Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet †
Abstract
1. Introduction
2. Materials and Methods
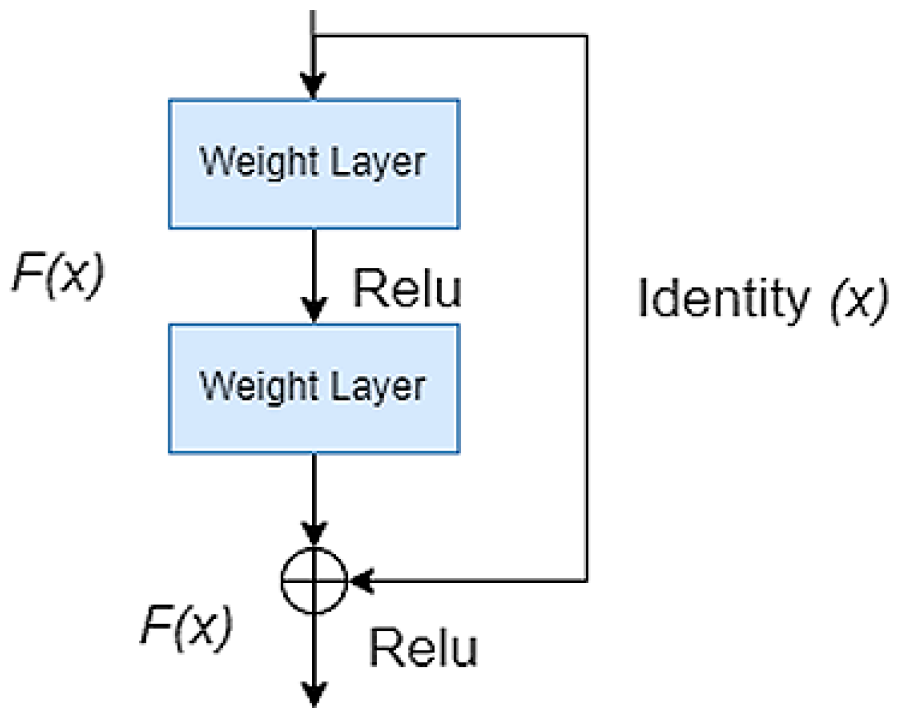
2.1. Pre-Trained Encoder
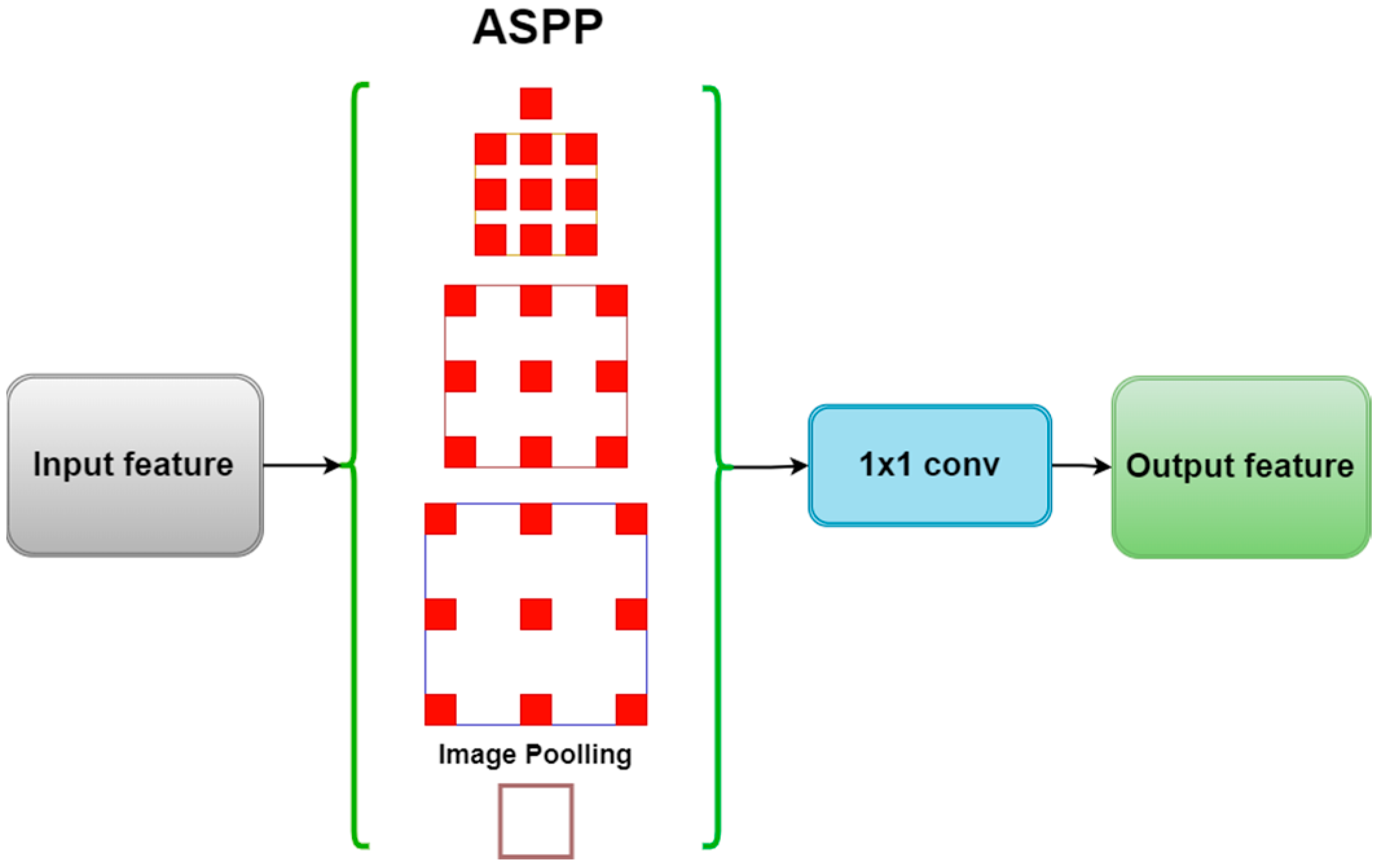
2.2. ASPP Module
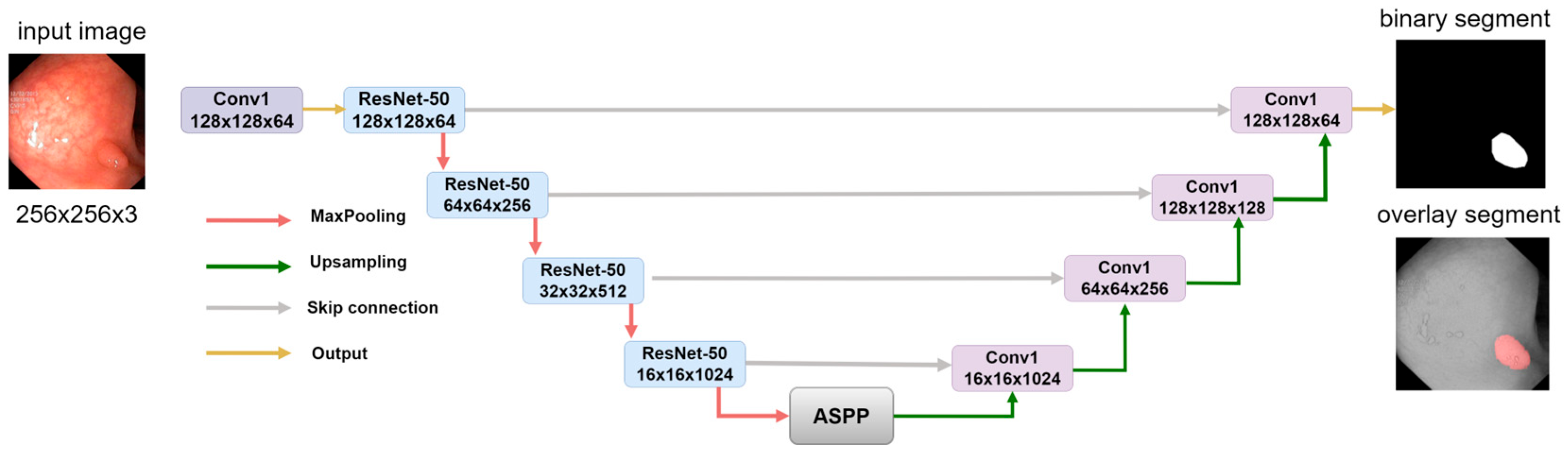
2.3. Model Decoder Path
2.4. Comparison of Segmentation Models
2.5. Evaluation Metrics
2.6. Data Preprocessing
2.7. Implementation Details
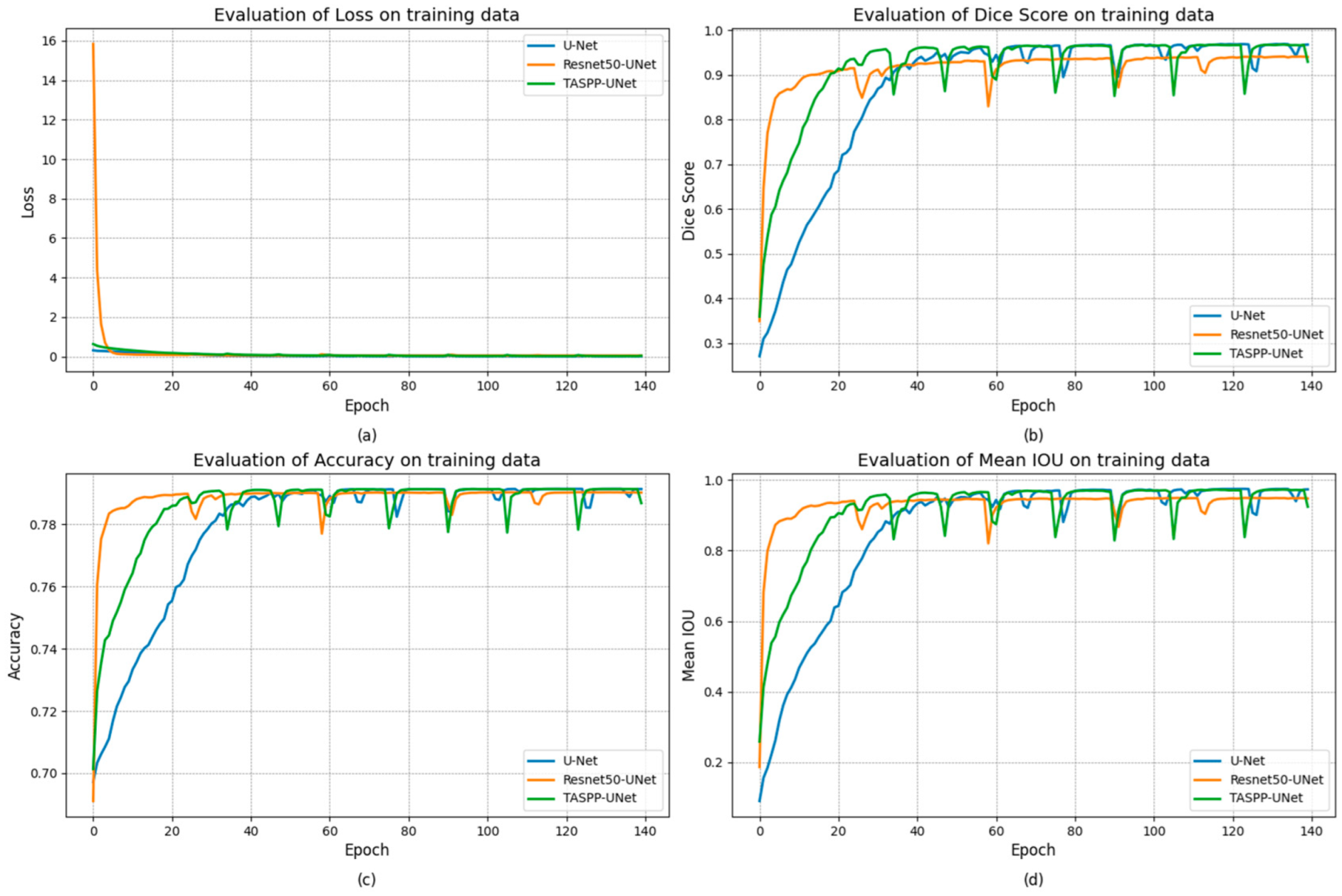
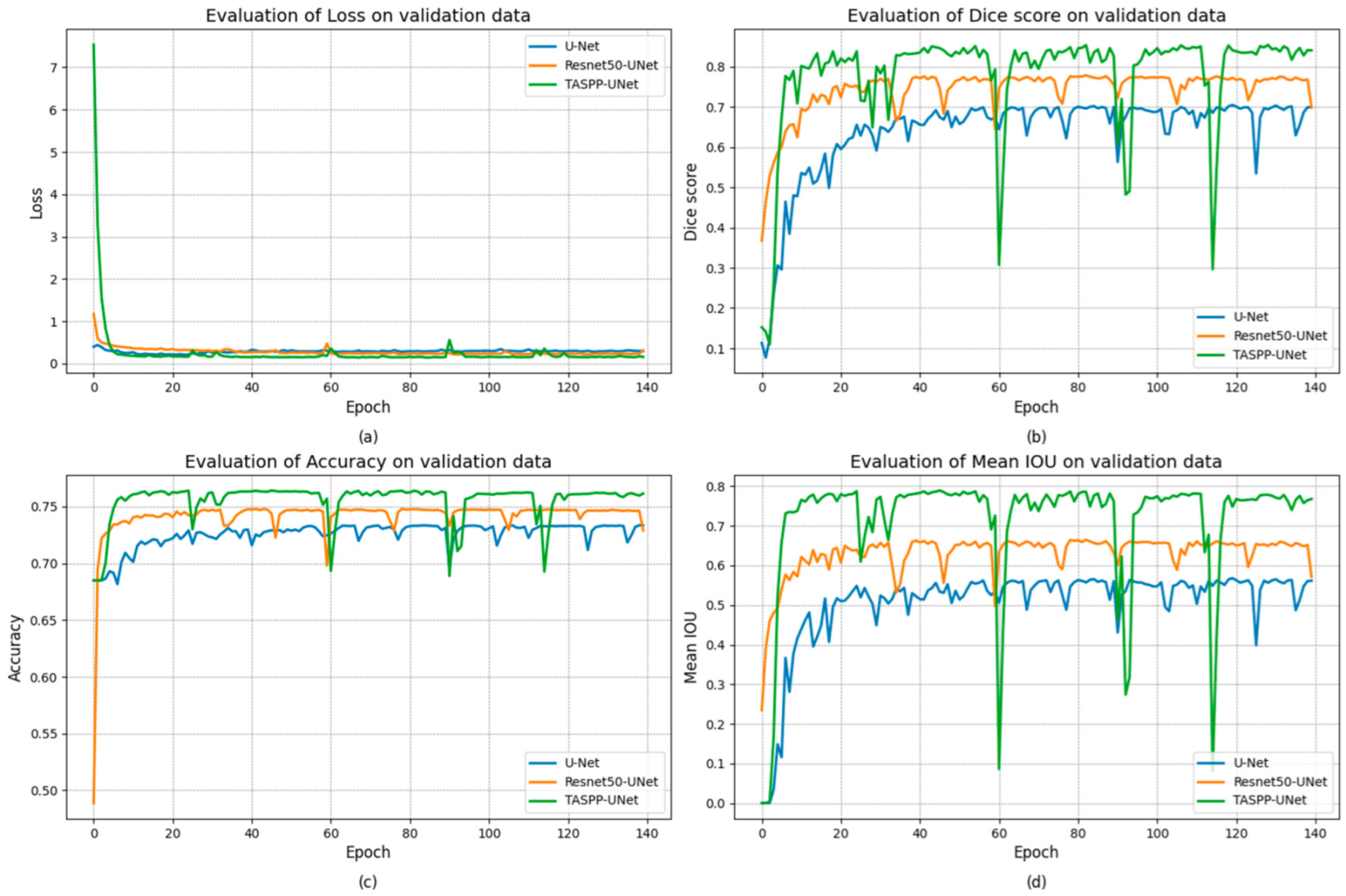
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Waldum, H.; Fossmark, R. Gastritis, Gastric Polyps and Gastric Cancer. Int. J. Mol. Sci. 2021, 22, 6548. [Google Scholar] [CrossRef] [PubMed]
- Maida, M.; Macaluso, F.S.; Ianiro, G.; Mangiola, F.; Sinagra, E.; Hold, G.; Maida, C.; Cammarota, G.; Gasbarrini, A.; Scarpulla, G. Screening of colorectal cancer: Present and future. Expert Rev. Anticancer Ther. 2017, 17, 1131–1146. [Google Scholar] [CrossRef] [PubMed]
- Mathews, A.A.; Draganov, P.V.; Yang, D. Endoscopic management of colorectal polyps: From benign to malignant polyps. World J. Gastrointest. Endosc. 2021, 13, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-H. Understanding colorectal polyps to prevent colorectal cancer. J. Korean Med. Assoc. 2023, 66, 626–631. [Google Scholar] [CrossRef]
- Jia, X.; Xing, X.; Yuan, Y.; Xing, L.; Meng, M.Q.-H. Wireless Capsule Endoscopy: A New Tool for Cancer Screening in the Colon With Deep-Learning-Based Polyp Recognition. Proc. IEEE 2020, 108, 178–197. [Google Scholar] [CrossRef]
- Kim, N.H.; Jung, Y.S.; Jeong, W.S.; Yang, H.J.; Park, S.K.; Choi, K.; Park, D.I. Miss rate of colorectal neoplastic polyps and risk factors for missed polyps in consecutive colonoscopies. Intestig. Res. 2017, 15, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Glissen Brown, J.R.; Mansour, N.M.; Wang, P.; Chuchuca, M.A.; Minchenberg, S.B.; Chandnani, M.; Liu, L.; Gross, S.A.; Sengupta, N.; Berzin, T.M. Deep Learning Computer-aided Polyp Detection Reduces Adenoma Miss Rate: A United States Multi-center Randomized Tandem Colonoscopy Study (CADeT-CS Trial). Clin. Gastroenterol. Hepatol. 2022, 20, 1499–1507.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, P.; Glissen Brown, J.R.; Berzin, T.M.; Zhou, G.; Lei, S.; Liu, X.; Li, L.; Xiao, X. Lower Adenoma Miss Rate of Computer-Aided Detection-Assisted Colonoscopy vs. Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology 2020, 159, 1252–1261.e5. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ghatwary, N.; Jha, D.; Isik-Polat, E.; Polat, G.; Yang, C.; Li, W.; Galdran, A.; Ballester, M.Á.G.; Thambawita, V.; et al. Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge. Sci. Rep. 2024, 14, 2032. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Springer: Cham, Switzerland, 2015; Volume 9351. [Google Scholar] [CrossRef]
- Zunair, H.; Ben Hamza, A. Sharp U-Net: Depthwise convolutional network for biomedical image segmentation. Comput. Biol. Med. 2021, 136, 104699. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Peralta, L.F.; Bote-Curiel, L.; Picon, A.; Sanchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, A.A.; Iglovikov, V.I.; Rakhlin, A.; Shvets, A.A. Medical Image Segmentation Using Deep Neural Networks with Pre-trained Encoders. In Deep Learning Applications: Advances in Intelligent Systems and Computing; Wani, M., Kantardzic, M., Sayed-Mouchaweh, M., Eds.; Springer: Singapore, 2020; Volume 1098. [Google Scholar] [CrossRef]
- Zamanoglu, E.S.; Erbay, S.; Cengil, E.; Kosunalp, S.; Tumen, V.; Demir, K. Land Cover Segmentation using DeepLabV3 and ResNet50. In Proceedings of the 2023 4th International Conference on Communications, Information, Electronic and Energy Systems (CIEES), Plovdiv, Bulgaria, 23–25 November 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Bengio, Y. Practical Recommendations for Gradient-Based Training of Deep Architectures. In Neural Networks: Tricks of the Trade; Lecture Notes in Computer Science; Montavon, G., Orr, G.B., Müller, K.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7700. [Google Scholar] [CrossRef]
- Jie, F.; Nie, Q.; Li, M.; Yin, M.; Jin, T. Atrous spatial pyramid convolution for object detection with encoder-decoder. Neurocomputing 2021, 464, 107–118. [Google Scholar] [CrossRef]
- Khan, S.H.; Hayat, M.; Porikli, F. Regularization of deep neural networks with spectral dropout. Neural Netw. 2019, 110, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, W.; Chen, X.; Liu, Y.; Niu, J. An End-to-End Atrous Spatial Pyramid Pooling and Skip-Connections Generative Adversarial Segmentation Network for Building Extraction from High-Resolution Aerial Images. Appl. Sci. 2022, 12, 5151. [Google Scholar] [CrossRef]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Yeung, M.; Rundo, L.; Nan, Y.; Sala, E.; Schönlieb, C.B.; Yang, G. Calibrating the Dice Loss to Handle Neural Network Overconfidence for Biomedical Image Segmentation. J. Digit. Imaging 2023, 36, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Popovic, A.; De la Fuente, M.; Engelhardt, M.; Radermacher, K. Statistical validation metric for accuracy assessment in medical image segmentation. Int. J. Comput. Assist. Radiol. Surg. 2007, 2, 169–181. [Google Scholar] [CrossRef]
- Berman, M.; Triki, A.R.; Blaschko, M.B. The lovász-softmax loss: A tractable surrogate for the optimization of the intersection-over-union measure in neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4413–4421. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D. Kvasir-SEG: A Segmented Polyp Dataset. In MultiMedia Modeling MMM 2020; Lecture Notes in Computer Science; Ro, Y., Cheng, W.-H., Kim, J., Chu, W.-T., Cui, P., Choi, J.-W., Hu, M.-C., De Neve, W., Eds.; Springer: Cham, Switzerland, 2020; Volume 11962. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Mukasheva, A.; Koishiyeva, D.; Suimenbayeva, Z.; Rakhmetulayeva, S.; Bolshibayeva, A.; Sadikova, G. Comparison evaluation of unet-based models with noise augmentation for breast cancer segmentation on ultrasound image. East.-Eur. J. Enterp. Technol. 2023, 5, 85–97. [Google Scholar] [CrossRef]
- Bock, S.; Weiß, M. A Proof of Local Convergence for the Adam Optimizer. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar] [CrossRef]


| Method | Features | Parameters | Size (MB) |
|---|---|---|---|
| TASPP-UNet | ASPP and ResNet-50 | 19,582,337 | 74.70 |
| Resnet50-UNet | ResNet-50 | 14,877,313 | 63.42 |
| U-Net | Skip connections | 7,531,521 | 28.73 |
| Method | Loss | Dice Score | Accuracy Score | Mean IOU |
|---|---|---|---|---|
| TASPP-U-Net | 0.2243 | 0.9147 | 0.7655 | 0.9276 |
| Resnet-50 U-Net | 0.0983 | 0.9176 | 0.7417 | 0.9128 |
| U-Net | 0.0608 | 0.8731 | 0.7256 | 0.8607 |
| Method | Loss | Dice Score | Accuracy Score | Mean IOU |
|---|---|---|---|---|
| TASPP-UNet | 0.2628 | 0.7798 | 0.7555 | 0.7141 |
| Resnet50-UNet | 0.2800 | 0.7430 | 0.7417 | 0.6311 |
| U-Net | 0.2877 | 0.6386 | 0.7256 | 0.5076 |
| Method | Loss | Dice Score | Accuracy Score | Mean IOU |
|---|---|---|---|---|
| TASPP-UNet | 0.0963 | 0.8967 | 0.5624 | 0.8789 |
| Resnet-50-UNet | 0.2248 | 0.7587 | 0.5406 | 0.6714 |
| U-Net | 0.1461 | 0.8234 | 0.5529 | 0.7667 |
| Method | Training Time | Epoch Duration | Training Speed (FPS) |
|---|---|---|---|
| TASPP-UNet | 1186 | 8.5 | 151.1 |
| Resnet50-UNet | 2123 | 15.2 | 84.4 |
| U-Net | 2269 | 16.2 | 79.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukasheva, A.; Koishiyeva, D.; Sergazin, G.; Sydybayeva, M.; Mukhammejanova, D.; Seidazimov, S. Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet. Eng. Proc. 2024, 70, 16. https://doi.org/10.3390/engproc2024070016
Mukasheva A, Koishiyeva D, Sergazin G, Sydybayeva M, Mukhammejanova D, Seidazimov S. Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet. Engineering Proceedings. 2024; 70(1):16. https://doi.org/10.3390/engproc2024070016
Chicago/Turabian StyleMukasheva, Assel, Dina Koishiyeva, Gani Sergazin, Madina Sydybayeva, Dinargul Mukhammejanova, and Syrym Seidazimov. 2024. "Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet" Engineering Proceedings 70, no. 1: 16. https://doi.org/10.3390/engproc2024070016
APA StyleMukasheva, A., Koishiyeva, D., Sergazin, G., Sydybayeva, M., Mukhammejanova, D., & Seidazimov, S. (2024). Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet. Engineering Proceedings, 70(1), 16. https://doi.org/10.3390/engproc2024070016







