7, 52. [Google Scholar] [CrossRef]
- Kaewbanjong, J.; Wan, S.H.P.; Boonme, P. Clotrimazole microemulsion and microemulsion-based gel: Evaluation of buccal drug delivery and irritancy using chick chorioallantoic membrane as the model. J. Pharm. Pharmacol. 2017, 69, 1716–1723. [Google Scholar] [CrossRef]
- Nonklang, S.; Abdel-Banat, B.M.; Cha-aim, K.; Moonjai, N.; Hoshida, H.; Limtong, S.; Yamada, M.; Akada, R. High-temperature ethanol fermentation and transformation with linear DNA in the thermotolerant yeast Kluyveromyces marxianus DMKU3-1042. Appl. Environ. Microbiol. 2008, 74, 7514–7521. [Google Scholar] [CrossRef]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Watanabe, T.; Watanabe, I.; Yamamoto, M.; Ando, A.; Nakamura, T. A UV-induced mutant of Pichia stipitis with increased ethanol production from xylose and selection of a spontaneous mutant with increased ethanol tolerance. Bioresour. Technol. 2011, 102, 1844–1848. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2008; Volume 1. [Google Scholar]
- Akuzawa, S.; Nagaoka, J.; Kanekatsu, M.; Kanesaki, Y.; Suzuki, T. Draft Genome Sequence of Oceanobacillus picturae Heshi-B3, Isolated from Fermented Rice Bran in a Traditional Japanese Seafood Dish. Genome Announc. 2016, 4, e01621-15. [Google Scholar] [CrossRef]
- Hirokawa, M.; Suzuki, A.; Miyauchi, A. Thyroid Fine-Needle Aspiration and Smearing Techniques. VideoEndocrinology 2018, 5, ve.2018.0119. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Lertwattanasakul, N.; Suprayogi; Murata, M.; Rodrussamee, N.; Limtong, S.; Kosaka, T.; Yamada, M. Essentiality of respiratory activity for pentose utilization in thermotolerant yeast Kluyveromyces marxianus DMKU 3-1042. Antonie Van Leeuwenhoek 2013, 103, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Heinisch, J.; Kohler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxP marker cassettes for Cre- mediated multiple gene knockouts in budding yeast. Nucl. Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.F.; de Souza, C.G., Jr.; de Morais, M.A., Jr. A simple and rapid method for lithium acetate-mediated transfor- mation of Kluyveromyces marxianus cells. World J. Microbiol. Biotechnol. 2000, 16, 653–654. [Google Scholar] [CrossRef]
- Billard, P.; Menart, S.; Blaisonneau, J.; Bolotin-Fukuhara, M.; Fukuhara, H.; Wesolowski-Louvel, M. Glucose uptake in Kluyveromyces lactis: Role of the HGT1 gene in glucose transport. J. Bacteriol. 1996, 178, 5860–5866. [Google Scholar] [CrossRef][Green Version]
- Weirich, J.; Goffrini, P.; Kuger, P.; Ferrero, I.; Breunig, K.D. Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur. J. Biochem. 1997, 249, 248–257. [Google Scholar] [CrossRef]
- Prior, C.; Mamessier, P.; Fukuhara, H.; Chen, X.; Wesolowski-Louvel, M. The hexokinase gene is required for transcriptional regulation of the glucose transporter gene RAG1 in Kluyveromyces lactis. Mol. Cell. Biol. 1993, 13, 3882–3889. [Google Scholar]
- Liu, P.; Lin, A.; Zhang, G.; Zhang, J.; Chen, Y.; Shen, T.; Zhao, J.; Wei, D.; Wang, W. Enhancement of cellulase production in Trichoderma reesei RUT-C30 by comparative genomic screening. Microb. Cell. Fact. 2019, 18, 81. [Google Scholar] [CrossRef]
- Patyshakuliyeva, A.; Arentshorst, M.; Allijn, I.E.; Ram, A.F.; de Vries, R.P.; Gelber, I.B. Improving cellulase production by Aspergillus niger using adaptive evolution. Biotechnol. Lett. 2016, 38, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Davison, S.A.; Den Haan, R.; van Zyl, W.H. Heterologous expression of cellulase genes in natural Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 2016, 100, 8241–8254. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Li, H.N.; Song, H.T.; Xiao, W.J.; Xia, W.C.; Liu, X.P.; Jiang, Z.B. Construction of a trifunctional cellulase and expression in Saccharomyces cerevisiae using a fusion protein. BMC. Biotechnol. 2018, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Wang, Y.; Kumagai, H.; Tamaki, H. Construction of thermotolerant yeast expressing thermostable cellulase genes. J. Biotechnol. 2007, 130, 114–123. [Google Scholar] [CrossRef]

| Strain/Plasmid | Relevant Properties | Strain/Plasmid Source/Reference |
|---|---|---|
| Kluyveromyces marxianus DMKU3-1042 | Parental strain | [27] |
| SY5-1-17 | Cellobiose-fermenting strain | This work |
| SY5-1-17-40 | Cellobiose-fermenting strain | This work |
| SY8 | Cellobiose-fermenting strain | This work |
| SY10 | Cellobiose-fermenting strain | This work |
| SY12 | Cellobiose-fermenting strain | This work |
| SY13 | Cellobiose-fermenting strain, 1 µM DSFR | This work |
| SY14 | Cellobiose-fermenting strain, 1 µM DSFR, 0.1% 2 DOGR | This work |
| SY15-1 | Cellobiose-fermenting strain, 1 µM DSFR, 0.1% 2 DOGR, 4 µg/mL CTZR | This work |
| SY15-2 | Cellobiose-fermenting strain, 10 µM DSFR 0.1% 2 DOGR | This work |
| SY15-3 | Cellobiose-fermenting strain, 1 µM DSFR 0.5% 2 DOGR | This work |
| pUC19 | ampR | [28] |
| pUC-KHT2-bleR | pUC19 with the KHT2 and bleR genes | This work |
| pUC-LAC12-bleR | pUC19 with the LAC12 and bleR genes | This work |
| Name | Sequence 5′ > 3′ |
|---|---|
| pUC19-up | GATCCCCGGGTACCGAGCTC |
| pUC19-down | GATCCTCTAGAGTCGACCTG |
| KHT2-up | CGGTACCCGGGGATCAAATTTTTCCACCCCGCTCT |
| KHT2-down | TGTGTGGGGGATCCCTGCCACATTTTAGAGGCCTT |
| LAC12-up | CGGTACCCGGGGATCAATTTTCCCCCCACTGA |
| LAC12-down | TGTGTGGGGGATCCCTTGATGTGAAGAATACTGGT |
| bleR-up | GGGATCCCCCACACACCATA |
| bleR-down | GGACTCTAGACCATCGCGTACACGCGTCTGTACAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murata, M.; Pattanakittivorakul, S.; Manabe, T.; Limtong, S.; Yamada, M. Mutants with Enhanced Cellobiose-Fermenting Ability from Thermotolerant Kluyveromyces marxianus DMKU 3-1042, Which Are Beneficial for Fermentation with Cellulosic Biomass. Fuels 2022, 3, 232-244. https://doi.org/10.3390/fuels3020015
Murata M, Pattanakittivorakul S, Manabe T, Limtong S, Yamada M. Mutants with Enhanced Cellobiose-Fermenting Ability from Thermotolerant Kluyveromyces marxianus DMKU 3-1042, Which Are Beneficial for Fermentation with Cellulosic Biomass. Fuels. 2022; 3(2):232-244. https://doi.org/10.3390/fuels3020015
Chicago/Turabian StyleMurata, Masayuki, Sornsiri Pattanakittivorakul, Toshiro Manabe, Savitree Limtong, and Mamoru Yamada. 2022. "Mutants with Enhanced Cellobiose-Fermenting Ability from Thermotolerant Kluyveromyces marxianus DMKU 3-1042, Which Are Beneficial for Fermentation with Cellulosic Biomass" Fuels 3, no. 2: 232-244. https://doi.org/10.3390/fuels3020015
APA StyleMurata, M., Pattanakittivorakul, S., Manabe, T., Limtong, S., & Yamada, M. (2022). Mutants with Enhanced Cellobiose-Fermenting Ability from Thermotolerant Kluyveromyces marxianus DMKU 3-1042, Which Are Beneficial for Fermentation with Cellulosic Biomass. Fuels, 3(2), 232-244. https://doi.org/10.3390/fuels3020015







