Abstract
Dd-cfDNA is a novel biomarker with many diagnostic applications in various areas of medicine. In this review of the literature, we investigate its role in the diagnosis of many complications that occur in liver transplantations. In our review, we retrieved data from the medical databases PubMed and Scopus. In our bibliography, many areas concerning the contributions of dd-cfDNA to the field of liver transplantation, such as in the diagnosis of complications that include signsof rejection or graft injury, are mentioned. Dd-cfDNA, which are correlated with other biomarkers such as liver enzymes, can have a high diagnostic value. Measurements of Dd-cfDNA also depend on the graft’s size and origin; therefore, these data should be taken into account for the estimation and explanation of dd-cfDNA values. Despite the utility of this novel diagnostic technique, it comes with some limitations and applicational exclusions, such as cases where there is a blood relation between the donor and recipient.
1. Introduction
Long-term survival beyond the first year after liver transplantation (LT) has not significantly improved in the past decades due to many factors, including the long-term effects of immunosuppression and graft dysfunction. An important obstacle to LT is the lack of a reliable and non-invasive biomarker with which to determine graft function and the general success of transplantation. In clinical practice, liver enzymes are usually used for the determination of graft function post LT [1]. Serum measurement of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) may be a non-invasive procedure, but it comes with certain limitations. Firstly, these enzymes are not liver-specific, and even when they are targeted towards the liver, they only reflect damage to specific cell types (hepatocytes or cholangiocytes). Moreover, they have long half-lives, which limits their ability to detect rapid damage [1,2]. Another and more accurate alternative procedure is surveillance liver graft biopsy (svLBxs). Although this method is more specific and accurate than the measurement of liver enzymes, when considering the complexity and large size of the liver, its sensitivity is far from 100%, which often leads to an inaccurate diagnosis. This, in combination with its high cost and the fact that it is an invasive procedure, have led to svLBxs’ exclusion from the routine examination of liver transplant recipients [3].
Donor-derived cell-free DNA (dd-cfDNA) has been heralded as a non-invasive, accurate biomarker with which to monitor liver transplant recipients. dd-cfDNA is a biomarker whose use has increased in the last several years, with potential applications including the early detection of cancer and graft dysfunction in various types of transplantation [4]. This biomarker is measured in serum and its assessment includes an evaluation of its quantity, but also an investigation of some quality characteristics, such as the size of the fragments and methylation patterns [5]. Particularly, for its detection in serum, various types of Polymerase Chain Reaction (PCR) are available; alternatively, an approach used for female recipients with male donors is the amplification of Y-chromosome specific genes [6]. The main principles for distinguishing the donor-derived cfDNA from the recipient-derived type include the following steps. Firstly, SNP (Single-nucleotide polymorphism) selection is conducted, followed by the targeted amplification and sequencing of the cfDNA samples. The next steps are the statistical determination of the recipient’s heterozygous cutoff, on whose basis the dd-cfDNA is estimated. Additionally, the value of the technique is increased by sample quality control and analytical validation [7]. dd-cfDNA is a biomarker with a short half-life (<1.5 h) that is released from necrotic or apoptotic cells in the transplanted organ [1]. The above characteristics result in the various potentially diagnostic applications of dd-cfDNA in liver transplantation, which include the earlier detection of common complications such as acute rejection (AR), the personalization of immunosuppression dose schemes, and the diagnosis of transplant-related infectious diseases [8,9]. In this review of the literature, we describe the various applications and certain usage limitations of dd-cfDNA as a biomarker in LT.
2. Materials and Methods
We conducted a literature review of the medical research databases PubMed and Scopus. We used the following key words for our search: Liver transplantation, dd-cfDNA, biomarkers in liver transplantation, acute rejection, and graft injury. Our research was limited to the period from 2000 to the present and includes studies written in the English language. Our exclusion criteria included articles in languages other than English, studies that were not human-related, and bibliographies that did not refer to dd-cfDNA usage in liver transplantation.
3. Results
The main applicational areas of dd-cfDNA in liver transplantation are listed below and in Figure 1. The main research studies concerning dd-cfDNA’s application in liver transplantation are listed in Table 1.
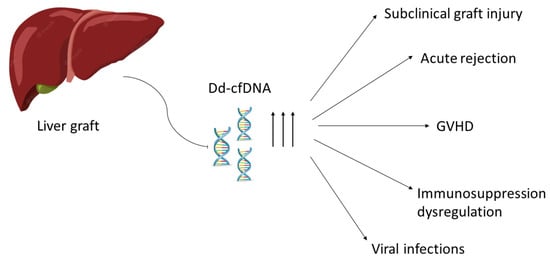
Figure 1.
Main areas of diagnostic application of dd-cfDNA (donor-derived cell-free DNA). GVHD: Graft-versus-Host disease.
dd-cfDNA Applications
- Subclinical graft injury:A subclinical graft injury is defined as significant histological inflammation with more or less normal liver enzymes, particularly those below 2 × ULN. With the aid of surveillance liver graft biopsies (svLBxs), relevant graft injuries that fulfill the criteria for T cell-mediated rejection (TCMR) have been found in more than 25% of LT recipients with normal liver enzymes [10,11]. Although svLBxs constitute a reliable diagnostic method for subclinical T cell-mediated rejection (SubTCMR), their use entails an invasive examination with potential complications, thus limiting their daily clinical use. dd-cfDNA can be used a noninvasive biomarker for the early detection of SubTCMR. Many studies have found a correlation between elevated levels of dd-cfDNA and SubTCMR. Specifically, Anna K. Baumann et al. reported significantly increased fractional dd-dfDNA levels in patients with SubTCMR compared to recipients without any graft damage. In the same study, no differences in absolute dd-cfDNA levels were found between the recipients with or without SubTCMR [12]. Moreover, Ekkehard Schutz et al., in their study concerning the personalization of immunosuppressive medication schemes, have found high levels of GcfDNA in a patient with subclinical draft damage due to insufficient (subtherapeutic) tacrolimus blood levels [11]. Short fractions of dd-cfDNA were also associated with graft damage in a study by Hoi Ioi Ng et al. [5]. Additionally, Oellerich et al. correlated dd-cfDNA levels with Tacrolimus levels and the incidence of graft injury [12].
- Acute rejection:In cases of acute rejection, the use of dd-cfDNA provides better sensitivity and diagnostic value than LFTs, allowing for the earlier and more sensitive diagnosis of this complication [3]. Fernández-Galán E et al., Zhao D et al., Schuetz E et al., and Gielis EM and J Levitsky have reported elevated serum dd-cfDNA during acute rejection among LT recipients in comparison to patients without rejection [2,7,9,13,14]. Moreover, dd-cfDNA is not only a non-invasive diagnostic method of acute rejection but also offers an earlier diagnosis compared to biopsy according to Schuetz E et al. [13]. A method for identifying the origin of dd-cfDNA and diagnosing the complication in the recipient is by determining the size of the fractions ratio. Notably, Fernández-Galán E et al. mention that patients with acute rejection show a higher short-fragments ratio than those with normal grafts [4].
- Graft vs. Host disease:It is known that Graft vs. Host disease (GVHD) has an asymptomatic phase, wherein the early detection and treatment through the adjustment of the immunosuppression dosage is possible [15]. Until now, the early detection of GVHD has been possible through molecular techniques for the detection of macro chimerism that are not cost-effective and not applicable in everyday clinical practice [16]. Duncan Lewis et al. reported a novel diagnostic method for the early detection of GVHD with dd-cfDNA. Specifically, the diagnosis of GVHD was made with the combination of elevated serum dd-cfDNA levels, high percentages of T and B cells of a donor origin, and donor-derived genomic-DNA (dd-gDNA) observed in skin biopsies. The above results indicate multisystemic GVHD and infection and may be potential non-invasive immune-monitoring tools to obtain earlier diagnoses of GVHD, thus combining diagnosis and prognosis with early treatment [8].
- Viral infection:Transplant recipients require lifelong immunosuppression and are at high risk of developing various opportunistic infections, such as EBV/CMV. This stresses the need for a non-invasive biomarker in order to investigate the possibility of viral infection in LT recipients. Dong Zhao et al. investigated the role of dd-cfDNA as a possible diagnostic method of the viral infection of the liver. In the results of their research, the authors mention that patients with EBV or CMV had a significantly higher dd-cfDNA fraction (%) and absolute quantification median dd-cfDNA (cp/mL); notably, the levels of dd-cfDNA were higher in EBV patients than those with CMV [14]. Moreover, Ekkehard Schuetz et al. investigated the possible diagnostic role of GcfDNA in LT recipients infected with HCV. In their results, it was determined that there is a slightly higher and more variable GcfDNA percentage in HCV+ patients than in those in a stable condition [13]. Oellerich, M et al. also show significantly elevated GcfDNA in HCV+ LT recipients in their results [17].

Table 1.
Main research articles that referred to dd-cfDNA applications. Type of study, area of application, sensitivity, and specificity of the method employed are mentioned in this table.
Table 1.
Main research articles that referred to dd-cfDNA applications. Type of study, area of application, sensitivity, and specificity of the method employed are mentioned in this table.
| Author | Year | Type of Study | Number of Patients | dd-cfDNA Application | dd-cfDNA Detection Method | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Anna K. Baumann et al. [7] | 2022 | Cross-sectional study | 108 | Subclinical graft injury detection | cfDNA was isolated from cryo-conserved plasma and subjected to allele-specific droplet digital PCR | 81% | 91% |
| Esther Fernández-Galán et al. [4] | 2022 | pilot study | 27 | Early detection of acute rejection | The cfDNA was extracted from plasma; the total cfDNA concentration in plasma was determined using fluorometry | 85.7% | 63.3% |
| Hoi Ioi Ng et al. [5] | 2019 | Original study | 11 | Early detection of graft injury | DNA fragments from cell-free plasma were extracted and then distributed in different sizes of DNA fragments | NA 1 | NA 1 |
| Duncan Lewis et al. [6] | 2021 | Case report | 1 | Graft-versus-host-disease early diagnosis | dd-cfDNA serum and skin levels were counted using a targeted, next-generation sequencing assay | NA 1 | NA 1 |
| Ekkehard Schu¨tz et al. [11] | 2017 | Prospective, observational, multicenter cohort study | 115 | Early detection of acute rejection | In cfDNA from plasma samples, graft cfDNA/total cfDNA percentage was measured using droplet digital PCR based on a limited number of predefined SNPs | 89.3 | 95.7 |
| Dong Zhao et al. [13] | 2021 | Prospective diagnostic study | 49 | Early detection of acute rejection | Blood was drawn into cfDNA blood collection tubes. DNA samples were used for library construction, target region capture sequencing, bioinformatics, and dd-cfDNA quantification | For dd-cfDNA% ≥ 28.7%: 72.7%, and for dd-cfDNA (cp/ mL) ≥ 2076 cp/mL: 81.8%, | For dd-cfDNA% ≥ 28.7%: 94.7% and for dd-cfDNA (cp/ mL) ≥ 2076 cp/mL: 81.9%. |
| M. Oellerich et al. [12] | 2014 | Prospective diagnostic study | 12 | Graft dysfunction and graft injury | GcfDNA, isolated from plasma samples, was measured using ddPCR assay | NA 1 | NA 1 |
| Kanzow, P. et al. [18] | 2014 | Case report | 1 | HELLP syndrome and immunosuppression monitoring | Serum GcfDNA measurements were taken using ddPCR | NA 1 | NA 1 |
| Josh Levitsky et al. [19] | 2022 | Multicenter study | 219 | Early detection of acute rejection and graft injury | dd-cfDNA was derived from plasma samples; dsDNA was used for the construction of libraries and SNP sequencing. | 100% | 80% |
1 NA: not applicable. dd-cfDNA: Donor-derived cell-free DNA; PCR: Polymerase chain reaction; dsDNA: double-stranded DNA; SNP: single-nucleotide polymorphism; ddPCR: droplet digital polymerase chain reaction; GcfDNA: graft-derived cell-free DNA.
4. Discussion
Dd-cfDNA appears to be an easily accessible biomarker with various diagnostic applications in liver transplantation. Its non-invasive character in combination with its reliability and conduciveness towards fast measurement may render it the ‘golden standard’ for graft monitoring in the future. Particularly, curves with measurements of dd-cfDNA, in correlation with other biomarkers such as liver enzymes, can prove very useful with respect to the monitoring of LT recipients [15]. The diagnostic value of these curves is based on time-dependent percentages of dd-cfDNA in liver transplant patients without rejection, complications, or infection [11]. It is important to remember that measurements of dd-cfDNA correlate not only with pathological events, such as graft injury or acute rejection, but also with the size of the graft and its origin. Particularly, Zhao D et al. reported higher peaks in the dd-cfDNA curves in LT recipients when the origin of the graft was a deceased donor than those with transplants from living donor transplantation [12]. This is most likely explained by ischemia–reperfusion injury, which is higher in grafts from deceased donors, but also by the fact that elevations in dd-cfDNA are more dramatic in larger grafts from deceased donors than from partial grafts in living donors [16,17].
Despite the benefits and potential diagnostic uses that dd-cfDNA offer, there are certainly limitations to its use. Firstly, LT recipients due to immunosuppression or other pathological situations are prone to leukopenia, leukocytosis, and inflammatory illness, which may influence fractional dd-cfDNA determination. Moreover, as stated above, dd-cfDNA is a potential biomarker for many LT complications related to graft injury, and not only those related to rejection [18]. There are also certain cases of LT where dd-cfDNA cannot be a potential biomarker, including instances involving an identical twin donor/recipient pairs and donor/recipient siblings from consanguineous marriages. Lastly, dual organ transplants from a single donor and multiple organ transplants from different donors also pose some limitations concerning the application of dd-cfDNA as a diagnostic method [20]. An increase in the number and types οf SNPs used for dd-cfDNA detection and measurement, or the use of epigenetic pattern differences as a method of detection, could be potential solutions to the limitations mentioned above [21].
5. Conclusions
Our review highlights the importance of dd-cfDNA as a potential biomarker for the monitoring of LT recipients and the detection of the most commonly found complications. It provides up-to-date information that can benefit research conducted for dd-cfDNA diagnostic usage in liver transplantation. As far as we know, this is the only review of the literature that highlights all the possible diagnostic applications of dd-cfDNA. There are certain limitations in our review of the literature, including a lack of variability of the research papers due to the novelty of the technique as well as language limitations. These difficulties only enhance the need for more detailed research in this new and exciting field.
Author Contributions
E.A. conceptualized the idea; visualized, collected, and analyzed the data; and wrote the manuscript. S.V. provided resources and critically reviewed the manuscript; N.A. critically reviewed the manuscript; G.K. critically reviewed the manuscript; A.K. critically reviewed the manuscript; K.-E.K. critically reviewed the manuscript; and G.T. supervised, assisted with the data curation, and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lehmann-Werman, R.; Magenheim, J.; Moss, J.; Neiman, D.; Abraham, O.; Piyanzin, S.; Zemmour, H.; Fox, I.; Dor, T.; Grompe, M.; et al. Monitoring liver damage using hepatocyte-specific methylation markers in cell-free circulating DNA. JCI Insight 2018, 3, e120687. [Google Scholar] [CrossRef]
- Bardhi, E.; McDaniels, J.; Rousselle, T.; Maluf, D.G.; Mas, V.R. Nucleic acid biomarkers to assess graft injury after liver transplantation. JHEP Rep. 2022, 4, 100439. [Google Scholar] [CrossRef]
- Puliyanda, D.P.; Swinford, R.; Pizzo, H.; Garrison, J.; De Golovine, A.M.; Jordan, S.C. Donor-derived cell-free DNA (dd-cfDNA) for detection of allograft rejection in pediatric kidney transplants. Pediatr. Transplant. 2021, 25, e13850. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Galán, E.; Badenas, C.; Fondevila, C.; Jiménez, W.; Navasa, M.; Puig-Butillé, J.A.; Brunet, M. Monitoring of Donor-Derived Cell-Free DNA by Short Tandem Repeats: Concentration of Total Cell-Free DNA and Fragment Size for Acute Rejection Risk Assessment in Liver Transplantation. Liver Transplant. 2022, 28, 257–268. [Google Scholar] [CrossRef]
- Ng, H.I.; Zhu, X.; Xuan, L.; Long, Y.; Mao, Y.; Shi, Y.; Sun, L.; Liang, B.; Scaglia, F.; Choy, K.W.; et al. Analysis of fragment size distribution of cell-free DNA: A potential non-invasive marker to monitor graft damage in living-related liver transplantation for inborn errors of metabolism. Mol. Genet. Metab. 2019, 127, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.; Glehn-Ponsirenas, R.; Gulbahce, N.; Hooey, L.J.; Chaffin, J.M.; Miles, J.; Woodward, R.; Duarte, S.; Beduschi, T.; Zarrinpar, A. High levels of donor-derived cell-free DNA in a case of graft-versus-host-disease following liver transplantation. Am. J. Transplant. 2022, 22, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.K.; Beck, J.; Kirchner, T.; Hartleben, B.; Schütz, E.; Oellerich, M.; Wedemeyer, H.; Jaeckel, E.; Taubert, R. Elevated fractional donor-derived cell-free DNA during subclinical graft injury after liver transplantation. Liver Transpl. 2022, 28, 1911–1919. [Google Scholar] [CrossRef] [PubMed]
- Gielis, E.M.; Ledeganck, K.J.; De Winter, B.Y.; Del Favero, J.; Bosmans, J.-L.; Claas, F.H.J.; Abramowicz, D.; Eikmans, M. Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am. J. Transplant. 2015, 15, 2541–2551. [Google Scholar] [CrossRef] [PubMed]
- Höfer, A.; Jonigk, D.; Hartleben, B.; Verboom, M.; Hallensleben, M.; Manns, M.P.; Jaeckel, E.; Taubert, R. Non-invasive screening for subclinical liver graft injury in adults via donor-specific anti-HLA antibodies. Sci. Rep. 2020, 10, 14242. [Google Scholar] [CrossRef]
- Saunders, E.A.; Engel, B.; Höfer, A.; Hartleben, B.; Vondran, F.W.; Richter, N.; Potthoff, A.; Zender, S.; Wedemeyer, H.; Jaeckel, E.; et al. Outcome and safety of a surveillance biopsy guided personalized immunosuppression program after liver transplantation. Am. J. Transplant. 2022, 22, 519–531. [Google Scholar] [CrossRef]
- Schütz, E.; Fischer, A.; Beck, J.; Harden, M.; Koch, M.; Wuensch, T.; Stockmann, M.; Nashan, B.; Kollmar, O.; Matthaei, J.; et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med. 2017, 14, e1002286. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Schütz, E.; Kanzow, P.; Schmitz, J.; Beck, J.; Kollmar, O.; Streit, F.; Walson, P.D. Use of Graft-Derived Cell-Free DNA as an Organ Integrity Biomarker to Reexamine Effective Tacrolimus Trough Concentrations After Liver Transplantation. Ther. Drug Monit. 2014, 36, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhou, T.; Luo, Y.; Wu, C.; Xu, D.; Zhong, C.; Cong, W.; Liu, Q.; Zhang, J.; Xia, Q. Preliminary clinical experience applying donor-derived cell-free DNA to discern rejection in pediatric liver transplant recipients. Sci. Rep. 2021, 11, 1138. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.L.; Gibbs, P.; Sudhindran, S.; Key, T.; Goodman, R.S.; Morgan, C.H.; Watson, C.J.E.; Delriviere, L.; Alexander, G.J.; Jamieson, N.V.; et al. Monitoring systemic donor lymphocyte macrochimerism to aid the diagnosis of graftversus-host disease after liver transplantation. Transplantation 2004, 77, 441–445. [Google Scholar] [CrossRef]
- Taylor, A.L.; Gibbs, P.; Bradley, J.A. Acute Graft Versus Host Disease Following Liver Transplantation: The Enemy Within. Am. J. Transplant. 2004, 4, 466–474. [Google Scholar] [CrossRef]
- Oellerich, M.; Walson, P.D.; Beck, J.; Schmitz, J.; Kollmar, O.; Schütz, E. Graft-Derived Cell-Free DNA as a Marker of Transplant Graft Injury. Ther. Drug Monit. 2016, 38 (Suppl. S1), S75–S79. [Google Scholar] [CrossRef]
- Ng, H.-I.; Sun, L.-Y.; Zhu, Z.-J. Detecting Graft-Derived Cell-Free DNA Through Amplification Refractory Mutation System Polymerase Chain Reaction in Living-Donor Liver Transplantation: Report of 2 Cases. Transplant. Proc. 2019, 51, 820–822. [Google Scholar] [CrossRef]
- Kanzow, P.; Kollmar, O.; Schütz, E.; Oellerich, M.; Schmitz, J.; Beck, J.; Walson, P.D.; Slotta, J.E. Graf-derived cell-free DNA as an early organ integrity biomarker after transplantation of a marginal HELLP syndrome donor liver. Transplantation 2014, 98, e43–e45. [Google Scholar] [CrossRef]
- Levitsky, J.; Kandpal, M.; Guo, K.; Kleiboeker, S.; Sinha, R.; Abecassis, M. Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients. Am. J. Transplant. 2022, 22, 532–540. [Google Scholar] [CrossRef]
- Oellerich, M.; Budde, K.; Osmanodja, B.; Bornemann-Kolatzki, K.; Beck, J.; Schütz, E.; Walson, P.D. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front. Genet. 2022, 13, 1031894. [Google Scholar] [CrossRef]
- Chen, L.; Wang, J.; Tan, L.; Lu, C.; Fu, G.; Fu, L.; Zhang, X.; Wang, Q.; Ma, C.; Cong, B.; et al. Highly accurate mtGenome haplotypes from long-read SMRT sequencing can distinguish between monozygotic twins. Forensic Sci. Int. Genet. 2020, 47, 102306. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).