AI Innovations in Liver Transplantation: From Big Data to Better Outcomes
Abstract
:1. Introduction
2. Materials and Methods
3. Results
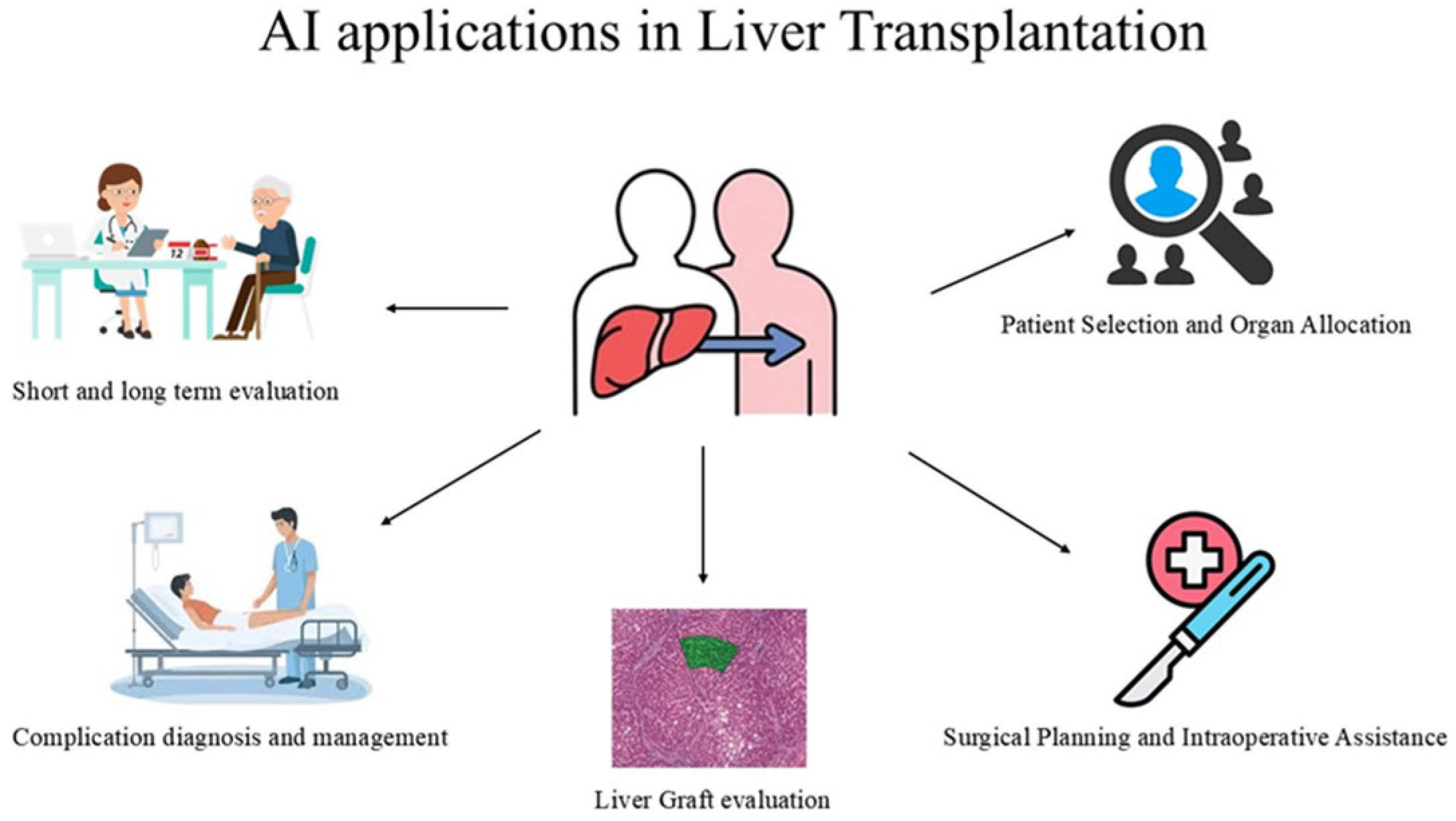
3.1. Applications of AI During LT
3.1.1. Pre-Transplant Stage
Patient Selection and Organ Allocation
3.1.2. Transplant Operation
Surgical Planning and Intraoperative Assistance
Graft Evaluation
3.1.3. Post-Transplantation
Complication Diagnosis and Management
Short and Long Term Graft Evaluation
| Authors | Year | N of Patients | Stage of Transplant Process | AI Application | Description | Key Benefits | Results |
|---|---|---|---|---|---|---|---|
| Parmanto et al. [63] | 2001 | NA | Post-Transplant | RNN | Prediction of graft failure based on clinical observations. | Use of RNN to accurately model the temporal sequence of clinical observations | 90% correct classification on the learning set and 78% on the test set. |
| Hughes et al. [73] | 2001 | 117 | Post-Transplant | ANN | ML model to monitor and detect acute rejection in LT. | This model uses individual clinical and biochemical variables and can be used for the monitoring of LT recipients in the early post-LT period. | AUROC = 0.902, sensitivity = 80.0% specificity = 90.1% |
| Rodriguez-Luna et al. [35] | 2005 | 19 | Post-Transplant | ANN | ANN analysis performed to prognosticate the risk of HCC recurrence. | The first AI algorithm able to predict the HCC reoccurrence. | This algorithm had a high discriminatory power (17/19, 89.5%), accurately predicting HCC recurrence. |
| Cucchetti et al. [36] | 2010 | 250 | Post-Transplant | ANN and LR | ANN model able to predict tumor grade and MVI. | Preoperative serum alpha-fetoprotein (AFP), tumor number, size, and volume were the used variables. | ANN correctly identified 93.3% of tumor grades and 91% of MVI. |
| Briceño et al. [64] | 2014 | 1031 | Post-Transplant | ANN | ANNs for organ allocation should be based on the concept of survival benefit. | Predicting the 3-month outcome based on donor–recipient matching and 57 variables. | ANNs showed great performance in predicting the probability of graft survival (90.79%) and loss (71.42%). |
| Dorado-Moreno et al. [66] | 2017 | 1406 | Post-Transplant | ANN | ML model for prediction of graft failure. | The model aids healthcare professionals in the donor–recipient matching process. | Accuracy = 73.57%, |
| Lau et al. [67] | 2017 | 15,401 | Post-Transplant | ANN and RF | ML model for prediction of graft failure. | Identification of 15 significant donor, recipient, and transplant factors influencing graft failure within 30 days post-transplant. | AUC = 0.818 |
| Zare et al. [74] | 2017 | 148 | Post-Transplant | ANN | ML model able to monitor and detect acute rejection in LT. | This model can provide efficiency in clinical decision making based on routine laboratory data. | Accuracy = 90%, sensitivity = 87%, and specificity = 90% |
| Lee et al. [50] | 2018 | 1211 | Post-Transplant | DT, RF, GVM, SVM, naïve Bayes, multilayer perceptron, and DBN | ML-based algorithms for prediction of AKI. | An internet-based risk estimator was developed based on the GBM model. | GBM had the best performance with AUROC 0.90 |
| Bhat et al. [52] | 2018 | 61,677 | Post-Transplant | RF and LR | Ml algorithm able to identify key predictors and survival outcomes of new-onset diabetes post-LT. | Sirolimus use and Black race were some variables associated with new-onset diabetes. | Various variables were investigated with both models. |
| Ayllon et al. [68] | 2018 | 822 | Post-Transplant | ANN | ANN model for donor–recipient matching. | ANN model demonstrated excellent predictive capabilities for both 3-month and 12-month graft survival, outperforming traditional scoring systems. | 3 month predictive capability AUC = 0.94 and 12 month AUC = 0.78 |
| Moccia et al. [29] | 2020 | 40 | Transplant Operation | SVM, RF, and MIL | ML algorithm for the analysis of liver graft texture. | This is the first model able to assist surgeons in liver graft assessment inside the OR using RGB images. | Sensitivity = 95, specificity = 81, and accuracy = 88% |
| Cesaretti et al. [30] | 2020 | 117 | Transplant Operation | SVM-SIL and FCNN | ML algorithm assessing liver steatosis based on smartphone images. | Smartphone cameras are widely available among surgery team members. In the future, better camera quality could lead to better performance of this algorithm. | Automatic liver graft segmentation from smartphone images achieved an accuracy of 98%, whereas the analysis of the liver graft features (cropped picture and donor data) showed an accuracy of 89% in graft classification. |
| Guo et al. [11] | 2021 | 34,575 | Pre-Transplant | DNN, LR, and RF | ML model used for early mortality prediction in patients on LT waiting list. | Variables such as ALP, ALT, and hemoglobin were also top informative features besides the 4 MELD-Na variables. | The performance of models comprising all variables outperformed those with 4 MELD-NA variables for all prediction cases and the DNN model outperformed the LR and RF models. |
| Pérez-Sanz et al. [21] | 2021 | 20 | Transplant Operation | CV ML | ML algorithm for the measurement of steatosis. | The model was able to easily differentiate specific fat staining from artifacts related to the staining procedure. | Accuracy for all classifiers (>0.99) with sensitivity > 0.8 and specificity > 0.9. Regarding speed, KNN and naïve Bayes were the fastest algorithms. |
| He et al. [37] | 2021 | 109 | Post-Transplant | DL | ML model able to distinguish post-LT recipients of high recurrence risk. | The variables used included clinical features, MRI images, and pathology images | The combined model showed 80% recall and 89% precision. |
| Chen et al. [42] | 2021 | 591 | Post-Transplant | LR, SVM, RF, ADABoost, XGBoost, and GBM | ML algorithm for prediction of postoperative pneumonia on LT recipients. | Pneumonia was associated with 14 features: INR, HCT, PLT, ALB, ALT, FIB, WBC, PT, serum Na+, TBIL, anesthesia time, preoperative length of stay, total fluid transfusion, and operation time. | XGBoost model performed best (sensitivity: 52.6%; specificity: 77.5%) |
| Jain et al. [45] | 2021 | 1459 | Post-Transplant | LR, RF, SVM, GBM, and XGBoost | ML model used to predict major adverse cardiovascular events, all-cause mortality, and cardiovascular mortality post-LT. | The top influential factors for postoperative cardiovascular adverse events were age at transplantation, diabetes, serum creatinine, cirrhosis caused by non-alcoholic steatohepatitis, right ventricular systolic pressure, and left ventricular ejection fraction. | GBM model XGBoost achieved the highest performance, with AUROC = 0.71 |
| He et al. [49] | 2021 | 493 | Post-Transplant | RF, SVM, CDT, and CIT | ML-based algorithms for the prediction of AKI | RF models can be a useful tool for early clinical intervention of AKI to improve patient survival. | The RF model demonstrated the highest prediction accuracy of 0.79 with an AUC of 0.850 |
| Zhang et al. [51] | 2021 | 780 | Post-Transplant | GBM and ADA | ML-based algorithms for early prediction of AKI. | High preoperative indirect bilirubin, low intraoperative urine output, long anesthesia time, low preoperative platelets, and graft steatosis graded 1 were the top predictors for AKI. | The GBM model achieved the highest AUC 0.76 |
| Guijo-Rubio et al., [69] | 2021 | 39,189 | Post-Transplant | MLP, RF, GB, and SVM | ML techniques for donor–recipient matching in LT. | ML methods did not improve liver allocation performance. | LR outperformed several machine learning techniques across all endpoints achieving an AUC of 0.64 |
| Nagai et al. [10] | 2022 | 105,140 | Pre-Transplant | NN | NN models that can predict LT waitlist mortality. | The 90-day mortality model specifically identified more waitlist deaths with a higher recall, having the potential to decrease waitlist mortality and lead to more equitable allocation systems | The NN 90-day mortality model outperformed MELD-based models across all the subsets in predicting mortality. |
| Kwong et al. [14] | 2022 | 18,920 | Pre-Transplant | NA | An ML model predicted 3-, 6-, and 12-month waitlist dropouts among patients with HCC. | An online calculator was created for clinical use | This ML model predicted 3-, 6-, and 12-month waitlist dropouts among patients with HCC with a c-statistic of 0.74 |
| Park et al. [16] | 2022 | 581 | Transplant Operation | DL | DL-assisted CT volumetry to evaluate graft weight | This algorithm offers a time-efficient graft weight evaluation results derived from the DL model not requiring additional correction in approximately 70% of donors. A graft volume-to-weight conversion formula was also developed. | The CCC for the agreement between the estimated and measured graft weights was 0.834 |
| Narayan et al. [19] | 2022 | 90 | Transplant Operation | CVAI | A CVAI platform able to calculate donor liver steatosis and pred EAD. | The difference in the CVAI steatosis scores between the grafts developing EAD and those that did not was statistically significant. | CVAI steatosis scores were lower than pathologist scores (median 3% vs. 20%, p < 0.001). |
| Sun et al. [20] | 2022 | 91 | Transplant Operation | DL CNN | ML model generating steatosis probability map from an input WSI | This algorithm provides fast, accurate, and reproducible donor liver evaluation. | The model had good correlation and agreement with the annotation in both the training set (r = 0.88, ICC = 0.88) and novel input test sets (r = 0.85 and ICC = 0.85). |
| Chen et al. [53] | 2022 | 1239 | Post-Transplant | CatBoost | An ML algorithm for the prediction of intraoperative massive blood transfusion | 15 variables were screened out, including age, weight, hemoglobin, platelets, white blood cell count, activated partial thromboplastin time, prothrombin time, thrombin time, direct bilirubin, aspartate aminotransferase, total protein, albumin, globulin, creatinine, and urea. | AUROC: 0.810 |
| Lee et al. [58] | 2022 | 116 | Post-Transplant | XGBoost | ML model predicting harmful alcohol use post-LT. | 13 psychosocial variables were used. | AUC = 0.692 and positive predictive value = 0.82 |
| Cooper et al. [75] | 2022 | 1938 | Post-Transplant | LR, NN, and GBM | Predictive model for GVHD | Variables used included ABO matching, CMV and EBV, serostatus matching, age differences, and donor race/ethnicity to recipient race/ethnicity matching. | The C5.0 model had the best performance with AUROC 0.86, sensitivity 0.80, and detection prevalence 0.21. |
| Börner et al. [65] | 2022 | 529 | Post-Transplant | DL | ML techniques for donor–recipient matching and survival prediction in LT. | This NN utilizes transparent and easily interpretable data to predict the outcome after LT. | Accuracy = 95.8%, AUC = 0.940, and F1 = 0.899 |
| Giglio et al. [17] | 2023 | 872 | Transplant Operation | ML | ML able to predict GW. | The following information was used: donor’s age, sex, height, weight, body mass index, graft type, computed tomography estimated graft volume, and total liver volume | MAE value of 50 ± 62 g in predicting GW, with a mean error of 10.3%. |
| Jiao et al. [22] | 2023 | 88 | Transplant Operation | SAM | ML 3-step model able to detect LDF | The AI model is designed to adhere to the Banff consensus recommendations, ensuring clinical relevance and accuracy. | Correlation coefficients between pathologist and computer-assisted manual quantification, between computer-assisted manual quantification and the AI model, and between the AI model and pathologist were 0.94, 0.88, and 0.81, respectively. |
| Ivanics et al. [38] | 2023 | 66.059 | Post-Transplant | ML | ML-based models for predicting 90-day post-LT mortality across three international registries. | Standardization of registry-based variables could facilitate the added value of MLA-based models. | The best model performance was obtained in Canada (LightGBM: AUROC, 0.42; range, 0.25–0.55) and the US |
| Liu et al. [39] | 2023 | 315 | Post-Transplant | MLP | ML model for the prediction of HCC recurrence | A web calculator based on the MLP model was also developed. | γ-glutamyl transpeptidase (GGT), fibrinogen, neutrophil, aspartate aminotransferase (AST), and total bilirubin (TB) were the top five important factors for the recurrence risk of HCC. |
| Zaver et al. [46] | 2023 | 300 | Post-Transplant | CNN | AI-ECG algorithm in predicting cardiac factors for post-LT cardiovascular complications. | AI-ECG trained to recognize patterns from a standard 12-lead ECG in order to identify the presence of left ventricular systolic dysfunction and atrial fibrillation. | ECG in sinus rhythm had an AUROC = 0.69 for prediction of de novo post-transplant atrial fibrillation. |
| Zabara et al. [59] | 2023 | 90 | Post-Transplant | DL | An ML model to predict postoperative complications following LT in hepatitis C patients. | This model can be used as a tool to guide a more intensive follow-up protocol in high-risk patients. | Accuracy > 99.76% |
| Tang et al. [23] | 2024 | 95 | Transplant Operation | SAM | ML model able to detect LDF hepatocytes in liver biopsies. | Additional algorithms can be applied to filter the false positive results of this model. | The model showed high sensitivity but low specificity due to similarities with other structures. |
| Soldera et al. [44] | 2024 | 575 | Post-Transplant | XGBoost | ML model used to predict major adverse cardiovascular events post-LT. | The modeling dataset included 83 features, encompassing patient and laboratory data, cirrhosis complications, and pre-LT cardiac assessments. | AUROC = 0.89, precision = 0.89, recall = 0.80, and F1-score = 0.84. |
| Li et al. [60] | 2024 | 160,360 | Post-Transplant | DNN | An ML model predicting post-LT risk factors. | The model significantly reduced the task discrepancy by 39%. | Multi-task learning outperform single-task prediction. |
| Börner et al. [70] | 2024 | 1066 | Post-Transplant | DL | This model enables continuous, risk-adjusted monitoring of in-hospital mortality rates after LT. | This approach aims to promptly detect deviations in surgical outcomes, thereby facilitating timely interventions. | DL AUC = 0.857, surpassing traditional risk scores |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69S, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Kalton, A.; Falconer, E.; Docherty, J.; Alevras, D.; Brann, D.; Johnson, K. Multi-Agent-Based Simulation of a Complex Ecosystem of Mental Health Care. J. Med. Syst. 2016, 40, 39. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Upton, R.; Mumith, A.; Beqiri, A.; Parker, A.; Hawkes, W.; Gao, S.; Porumb, M.; Sarwar, R.; Marques, P.; Markham, D.; et al. Automated Echocardiographic Detection of Severe Coronary Artery Disease Using Artificial Intelligence. JACC Cardiovasc. Imaging 2022, 15, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Rai, R. Liver transplantatation—An overview. Indian. J. Surg. 2013, 75, 185–191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bunnik, E.M. Ethics of allocation of donor organs. Curr. Opin. Organ. Transpl. 2023, 28, 192–196. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ruf, A.; Dirchwolf, M.; Freeman, R.B. From Child-Pugh to MELD score and beyond: Taking a walk down memory lane. Ann. Hepatol. 2022, 27, 100535. [Google Scholar] [CrossRef] [PubMed]
- Atiemo, K.; Skaro, A.; Maddur, H.; Zhao, L.; Montag, S.; VanWagner, L.; Goel, S.; Kho, A.; Ho, B.; Kang, R.; et al. Mortality Risk Factors Among Patients With Cirrhosis and a Low Model for End-Stage Liver Disease Sodium Score (≤15): An Analysis of Liver Transplant Allocation Policy Using Aggregated Electronic Health Record Data. Am. J. Transpl. 2017, 17, 2410–2419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kwong, A.J.; Lai, J.C.; Dodge, J.L.; Roberts, J.P. Outcomes for liver transplant candidates listed with low model for end-stage liver disease score. Liver Transpl. 2015, 21, 1403–1409. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bertsimas, D.; Kung, J.; Trichakis, N.; Wang, Y.; Hirose, R.; Vagefi, P.A. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am. J. Transpl. 2019, 19, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Nallabasannagari, A.R.; Moonka, D.; Reddiboina, M.; Yeddula, S.; Kitajima, T.; Francis, I.; Abouljoud, M. Use of neural network models to predict liver transplantation waitlist mortality. Liver Transpl. 2022, 28, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Mazumder, N.R.; Ladner, D.P.; Foraker, R.E. Predicting mortality among patients with liver cirrhosis in electronic health records with machine learning. PLoS ONE 2021, 16, e0256428. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, H.; Tehreem, A.; Ting, P.S.; Gurakar, M.; Li, S.Y.; Simsek, C.; Alqahtani, S.A.; Kim, A.K.; Kohli, R.; Gurakar, A. Hepatocellular Carcinoma and the Role of Liver Transplantation: A Review. J. Clin. Transl. Hepatol. 2021, 9, 738–748. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996, 334, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.; Hameed, B.; Syed, S.; Ho, R.; Mard, H.; Arshad, S.; Mehta, N. Machine learning to predict waitlist dropout among liver transplant candidates with hepatocellular carcinoma. Cancer Med. 2022, 11, 1535–1541. [Google Scholar] [CrossRef]
- Wang, W.C.; Wu, T.H.; Hung, H.C.; Lee, J.C.; Cheng, C.H.; Wang, Y.C.; Lee, C.F.; Wu, T.J.; Chou, H.S.; Chan, K.M.; et al. Liver regeneration of living donor after liver donation for transplantation: Disparity in the left and right remnant liver. Medicine 2024, 103, e37632. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, R.; Lee, S.; Sung, Y.; Yoon, J.; Suk, H.I.; Kim, H.; Choi, S. Accuracy and Efficiency of Right-Lobe Graft Weight Estimation Using Deep-Learning-Assisted CT Volumetry for Living-Donor Liver Transplantation. Diagnostics 2022, 12, 590. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giglio, M.C.; Zanfardino, M.; Franzese, M.; Zakaria, H.; Alobthani, S.; Zidan, A.; Ayoub, I.I.; Shoreem, H.A.; Lee, B.; Han, H.S.; et al. Machine learning improves the accuracy of graft weight prediction in living donor liver transplantation. Liver Transpl. 2023, 29, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Croome, K.P.; Shalev, J.A.; Musto, K.R.; Sharma, M.; Keaveny, A.P.; Taner, C.B. Early allograft dysfunction after liver transplantation: An intermediate outcome measure for targeted improvements. Ann. Hepatol. 2016, 15, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Narayan, R.R.; Abadilla, N.; Yang, L.; Chen, S.B.; Klinkachorn, M.; Eddington, H.S.; Trickey, A.W.; Higgins, J.P.; Melcher, M.L. Artificial intelligence for prediction of donor liver allograft steatosis and early post-transplantation graft failure. HPB 2022, 24, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Marsh, J.N.; Matlock, M.K.; Chen, L.; Gaut, J.P.; Brunt, E.M.; Swamidass, S.J.; Liu, T.C. Deep learning quantification of percent steatosis in donor liver biopsy frozen sections. EBioMedicine 2020, 60, 103029. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pérez-Sanz, F.; Riquelme-Pérez, M.; Martínez-Barba, E.; de la Peña-Moral, J.; Salazar Nicolás, A.; Carpes-Ruiz, M.; Esteban-Gil, A.; Legaz-García, M.D.C.; Parreño-González, M.A.; Ramírez, P.; et al. Efficiency of Machine Learning Algorithms for the Determination of Macrovesicular Steatosis in Frozen Sections Stained with Sudan to Evaluate the Quality of the Graft in Liver Transplantation. Sensors 2021, 21, 1993. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Tang, H.; Sun, N.; Zhang, X. Artificial intelligence–aided steatosis assessment in donor livers according to the Banff consensus recommendations. Am. J. Clin. Pathol. 2024, 162, 401–407. [Google Scholar] [CrossRef]
- Tang, H.; Jiao, J.; Lin, J.D.; Zhang, X.; Sun, N. Detection of Large-Droplet Macrovesicular Steatosis in Donor Livers Based on Segment-Anything Model. Lab. Invest. 2024, 104, 100288. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Wang, F.; Vo, H.; Teng, D.; Teodoro, G.; Farris, A.B.; Castillo-Leon, E.; Vos, M.B.; Kong, J. Deep-learning-based accurate hepatic steatosis quantification for histological assessment of liver biopsies. Lab. Invest 2020, 100, 1367–1383. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mikolasevic, I.; Filipec-Kanizaj, T.; Mijic, M.; Jakopcic, I.; Milic, S.; Hrstic, I.; Sobocan, N.; Stimac, D.; Burra, P. Nonalcoholic fatty liver disease and liver transplantation—Where do we stand? World J. Gastroenterol. 2018, 24, 1491–1506. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Preechathammawong, N.; Charoenpitakchai, M.; Wongsason, N.; Karuehardsuwan, J.; Prasoppokakorn, T.; Pitisuttithum, P.; Sanpavat, A.; Yongsiriwit, K.; Aribarg, T.; Chaisiriprasert, P.; et al. Development of a diagnostic support system for the fibrosis of nonalcoholic fatty liver disease using artificial intelligence and deep learning. Kaohsiung J. Med. Sci. 2024, 40, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Van Ha, T.G. Liver biopsy in liver transplant recipients. Semin. Intervent Radiol. 2004, 21, 271–274. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kurakawa, K.I.; Okada, A.; Bessho, K.; Jo, T.; Ono, S.; Michihata, N.; Kumazawa, R.; Matsui, H.; Fushimi, K.; Yamaguchi, S.; et al. Major complications after percutaneous biopsy of native or transplanted liver in pediatric patients: A nationwide inpatient database study in Japan. BMC Gastroenterol. 2022, 22, 395. [Google Scholar] [CrossRef]
- Moccia, S.; Mattos, L.S.; Patrini, I.; Ruperti, M.; Poté, N.; Dondero, F.; Cauchy, F.; Sepulveda, A.; Soubrane, O.; De Momi, E.; et al. Computer-assisted liver graft steatosis assessment via learning-based texture analysis. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Cesaretti, M.; Brustia, R.; Goumard, C.; Cauchy, F.; Poté, N.; Dondero, F.; Paugam-Burtz, C.; Durand, F.; Paradis, V.; Diaspro, A.; et al. Use of Artificial Intelligence as an Innovative Method for Liver Graft Macrosteatosis Assessment. Liver Transpl. 2020, 26, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Agostini, C.; Buccianti, S.; Risaliti, M.; Fortuna, L.; Tirloni, L.; Tucci, R.; Bartolini, I.; Grazi, G.L. Complications in Post-Liver Transplant Patients. J. Clin. Med. 2023, 12, 6173. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Craig, E.V.; Heller, M.T. Complications of liver transplant. Abdom. Radiol. 2021, 46, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Azzam, A.Z. Liver transplantation as a management of hepatocellular carcinoma. World J. Hepatol. 2015, 7, 1347–1354. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sposito, C.; Citterio, D.; Virdis, M.; Battiston, C.; Droz Dit Busset, M.; Flores, M.; Mazzaferro, V. Therapeutic strategies for post-transplant recurrence of hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 4929–4942. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodriguez-Luna, H.; Vargas, H.E.; Byrne, T.; Rakela, J. Artificial neural network and tissue genotyping of hepatocellular carcinoma in liver-transplant recipients: Prediction of recurrence. Transplantation 2005, 79, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Cucchetti, A.; Piscaglia, F.; Grigioni, A.D.; Ravaioli, M.; Cescon, M.; Zanello, M.; Grazi, G.L.; Golfieri, R.; Grigioni, W.F.; Pinna, A.D. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: A pilot study. J. Hepatol. 2010, 52, 880–888. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Fong, J.N.; Moore, L.W.; Ezeana, C.F.; Victor, D.; Divatia, M.; Vasquez, M.; Ghobrial, R.M.; Wong, S.T.C. An imageomics and multi-network based deep learning model for risk assessment of liver transplantation for hepatocellular cancer. Comput. Med. Imaging Graph. 2021, 89, 101894. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ivanics, T.; So, D.; Claasen, M.P.A.W.; Wallace, D.; Patel, M.S.; Gravely, A.; Choi, W.J.; Shwaartz, C.; Walker, K.; Erdman, L.; et al. Machine learning-based mortality prediction models using national liver transplantation registries are feasible but have limited utility across countries. Am. J. Transpl. 2023, 23, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, S.; Yu, H.Y.; Zeng, K.; Liang, Z.; Li, S.; Hu, Y.; Yang, Y.; Ye, L. Prediction model for hepatocellular carcinoma recurrence after hepatectomy: Machine learning-based development and interpretation study. Heliyon 2023, 9, e22458. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hernandez Mdel, P.; Martin, P.; Simkins, J. Infectious Complications After Liver Transplantation. Gastroenterol. Hepatol. 2015, 11, 741–753. [Google Scholar] [PubMed] [PubMed Central]
- Singh, N.; Limaye, A.P. Infections in Solid-Organ Transplant Recipients. Mand. Douglas Bennett’s Princ. Pract. Infect. Diseases 2015, 22, 3440–3452. [Google Scholar] [CrossRef] [PubMed Central]
- Chen, C.; Yang, D.; Gao, S.; Zhang, Y.; Chen, L.; Wang, B.; Mo, Z.; Yang, Y.; Hei, Z.; Zhou, S. Development and performance assessment of novel machine learning models to predict pneumonia after liver transplantation. Respir. Res. 2021, 22, 94. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luca, L.; Westbrook, R.; Tsochatzis, E.A. Metabolic and cardiovascular complications in the liver transplant recipient. Ann. Gastroenterol. 2015, 28, 183–192. [Google Scholar] [PubMed] [PubMed Central]
- Soldera, J.; Corso, L.L.; Rech, M.M.; Ballotin, V.R.; Bigarella, L.G.; Tomé, F.; Moraes, N.; Balbinot, R.S.; Rodriguez, S.; Brandão, A.B.M.; et al. Predicting major adverse cardiovascular events after orthotopic liver transplantation using a supervised machine learning model: A cohort study. World J. Hepatol. 2024, 16, 193–210. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jain, V.; Bansal, A.; Radakovich, N.; Sharma, V.; Khan, M.Z.; Harris, K.; Bachour, S.; Kleb, C.; Cywinski, J.; Argalious, M.; et al. Machine Learning Models to Predict Major Adverse Cardiovascular Events After Orthotopic Liver Transplantation: A Cohort Study. J. Cardiothorac. Vasc. Anesth. 2021, 35, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Zaver, H.B.; Mzaik, O.; Thomas, J.; Roopkumar, J.; Adedinsewo, D.; Keaveny, A.P.; Patel, T. Utility of an Artificial Intelligence Enabled Electrocardiogram for Risk Assessment in Liver Transplant Candidates. Dig. Dis. Sci. 2023, 68, 2379–2388. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thongprayoon, C.; Kaewput, W.; Thamcharoen, N.; Bathini, T.; Watthanasuntorn, K.; Lertjitbanjong, P.; Sharma, K.; Salim, S.A.; Ungprasert, P.; Wijarnpreecha, K.; et al. Incidence and Impact of Acute Kidney Injury after Liver Transplantation: A Meta-Analysis. J. Clin. Med. 2019, 8, 372. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fabrizi, F.; Donato, M.F.; Cerutti, R.; Invernizzi, F.; Porata, G.; Frontini, G.; Raffiotta, F.; De Feo, T.; Alfieri, C.M.; Lampertico, P.; et al. Acute kidney injury and chronic kidney disease after liver transplant: A retrospective observational study. Nefrologia 2022, 42, 41–49. [Google Scholar] [CrossRef] [PubMed]
- He, Z.L.; Zhou, J.B.; Liu, Z.K.; Dong, S.Y.; Zhang, Y.T.; Shen, T.; Zheng, S.S.; Xu, X. Application of machine learning models for predicting acute kidney injury following donation after cardiac death liver transplantation. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Yoon, S.B.; Yang, S.-M.; Kim, W.H.; Ryu, H.-G.; Jung, C.-W.; Suh, K.-S.; Lee, K.H. Prediction of Acute Kidney Injury after Liver Transplantation: Machine Learning Approaches vs. Logistic Regression Model. J. Clin. Med. 2018, 7, 428. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, D.; Liu, Z.; Chen, C.; Ge, M.; Li, X.; Luo, T.; Wu, Z.; Shi, C.; Wang, B.; et al. An explainable supervised machine learning predictor of acute kidney injury after adult deceased donor liver transplantation. J. Transl. Med. 2021, 19, 321. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhat, V.; Tazari, M.; Watt, K.D.; Bhat, M. New-Onset Diabetes and Preexisting Diabetes Are Associated With Comparable Reduction in Long-Term Survival After Liver Transplant: A Machine Learning Approach. Mayo Clin. Proc. 2018, 93, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, L.P.; Wang, Y.J.; Zhou, X.H.; Dong, H.; Chen, Z.W.; Wu, J.; Gui, R.; Zhao, Q.Y. Advancing Prediction of Risk of Intraoperative Massive Blood Transfusion in Liver Transplantation With Machine Learning Models. A Multicenter Retrospective Study. Front. Neuroinform. 2022, 16, 893452, Erratum in Front. Neuroinform. 2023, 17, 1161475. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marroni, C.A.; Fleck, A.M., Jr.; Fernandes, S.A.; Galant, L.H.; Mucenic, M.; de Mattos Meine, M.H.; Mariante-Neto, G.; Brandão, A.B.M. Liver transplantation and alcoholic liver disease: History, controversies, and considerations. World J. Gastroenterol. 2018, 24, 2785–2805. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bramstedt, K.A.; Jabbour, N. When alcohol abstinence criteria create ethical dilemmas for the liver transplant team. J. Med. Ethics. 2006, 32, 263–265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shinde, S.S.; Chakole, S.; Humane, S. Understanding Alcohol Relapse in Liver Transplant Patients With Alcohol-Related Liver Disease: A Comprehensive Review. Cureus 2024, 16, e54052. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Young, K.; Patel, Y.A.; Hoffman, B.; Peskoe, S.; Chow, S.C.; Erhart, K.; Jackson, J.; Garbarino, S. Alcohol Relapse after Liver Transplantation: Risk Factors, Outcomes, and a Comparison of Risk Stratification Models. Gastro. Hep Adv. 2025, 4, 100550. [Google Scholar] [CrossRef]
- Lee, B.P.; Roth, N.; Rao, P.; Im, G.Y.; Vogel, A.S.; Hasbun, J.; Roth, Y.; Shenoy, A.; Arvelakis, A.; Ford, L.; et al. Artificial intelligence to identify harmful alcohol use after early liver transplant for alcohol-associated hepatitis. Am. J. Transpl. 2022, 22, 1834–1841. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zabara, M.L.; Popescu, I.; Burlacu, A.; Geman, O.; Dabija, R.A.C.; Popa, I.V.; Lupascu, C. Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience. Sensors 2023, 23, 2149. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, C.; Jiang, X.; Zhang, K. A transformer-based deep learning approach for fairly predicting post-liver transplant risk factors. J. Biomed. Inform. 2024, 149, 104545. [Google Scholar] [CrossRef] [PubMed]
- Durand, F. How to improve long-term outcome after liver transplantation? Liver Int. 2018, 38 (Suppl. S1), 134–138. [Google Scholar] [CrossRef] [PubMed]
- Fuochi, E.; Anastasio, L.; Lynch, E.N.; Campani, C.; Dragoni, G.; Milani, S.; Galli, A.; Innocenti, T. Main factors influencing long-term outcomes of liver transplantation in 2022. World J. Hepatol. 2023, 15, 321–352. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parmanto, B.; Doyle, H.R. Recurrent neural networks for predicting outcomes after liver transplantation: Representing temporal sequence of clinical observations. Methods Inf. Med. 2001, 40, 386–391. [Google Scholar] [PubMed]
- Briceño, J.; Cruz-Ramírez, M.; Prieto, M.; Navasa, M.; Ortiz de Urbina, J.; Orti, R.; Gómez-Bravo, M.Á.; Otero, A.; Varo, E.; Tomé, S.; et al. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: Results from a multicenter Spanish study. J. Hepatol. 2014, 61, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Börner, N.; Schoenberg, M.B.; Pöschke, P.; Heiliger, C.; Jacob, S.; Koch, D.; Pöllmann, B.; Drefs, M.; Koliogiannis, D.; Böhm, C.; et al. A Novel Deep Learning Model as a Donor-Recipient Matching Tool to Predict Survival after Liver Transplantation. J. Clin. Med. 2022, 11, 6422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dorado-Moreno, M.; Pérez-Ortiz, M.; Gutiérrez, P.A.; Ciria, R.; Briceño, J.; Hervás-Martínez, C. Dynamically weighted evolutionary ordinal neural network for solving an imbalanced liver transplantation problem. Artif. Intell. Med. 2017, 77, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.; Kankanige, Y.; Rubinstein, B.; Jones, R.; Christophi, C.; Muralidharan, V.; Bailey, J. Machine-Learning Algorithms Predict Graft Failure After Liver Transplantation. Transplantation 2017, 101, e125–e132. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ayllón, M.D.; Ciria, R.; Cruz-Ramírez, M.; Pérez-Ortiz, M.; Gómez, I.; Valente, R.; O’Grady, J.; de la Mata, M.; Hervás-Martínez, C.; Heaton, N.D.; et al. Validation of artificial neural networks as a methodology for donor-recipient matching for liver transplantation. Liver Transpl. 2018, 24, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Guijo-Rubio, D.; Briceño, J.; Gutiérrez, P.A.; Ayllón, M.D.; Ciria, R.; Hervás-Martínez, C. Statistical methods versus machine learning techniques for donor-recipient matching in liver transplantation. PLoS ONE 2021, 16, e0252068. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Börner, N.; Schoenberg, M.B.; Pöllmann, B.; Pöschke, P.; Böhm, C.; Koch, D.; Drefs, M.; Koliogiannis, D.; Andrassy, J.; Werner, J.; et al. Deep Learning-Adjusted Monitoring of In-Hospital Mortality after Liver Transplantation. J. Clin. Med. 2024, 13, 6046. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Choudhary, N.S.; Saigal, S.; Bansal, R.K.; Saraf, N.; Gautam, D.; Soin, A.S. Acute and Chronic Rejection After Liver Transplantation: What A Clinician Needs to Know. J. Clin. Exp. Hepatol. 2017, 7, 358–366. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alarcon-Zurita, A.; Ladefoged, J. Treatment of acute allograft rejection with high doses of corticosteroids. Kidney Int. 1976, 9, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Hughes, V.F.; Melvin, D.G.; Niranjan, M.; Alexander, G.A.; Trull, A.K. Clinical validation of an artificial neural network trained to identify acute allograft rejection in liver transplant recipients. Liver Transpl. 2001, 7, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Zare, A.; Zare, M.A.; Zarei, N.; Yaghoobi, R.; Zare, M.A.; Salehi, S.; Geramizadeh, B.; Malekhosseini, S.A.; Azarpira, N. A neural network approach to predict acute allograft rejection in liver transplant recipients using routine laboratory data. Hepat. Mon. 2017, 17, e55092. [Google Scholar] [CrossRef]
- Cooper, J.P.; Perkins, J.D.; Warner, P.R.; Shingina, A.; Biggins, S.W.; Abkowitz, J.L.; Reyes, J.D. Acute Graft-Versus-Host Disease After Orthotopic Liver Transplantation: Predicting This Rare Complication Using Machine Learning. Liver Transpl. 2022, 28, 407–421. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Balsano, C.; Burra, P.; Duvoux, C.; Alisi, A.; Piscaglia, F.; Gerussi, A.; Special Interest Group (SIG). Artificial Intelligence and Liver Disease; Italian Association for the Study of Liver (AISF). Artificial Intelligence and liver: Opportunities and barriers. Dig. Liver Dis. 2023, 55, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Birks, S.; Gray, J.; Darling-Pomranz, C. Using artificial intelligence to provide a ’flipped assessment’ approach to medical education learning opportunities. Med. Teach. 2024, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; Sfikakis, P.P.; et al. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimization paradigm on a weak training brain magnetic resonance imaging datasets: A supercomputer application. Med. Biol. Eng. Comput. 2021, 59, 511–533. [Google Scholar] [CrossRef] [PubMed]
- Gerussi, A.; Scaravaglio, M.; Cristoferi, L.; Verda, D.; Milani, C.; De Bernardi, E.; Ippolito, D.; Asselta, R.; Invernizzi, P.; Kather, J.N.; et al. Artificial intelligence for precision medicine in autoimmune liver disease. Front. Immunol. 2022, 13, 966329. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silveira, M.G.; Talwalkar, J.A.; Lindor, K.D.; Wiesner, R.H. Recurrent primary biliary cirrhosis after liver transplantation. Am. J. Transpl. 2010, 10, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Faisal, N.; Renner, E.L. Recurrence of autoimmune liver diseases after liver transplantation. World J. Hepatol. 2015, 7, 2896–2905. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, X.; Geng, Q.; Lin, L.; Zhang, L.; Shi, C.; Liu, B.; Yan, L.; Cao, Z.; Li, L.; Lu, P.; et al. Insights gained into the injury mechanism of drug and herb induced liver injury in the hepatic microenvironment. Toxicology 2024, 507, 153900. [Google Scholar] [CrossRef] [PubMed]
- Teschke, R.; Danan, G. Idiosyncratic Drug-Induced Liver Injury (DILI) and Herb-Induced Liver Injury (HILI): Diagnostic Algorithm Based on the Quantitative Roussel Uclaf Causality Assessment Method (RUCAM). Diagnostics 2021, 11, 458. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teschke, R. DILI, HILI, RUCAM Algorithm, and, A.I.; the Artificial Intelligence: Provocative issues, Progress, and Proposals. Arch. Gastroenterol. Res. 2020, 1, 4–11. [Google Scholar]
- Mohamed, I.B.; Aloor, F.Z.; Jalal, P.K. Strategies to Improve Immune Suppression Post-Liver Transplantation: A Review. Transplantology 2021, 2, 441–454. [Google Scholar] [CrossRef]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting Black-Box Models: A Review on Explainable Artificial Intelligence. Cogn. Comput. 2024, 16, 45–74. [Google Scholar] [CrossRef]
- Mirmozaffari, M.; Kamal, N. The Application of Data Envelopment Analysis to Emergency Departments and Management of Emergency Conditions: A Narrative Review. Healthcare 2023, 11, 2541. [Google Scholar] [CrossRef]
- Mirmozaffari, M.; Kamal, N. A data envelopment analysis model for optimizing transfer time of ischemic stroke patients under endovascular thrombectomy. Healthc. Anal. 2024, 6, 100364. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avramidou, E.; Todorov, D.; Katsanos, G.; Antoniadis, N.; Kofinas, A.; Vasileiadou, S.; Karakasi, K.-E.; Tsoulfas, G. AI Innovations in Liver Transplantation: From Big Data to Better Outcomes. Livers 2025, 5, 14. https://doi.org/10.3390/livers5010014
Avramidou E, Todorov D, Katsanos G, Antoniadis N, Kofinas A, Vasileiadou S, Karakasi K-E, Tsoulfas G. AI Innovations in Liver Transplantation: From Big Data to Better Outcomes. Livers. 2025; 5(1):14. https://doi.org/10.3390/livers5010014
Chicago/Turabian StyleAvramidou, Eleni, Dominik Todorov, Georgios Katsanos, Nikolaos Antoniadis, Athanasios Kofinas, Stella Vasileiadou, Konstantina-Eleni Karakasi, and Georgios Tsoulfas. 2025. "AI Innovations in Liver Transplantation: From Big Data to Better Outcomes" Livers 5, no. 1: 14. https://doi.org/10.3390/livers5010014
APA StyleAvramidou, E., Todorov, D., Katsanos, G., Antoniadis, N., Kofinas, A., Vasileiadou, S., Karakasi, K.-E., & Tsoulfas, G. (2025). AI Innovations in Liver Transplantation: From Big Data to Better Outcomes. Livers, 5(1), 14. https://doi.org/10.3390/livers5010014







