An Alternative Method to Measure Glucose and Lactic Acid as Biomarkers of the Postmortem Interval (PMI)
Abstract
1. Introduction
2. Materials and Methods
2.1. Donor Samples
2.2. Glucose and Lactic Acid Colorimetric Test
2.3. Population Model for the Kinetics of the Two Markers
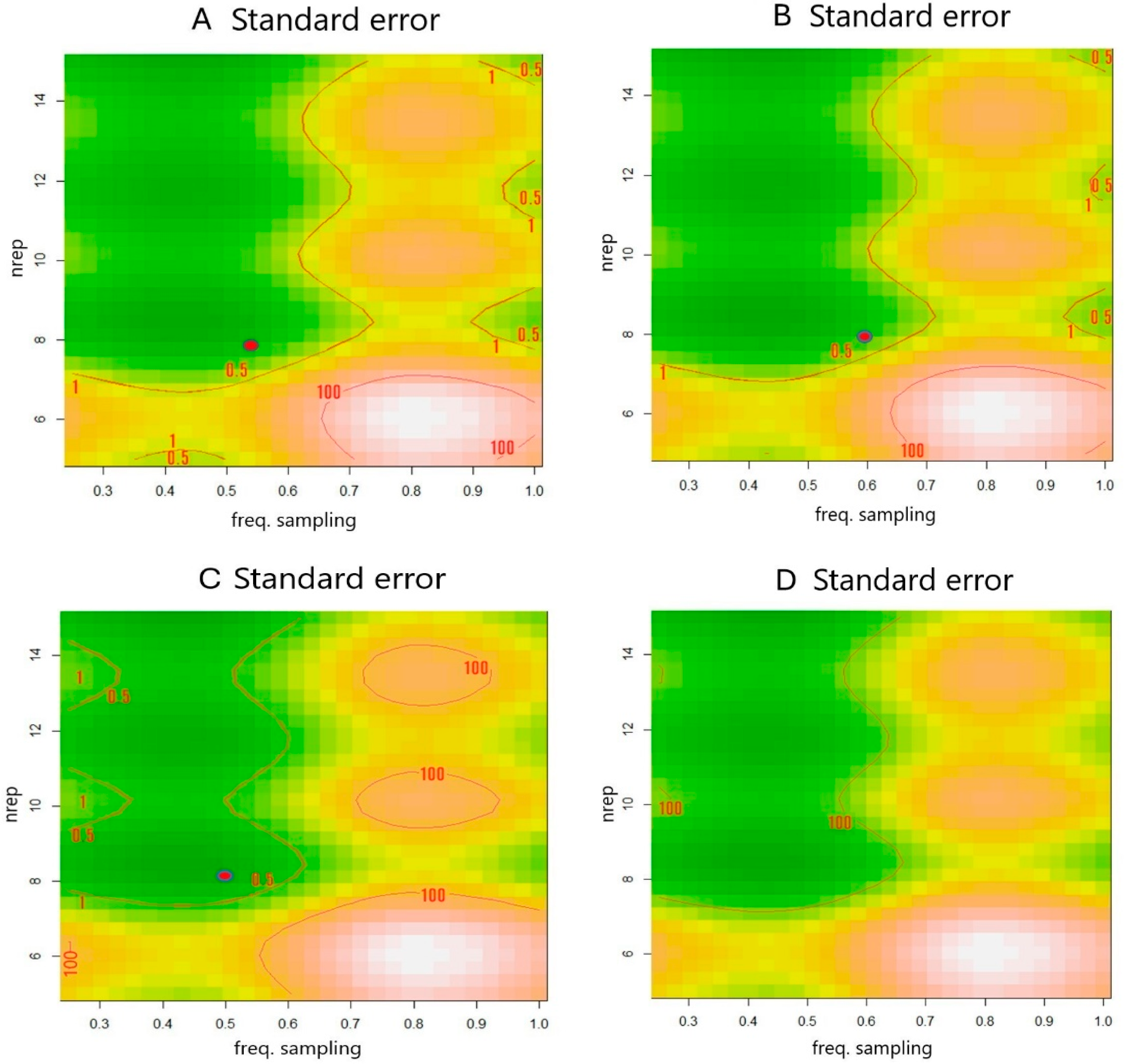
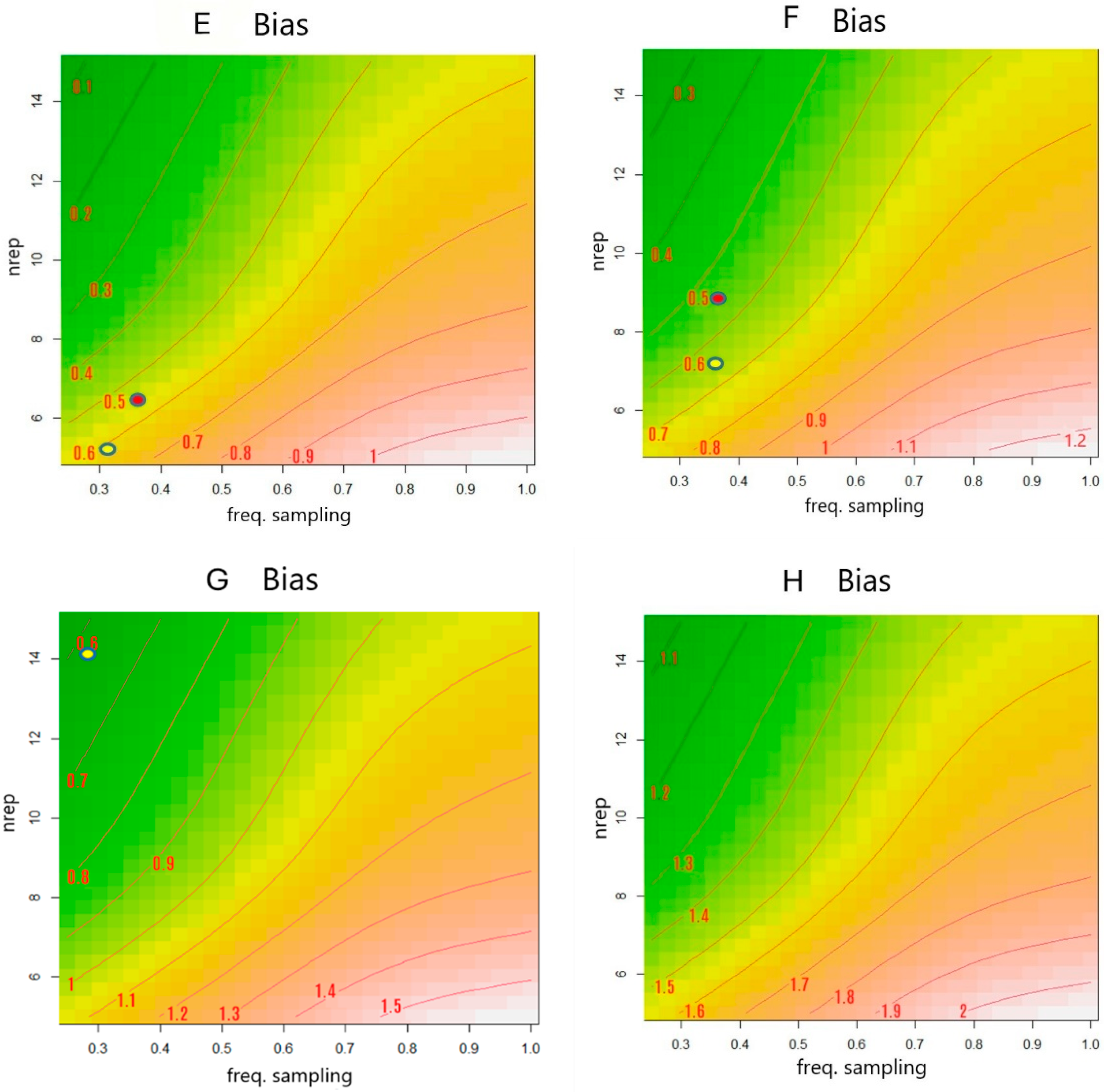
2.4. Effect of the Postmortem Sampling Design in the Accuracy and Precision of the PMI Estimate
2.5. Statistics
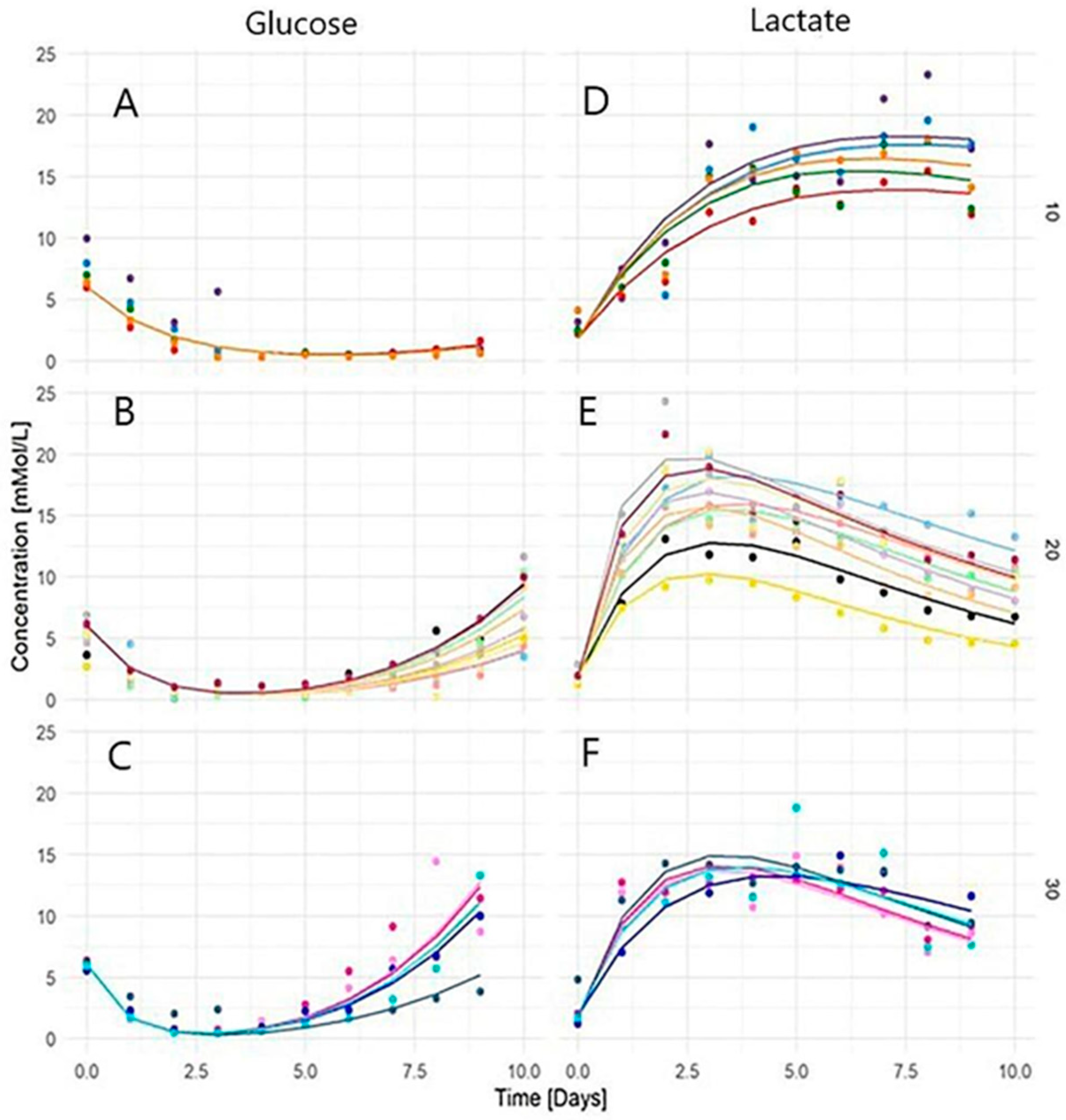
3. Results
3.1. Glucose and Lactic Acid Kinetics and Temperature Dependence
3.2. Parameters of Population Model
3.3. Best Set of Multiple Measurements That Delivered the Least Accuracy and Best Precision
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaliszan, M.; Hauser, R.; Kernbach-Wighton, G. Estimation of the time of death based on the assessment of post mortem processes with emphasis on body cooling. Legal Med. 2009, 11, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Kanchan, T.; Krishan, K. Methods of Estimation of Time Since Death. In StatPearls [Internet]; StatPearls: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK549867/ (accessed on 6 June 2024).
- Saukko, P.; Knight, B. Knight’s Forensic Pathology, 3rd ed.; Hodder Arnold: London, UK, 2004; p. 64. [Google Scholar]
- Swain, R.; Kumar, A.; Sahoo, J.; Lakshmy, R.; Gupta, S.K.; Bhardwaj, D.N.; Pandey, R.M. Estimation of post-mortem interval: A comparison between cerebrospinal fluid and vitreous humour chemistry. J. Forensic Leg. Med. 2015, 36, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Sacco, M.A.; Cordasco, F.; Scalise, C.; Ricci, P.; Aquila, I. Systematic Review on Post-Mortem Protein Alterations: Analysis of Experimental Models and Evaluation of Potential Biomarkers of Time of Death. Diagnostics 2022, 12, 1490. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Cunha, E. Can we infer post mortem interval on the basis of decomposition rate? A case from a Portuguese cemetery. Forensic Sci. Int. 2013, 226, 298.e1–298.e6. [Google Scholar] [CrossRef]
- Nolan, A.-N.; Mead, R.J.; Maker, G.; Speers, S.J. A review of the biochemical products produced during mammalian decomposition with the purpose of determining the post-mortem interval. Aust. J. Forensic Sci. 2019, 52, 477–488. [Google Scholar] [CrossRef]
- Yang, T.; You, M.; Zhang, H.; Zhang, F.; Bai, R.; Chang, L.; Di, S.; Guo, Z.; Lu, D.; Wang, X.; et al. Estimation of the Postmortem Interval by Measuring Blood Oxidation-reduction Potential Values. J. Forensic Sci. Med. 2016, 2, 8–11. [Google Scholar] [CrossRef]
- Kumar, S.; Verma, A.K. Estimation of postmortem interval using the data of insulin level in the cadaver’s blood. Data Brief 2016, 7, 354–356. [Google Scholar] [CrossRef]
- Nishida, A.; Funaki, H.; Kobayashi, M.; Tanaka, Y.; Akasaka, Y.; Kubo, T.; Ikegaya, H. Blood creatinine level in postmortem cases. Sci. Justice 2015, 55, 195–199. [Google Scholar] [CrossRef]
- Castillo-Peinado, L.S.; Luque de Castro, M.D. An overview on forensic analysis devoted to analytical chemists. Talanta 2017, 167, 181–192. [Google Scholar] [CrossRef]
- Whitney, A.R.; Diehn, M.; Popper, S.J.; Alizadeh, A.A.; Boldrick, J.C.; Relman, D.A.; Brown, P.O. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA 2003, 100, 1896–1901. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Y.; He, J.; Zhang, Y.; Wang, T.; Wu, L.; Sun, N.; Fang, T.; Mao, H.; Tang, N.J.; et al. Effects of heat waves and cold spells on blood parameters: A cohort study of blood donors in Tianjin, China. Environ. Health Prev. Med. 2024, 29, 25. [Google Scholar] [CrossRef] [PubMed]
- Matrai, A.A.; Varga, G.; Tanczos, B.; Barath, B.; Varga, A.; Horvath, L.; Bereczky, Z.; Deak, A.; Nemeth, N. In vitro effects of temperature on red blood cell deformability and membrane stability in human and various vertebrate species. Clin. Hemorheol. Microcirc. 2021, 78, 291–300. [Google Scholar] [CrossRef]
- Meurs, J.; Krap, T.; Duijst, W. Evaluation of postmortem biochemical markers: Completeness of data and assessment of implication in the field. Sci. Justice 2019, 59, 177–180. [Google Scholar] [CrossRef]
- Kost, G.J.; Nguyen, T.H.; Tang, Z. Whole-blood glucose and lactate: Trilayer biosensors, drug interference, metabolism, and practice guidelines. Arch. Pathol. Lab. Med. 2000, 124, 1128–1134. [Google Scholar] [CrossRef]
- Costa, I.; Carvalho, F.; Magalhães, T.; Guedes de Pinho, P.; Silvestre, R.; Dinis-Oliveira, R.J. Promising blood-derived biomarkers for estimation of the postmortem interval. Toxicol. Res. 2015, 4, 1443–1452. [Google Scholar] [CrossRef]
- I Muñoz, J.; Costas, E.; Rodrĺguez-Calvo, M.S.; Suárez-Peñaranda, J.M.; López-Rivadulla, M.; Concheiro, L. A high-performance liquid chromatography method for hypoxanthine determination in vitreous humour: Application to estimation of post mortem interval. Hum. Exp. Toxicol. 2006, 25, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, C.; Ordóñez-Mayán, L.; Lendoiro, E.; Febrero-Bande, M.; Vieira, D.N.; Muñoz-Barús, J.I. A reliable method for estimating the postmortem interval from the biochemistry of the vitreous humor, temperature and body weight. Forensic Sci. Int. 2019, 295, 157–168. [Google Scholar] [CrossRef]
- Rodrigo, M.R. A Nonlinear Least Squares Approach to Time of Death Estimation Via Body Cooling. J. Forensic Sci. 2016, 61, 230–233. [Google Scholar] [CrossRef]
- Weiser, M.; Roggendorf, M.; Schuck, J. Optimal design of experiments for estimating the time of death in forensic medicine. Inverse Probl. 2018, 34, 125005. [Google Scholar] [CrossRef]
- Collins, S.; Maestrini, L.; Hui, F.K.C.; Stuart, B.; Ueland, M. The use of generalized linear mixed models to investigate postmortem lipids in textiles. iScience 2023, 26, 107371. [Google Scholar] [CrossRef]
- Beneke, R.; Hütler, M.; Jung, M.; Leithäuser, R.M. Modeling the blood lactate kinetics at maximal short-term exercise conditions in children, adolescents, and adults. J. Appl. Physiol. 2005, 99, 499–504. [Google Scholar] [CrossRef]
- Flores-Bazán, F.; Hadjisavvas, N.; Lara, F.; Montenegro, I. First- and Second-Order Asymptotic Analysis with Applications in Quasiconvex Optimization. J. Optim. Theory Appl. 2016, 170, 372–393. [Google Scholar] [CrossRef]
- Murthy, D.N.P.; Xie, M.; Jiang, R. Weibull Models; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2004. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; EISPACK authors; Heisterkamp, S.; Van Willigen, B.J.; Ranke, J.; R Core Team. nlme: Linear and nonlinear mixed effects models. R Package Version 2017, 3, 274. [Google Scholar]
- Haralampu, S.G.; Saguy, I.; Karel, M. Estimation of Arrhenius model parameters using three least squares methods. J. Food Sci. 1985, 9, 129–143. [Google Scholar] [CrossRef]
- Schoenly, K.; Goff, M.L.; Wells, J.D.; Lord, W.D. Quantifying statistical uncertainty in succession-based entomological estimates of the postmortem interval in death scene investigations: A simulation study. Am. Entomol. 1996, 42, 106–112. [Google Scholar] [CrossRef]
- Padfield, D.; Matheson, G. nls.multstart: Robust Non-Linear Regression using AIC Scores. R Package Version 2018, 1, 1–5. Available online: https://CRAN.R-project.org/package=nls.multstart (accessed on 15 April 2025).
- Nahm, F.S. Nonparametric statistical tests for the continuous data: The basic concept and the practical use. Korean J. Anesthesiol. 2016, 69, 8–14. [Google Scholar] [CrossRef]
- Garg, S.P.; Garg, V. Serum enzymes changes after death & its correlation with time since death. J. Indian Acad. Forensic Med. 2010, 32, 355–357. [Google Scholar]
- Go, A.; Shim, G.; Park, J.; Hwang, J.; Nam, M.; Jeong, H.; Chung, H. Analysis of hypoxanthine and lactic acid levels in vitreous humor for the estimation of post-mortem interval (PMI) using LC–MS/MS. Forensic Sci. Int. 2019, 299, 135–141. [Google Scholar] [CrossRef]
- Rognum, T.; Holmen, S.; Musse, M.; Dahlberg, P.; Stray-Pedersen, A.; Saugstad, O.; Opdal, S. Estimation of time since death by vitreous humor hypoxanthine, potassium, and ambient temperature. Forensic Sci. Int. 2016, 262, 160–165. [Google Scholar] [CrossRef]
- Szeremeta, M.; Samczuk, P.; Pietrowska, K.; Kowalczyk, T.; Przeslaw, K.; Sieminska, J.; Kretowski, A.; Niemcunowicz-Janica, A.; Ciborowski, M. In vitro animal model for estimating the time since death with attention to early postmortem stage. Metabolites 2023, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, J.; Takinami, Y.; Kawai, Y.; Kimura-Kataoka, K.; Takeshita, H. Comparison of serum cell-free DNA between postmortem and living samples. Clin. Chim. Acta 2021, 519, 255–259. [Google Scholar] [CrossRef] [PubMed]
- El-Din, E.A.A.; Ahmed, S.M.; El Shafei, D.A.; Mostafa, H.E.-S. Implication of high-mobility group box-1 and skin post mortem changes in estimation of time passed since death: Animal and human study. Legal Med. 2021, 53, 101949. [Google Scholar] [CrossRef] [PubMed]
- Usumoto, Y.; Kudo, K.; Tsuji, A.; Ihama, Y.; Ikeda, N. Predictive equation to estimate post-mortem interval using spectrophotometric blood-colour values. Med. Sci. Law 2019, 59, 36–41. [Google Scholar] [CrossRef]
- Donaldson, A.E.; Lamont, I.L. Biochemistry changes that occur after death: Potential markers for determining post-mortem interval. PLoS ONE 2013, 8, 1–10. [Google Scholar] [CrossRef]
- Palmiere, C.; Mangin, P.; Augsburger, M. Postmortem biochemical investigation results in situations of fatal mechanical compression of the neck region. Legal Med. 2018, 30, 59–63. [Google Scholar] [CrossRef]
- Palmiere, C. Postmortem diagnosis of diabetes mellitus and its complications. Croat. Med. J. 2015, 56, 181–193. [Google Scholar] [CrossRef]
- Trivedi, D.; Narayana, K.S. Postmortem blood glucose level: How useful to envisage the time since death. Int. J. Clin. Biochem. Res. 2015, 2, 61–64. [Google Scholar]
- Woydt, L.; Bernhard, M.; Kirsten, H.; Burkhardt, R.; Hammer, N.; Gries, A.; Dreßler, J.; Ondruschka, B. Intra-individual alterations of serum markers routinely used in forensic pathology depending on increasing post-mortem interval. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Van Dongen, H.P.; Olofsen, E.; Dinges, D.F.; Maislin, G. Mixed-model regression analysis and dealing with interindividual differences. Methods Enzymol. 2004, 384, 139–171. [Google Scholar] [CrossRef]
- Anderson, B.J.; Allegaert, K.; Holford, N.H.G. Population clinical pharmacology of children: Modelling covariate effects. Eur. J. Pediatr. 2006, 165, 819–829. [Google Scholar] [CrossRef]
- Allegaert, K.; Peeters, M.Y.; Verbesselt, R.; Tibboel, D.; Naulaers, G.; de Hoon, J.N.; Knibbe, C.A. Inter-individual variability in propofol pharmacokinetics in preterm and term neonates. Br. J. Anaesth. 2007, 99, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, H.; Yang, T.; Ding, Z.; Wu, S.; Qiu, X.; Liu, Q. A study on the estimation of postmortem interval based on environmental temperature and concentrations of substances in vitreous humor. J. Forensic Sci. 2018, 63, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, G.M.M.; Hegazy, N.I.; Etewa, R.L.; Elmesallamy, G.E.A. Postmortem redistribution of drugs: A literature review. Forensic Sci. Med. Pathol. 2024, 20, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Zilg, B.; Alkass, K.; Berg, S.; Druid, H. Postmortem identification of hyperglycemia. Forensic Sci. Int. 2009, 185, 89–95. [Google Scholar] [CrossRef]
- Chen, J.H.; Michiue, T.; Inamori-Kawamoto, O.; Ikeda, S.; Ishikawa, T.; Maeda, H. Comprehensive investigation of postmortem glucose levels in blood and body fluids with regard to the cause of death in forensic autopsy cases. Leg. Med. 2015, 17, 475–482. [Google Scholar] [CrossRef]
| Parameter Estimates | |
| Lactate | |
| Lmax | 24.02 (1.86) ** |
| IK1 | −0.88 (0.16) ** |
| Lo | 1.88 (0.38) ** |
| IK2ref | −1.98 (0.16) ** |
| Ea_k2 | 35.43 (4.45) ** |
| Parameter Estimates | |
| Glucose | |
| IKelref | −0.15 (0.09) * |
| Ea_kel | 29.14 (8.21) ** |
| Glus0 | 6.03 (0.25) ** |
| ImIref | −5.93 (0.47) ** |
| Ea_ml | 79.80 (11.97) ** |
| s | 3.33 (0.22) ** |
| Random Effects | |
| σ s ID | 0.16 |
| σ Ik1 ID | 0.44 |
| σ Ik2 ID | 0.35 |
| AIC | 1662.79 |
| BIC | 1731.48 |
| Log likelihood | −814.40 |
| Num. obs. | 420 |
| Num. groups | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tersaruolo, C.; Frias, J.; Howe, O. An Alternative Method to Measure Glucose and Lactic Acid as Biomarkers of the Postmortem Interval (PMI). Forensic Sci. 2025, 5, 17. https://doi.org/10.3390/forensicsci5020017
Tersaruolo C, Frias J, Howe O. An Alternative Method to Measure Glucose and Lactic Acid as Biomarkers of the Postmortem Interval (PMI). Forensic Sciences. 2025; 5(2):17. https://doi.org/10.3390/forensicsci5020017
Chicago/Turabian StyleTersaruolo, Claudio, Jesus Frias, and Orla Howe. 2025. "An Alternative Method to Measure Glucose and Lactic Acid as Biomarkers of the Postmortem Interval (PMI)" Forensic Sciences 5, no. 2: 17. https://doi.org/10.3390/forensicsci5020017
APA StyleTersaruolo, C., Frias, J., & Howe, O. (2025). An Alternative Method to Measure Glucose and Lactic Acid as Biomarkers of the Postmortem Interval (PMI). Forensic Sciences, 5(2), 17. https://doi.org/10.3390/forensicsci5020017









