An Interactive Dashboard for Statistical Analysis of Intensive Care Unit COVID-19 Data
Abstract
1. Introduction
Goals and Structure of This Paper
2. Related Work
2.1. Preliminary Work on the PREMO Project—The Starting Point
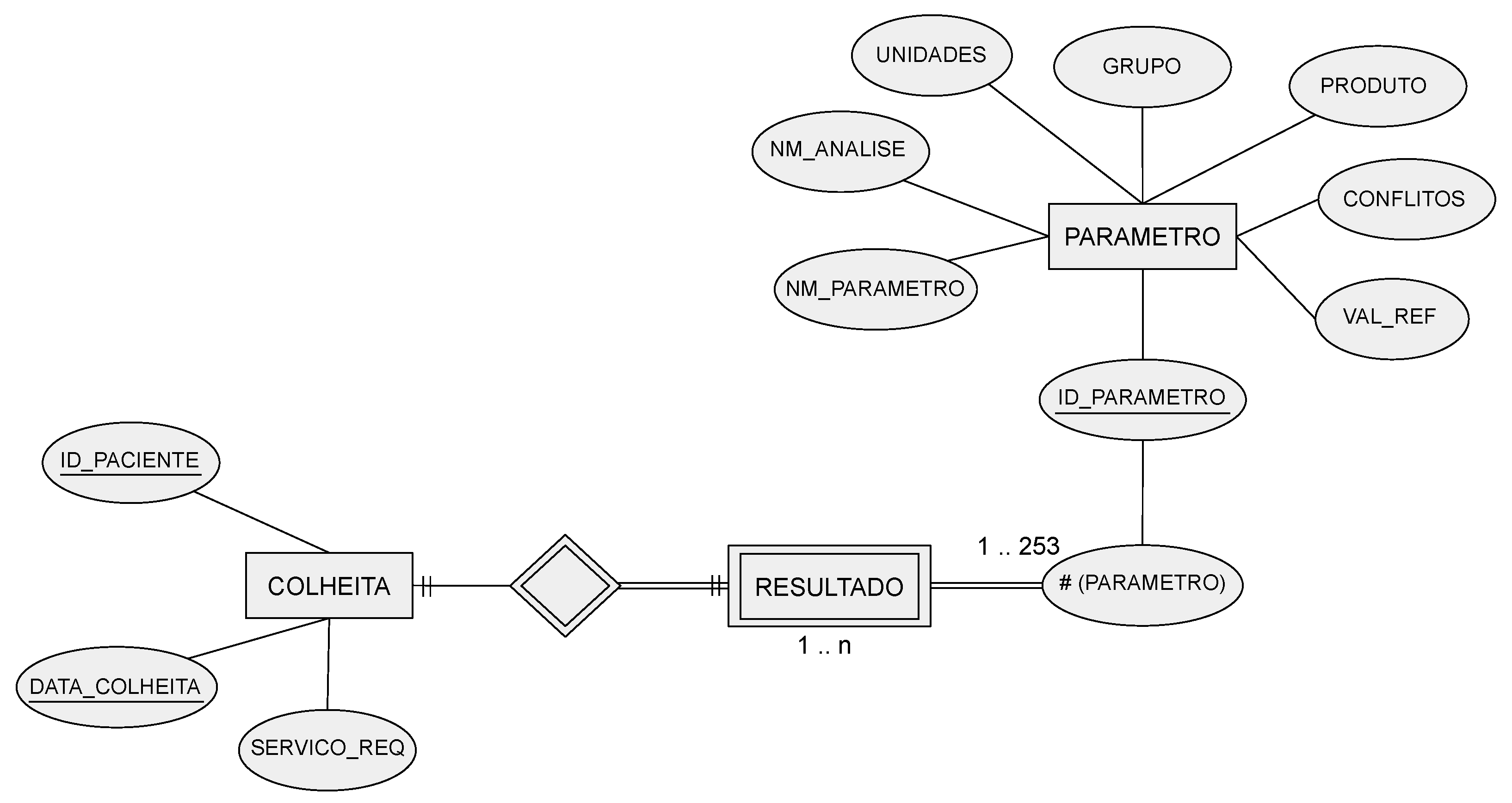
- To devise a relational database with the LD of the patients admitted to the ICU, using scripts written in Python. The database was set up dynamically allowing rapid adaptation depending on the number of biomarkers that may arise.
- To perform the necessary data transformations before being stored in the database, without providing any dashboard nor visualization capabilities.
- COLLECTION—identifies the patient, a collection date, and the service that requested the collection.
- PARAMETER—stores different variables (parameters), such as the name of the laboratory, the parameter name, and reference values, among others.
- RESULT—the patient’s analysis results for each parameter.
2.2. Dashboards for COVID-19 and Clinical Data
3. Proposed Solution—Web Application with a Dashboard
3.1. Application’s Functional and Nonfunctional Requirements and Key Entities
- Aggregate data from all the COVID-19 waves.
- Data import and export, using text files and graphic files.
- Data visualization through various types of statistical graphs, such as line, bar, pie, scatter plots, box plots, and survival curves.
- Download of graphics presented in various formats.
- Filling in forms to select the data to be displayed.
- Filter data by COVID-19 waves.
- Generate reports with statistical analysis.
- Users may have different roles for user management, such as adding, removing, and editing (available to users with the administrator role).
- Authentication system, with proper storage of passwords.
- Data processing, according to their type—cross-sectional or longitudinal.
- Provision of the solution through a web browser.
- Presentation tier: concerns the dashboard, provided by the Web server, which in turn provides the user interface.
- Logic tier: corresponds to the application server. This is in the intermediate layer, which has the business logic and the processing of user inputs. This also constitutes the connection between the two other layers.
- Data tier: uses a relational database server, in which the application’s persistent data are stored.
3.2. Data Processing Phases and Storage in the Database
3.3. Data Visualization Graphs and Diagrams, Importation, and Exportation
- Bar and circular graphs, to analyze the absolute and relative frequency for the categories of nominal variables.
- Box plot graphs, to visualize the distribution of quantitative variables. In addition, parallel box plots allow a data comparison between groups of patients. In both cases, a summary of descriptive statistics is provided.
- Line graphs, to analyze the evolution of a parameter across the ICU admission. It can be performed over a restricted group of patients.
- Scatter plots, to assess the correlation between two quantitative variables.
- Survival curves, to visualize the estimated survival probability of an individual living longer than a certain time.
4. Development of the Three Tiers of the Proposed Solution
4.1. Choice of Programming Languages and Libraries for Each Tier
4.2. Web Server Implementation’s Key Aspects and User Navigation
- An error code, used by all field components to display an error message.
- A handler invoked when there is a change in the field.
- A list of selected options and values, for drop-down lists.
- MyPlot, composed by ModalDownload and Plot.
- Controller, where each graphic has a specific field to control the data. All available graphics, except one, have one field that allows the user to choose a specific COVID-19 wave and another to separate the graph into many others where each one corresponds to a specific wave.
- Data, which refers to a list of objects, with the graphics to be drawn. These objects indicate the values on the XX-axis and on the YY-axis, type of graphics, content of legends, colors, and others.
- Layout, an object that holds information about the layout of the “page” in which the graphics are drawn, such as the dimension, title, name of the axes, borders, annotations (strings that can be placed together with the graphics), shapes (such as straight lines, that can be added to graphics) as well as other fields.
- Config, an object that allows one to define properties such as buttons in the menu and also interactivity with the graphics, such as scrolling to zoom in and out or add or remove buttons in the button menu, among other actions.
4.3. API Implementation and Module Organization
- Endpoints, with the application’s routes and handlers.
- Services, holding the contracts and specific implementations of the services that handle the application logic.
- Data access layer (DAL), with contracts and their specific implementations, to deal with the database access logic.
- Data transformation objects (DTO), having objects used to pass information between layers, between the business layer and the data access layer, or between servers.
- Exceptions, the exceptions that may be raised by running the application.
- Utils, which contains utility functions for the entire application.
4.4. Solution Deployment—Making the Solution Available
- Serve [54], a module written in JavaScript that allows applications to be made available in React.
- Nginx [55], in addition to serving applications, can also function as a load balancer and reverse proxy.
- Tomcat [56], which is an Apache project that acts as a Java Web server.
- Heroku [59], used to build, run, and operate applications in the cloud.
- Google App Engine [60], which is a cloud service to develop applications.
5. Data Visualization Graphics, Diagrams, and Statistical Analysis
5.1. Dashboard Landing Page and Request Input Page
5.2. Graphical Representations for Cross-Sectional Data
5.2.1. Pie Charts and Bar Charts—Observing Absolute and Relative Frequencies
5.2.2. Scatter Plots—Assessing Statistical Correlations
5.2.3. Box-Plot Diagrams—Comparing Groups of Patients
5.3. Graphical Representations for Longitudinal Data
5.3.1. Kaplan–Meier Survival Curves—Estimating Survival Probability
5.3.2. Line Graphics—Analysis and Comparison of Parameter Evolution
- One to analyze the evolution of a parameter for different patients, as shown in Figure 21.
5.4. Discussion on the Use of Reported Graphics
- Observe the frequency and percentage of the admission of patients to the ICU, among the COVID-19 waves. For instance, Figure 12 shows that the third wave was the one with more ICU admissions. On one hand, throughout the third wave, the Alpha variant was related with a larger rate of deaths in the ICU, when compared to the Delta variant, which probably led to a greater number of admissions to the ICU and more severe conditions of the disease, affecting the older population still not vaccinated. On the other hand, the number of ICU admissions in the fifth and sixth waves decreased significantly. This fact must be related to the appearance of other variants and the availability of vaccines to most of the Portuguese population [1].
- Check for correlations between parameters of the medical tests conducted on ICU patients. From the literature on COVID-19, we know that there exists a correlation between some parameters. By analyzing graphs such as the ones reported in Figure 13 and Figure 14, we can confirm existing known correlations or discover new ones. Additionally, we highlight that this tool allows users to calculate ratios between parameters, allowing them to explore their association with more severe events, in the context of COVID-19 patients. Available examples in our tool with a recognized clinical interest are neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios, found to be markers of inflammation and prognosis for more severe states of COVID-19 [64]. Moreover, with the ability to separate the data by wave, we can also look for correlations between parameters and their changes in different waves.
- Check for statistical significance between relevant data comparisons. This is a key feature of the developed application, since it provides a strong notion about the recorded analysis numbers and whether they are statistically significant or not. For instance, the box plots in Figure 15 and Figure 16 are examples of this case.
- Perform an analysis of the survival data. The analysis of the graphics in Figure 17 and Figure 18 allows one to observe and compare the survival probability of patients in both groups, along the ICU stay. For example, the clinician can visually compare the median survival time, by group, or understand which of the two groups of patients experiences a larger number of events in the first week of ICU admission.
- In Figure 19, we illustrate the possibility of visualizing and comparing survival curves that correspond to a stratification of the sample into groups defined by criteria of clinical interest, such as normality/non-normality values of certain biomarkers.
- Analyzing Figure 20, we gain a clear notion of how the survival probability drops with the increase in the number of days in the ICU, per gender and per wave. This may lead to a treatment adjustment and better resource management in the ICU. In addition, this graphic highlights the different characteristics of survival data per wave of COVID-19.
- The visualization of Figure 21, Figure 22 and Figure 23 allows the clinician to obtain information on the clinical practice biomarkers’ trajectory. Many studies address the importance of analyzing the association between death and certain biomarkers’ trajectory patterns. For example, Chen et al. [65] modeled the longitudinal trajectories of laboratory biomarkers and made dynamical predictions on individual prognoses.
6. Conclusions
Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Florensa, D.; Mateo, J.; Spaimoc, R.; Miret, C.; Godoy, S.; Solsona, F.; Godoy, P. Severity of COVID-19 cases in the months of predominance of the Alpha and Delta variants. Sci. Rep. 2022, 12, 15456. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Sohrabi, M.R.; Hannani, K. Five consecutive epidemiological waves of COVID-19: A population-based cross-sectional study on characteristics, policies, and health outcome. BMC Infect. Dis. 2023, 22, 906. [Google Scholar] [CrossRef] [PubMed]
- Abati, E.; Stellio, L.; Manini, A.; Moroni, F.; Azzalini, L.; Vilca, L. A cross-sectional survey study of the impact of COVID-19 pandemic on the training and quality of life of Italian medical residents in the Lombardy region. Ann. Med. 2022, 54, 2326–2339. [Google Scholar] [CrossRef]
- Collis, A.; Garimella, K.; Moehring, A.; Rahimian, M.A.; Babalola, S.; Gobat, N.H.; Shattuck, D.; Stolow, J.; Aral, S.; Eckles, D. Global survey on COVID-19 beliefs, behaviors and norms. Nat. Hum. Behav. 2022, 6, 1310–1317. [Google Scholar] [CrossRef]
- Novais, F.; Cordeiro, C.; Câmara Pestana, P.; Côrte-Real, B.; Reynolds Sousa, T.; Delerue Matos, A.; Telles-Correia, D. The Impact of COVID-19 in Older People in Portugal: Results from the Survey of Health, Ageing and Retirement (SHARE). Acta Médica Port. 2021, 34, 761–766. [Google Scholar] [CrossRef]
- Hart, W.; Miller, E.; Andrews, N.; Waight, P.; Maini, P.; Funk, S.; Thompson, R. Generation time of the alpha and delta SARS-CoV-2 variants: An epidemiological analysis. Lancet—Infect. Dis. 2022, 22, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Bahl, A.; Mielke, N.; Johnson, S.; Desai, A.; Qu, L. Severe COVID-19 outcomes in pediatrics: An observational cohort analysis comparing Alpha, Delta, and Omicron variants. Lancet Reg. Health—Am. 2023, 18, 100405. [Google Scholar] [CrossRef]
- Uemura, K.; Kanata, T.; Ono, S.; Michihata, N.; Yasunaga, H. The disease severity of COVID-19 caused by Omicron variants: A brief review. Ann. Clin. Epidemiol. 2023, 5, 31–36. [Google Scholar] [CrossRef]
- Petrone, D.; Mateo-Urdiales, A.; Sacco, C.; Riccardo, F.; Bella, A.; Ambrosio, L.; Presti, A.L.; Martino, A.D.; Ceccarelli, E.; Manso, M.D.; et al. Reduction of the risk of severe COVID-19 due to Omicron compared to Delta variant in Italy (November 2021–February 2022). Int. J. Infect. Dis. 2023, 129, 135–141. [Google Scholar] [CrossRef]
- Mondal, M.; Bharati, S.; Podder, P.; Kamruzzaman, J. Deep Learning and Federated Learning for Screening COVID-19: A Review. BioMedInformatics 2023, 3, 691–713. [Google Scholar] [CrossRef]
- Rocchi, E.; Peluso, S.; Sisti, D.; Carletti, M. A New Epidemic Model for the COVID-19 Pandemic: The θ-SI(R)D Model. BioMedInformatics 2022, 2, 398–404. [Google Scholar] [CrossRef]
- Rekowski, C. Development of Predictive Models for COVID-19 Prognosis Based on Patients’ Demographic and Clinical Data. Master’s Thesis, Instituto Superior de Engenharia de Lisboa, Lisboa, Portugal, 2022. [Google Scholar]
- Molenberghs, V.; Verbeke, V. Linear Mixed Models for Longitudinal Data, 3rd ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Kaplan, E.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Collett, D. Modelling Survival Data in Medical Research, 3rd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014. [Google Scholar] [CrossRef]
- Mantel, N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 1966, 50, 163–170. [Google Scholar]
- Cox, D. Regression Models and Life-tables (with discussion). J. R. Stat. Soc. 1972, 34, 187–220. [Google Scholar]
- Gehan, E. A Generalized Wilcoxon Test for Comparing Arbitrarily Singly-Censored Samples. Biometrika 1965, 52, 203–223. [Google Scholar] [CrossRef]
- Breslow, N. A generalized Kruskal-Wallis test for comparing K samples subject to unequal patterns of censorship. Biometrika 1970, 57, 579–594. [Google Scholar] [CrossRef]
- Daniel, W.; Cross, C. Biostatistics: A Foundation for Analysis in the Health Sciences, 10th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Ribeiro, D. Project Report—PREMO. Technical Report, Instituto Superior de Engenharia de Lisboa. 2022. Available online: www.isel.pt (accessed on 21 October 2023).
- Wexler, S.; Shaffer, J.; Cotgreave, A. The Big Book of Dashboards: Visualizing Your Data Using Real-World Business Scenarios; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Intezari, A.; Pauleen, D.J.; Taskin, N. The DIKW hierarchy and management decision-making. In Proceedings of the Hawaii International Conference on System Sciences (HICSS), Koloa, HI, USA, 5–8 January 2016; pp. 4193–4201. [Google Scholar]
- Rowley, J. The wisdom hierarchy: Representations of the DIKW hierarchy. J. Inf. Commun. Sci. 2007, 33, 163–180. [Google Scholar] [CrossRef]
- Zins, C. Conceptual approaches for defining data, information, and knowledge. J. Am. Soc. Inf. Sci. Technol. 2007, 58, 479–493. [Google Scholar] [CrossRef]
- Dong, E.; Ratcliff, J.; Goyea, T.D.; Katz, A.; Lau, R.; Ng, T.K.; Garcia, B.; Bolt, E.; Prata, S.; Zhang, D.; et al. The Johns Hopkins University Center for Systems Science and Engineering COVID-19 Dashboard: Data collection process, challenges faced, and lessons learned. Lancet Infect. Dis. 2022, 22, e370–e376. [Google Scholar] [CrossRef]
- Wissel, B.D.; Van Camp, P.; Kouril, M.; Weis, C.; Glauser, T.A.; White, P.S.; Kohane, I.S.; Dexheimer, J.W. An interactive online dashboard for tracking COVID-19 in US counties, cities, and states in real time. J. Am. Med. Inform. Assoc. 2020, 27, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.; Sorrell, S.; Nair, S.C.; Al Romaithi, A.; Al Mazrouei, S.; Kamour, A. Rapid development and utilization of a clinical intelligence dashboard for frontline clinicians to optimize critical resources during COVID-19. Acta Inform. Medica 2020, 28, 209. [Google Scholar] [CrossRef] [PubMed]
- Wijegunaratne, I.; Fernandez, G. The Three-Tier Application Architecture. In Distributed Applications Engineering: Building New Applications and Managing Legacy Applications with Distributed Technologies; Springer: London, UK, 1998; pp. 41–78. [Google Scholar] [CrossRef]
- IBM. What Is Three-Tier Architecture? Available online: https://www.ibm.com/topics/three-tier-architecture (accessed on 22 January 2024).
- AWS. Three-Tier Architecture Overview. Available online: https://docs.aws.amazon.com/whitepapers/latest/serverless-multi-tier-architectures-api-gateway-lambda/three-tier-architecture-overview.html (accessed on 22 January 2024).
- Witten, I.; Frank, E.; Hall, M.; Pal, C. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed.; Morgan Kauffmann: Burlington, MA, USA, 2016. [Google Scholar]
- Denney, M. Validating the extract, transform, load process used to populate a large clinical research database. Int. J. Med. Inform. 2016, 94, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Mozilla. JavaScript|MDN. Available online: https://developer.mozilla.org/en-US/docs/Web/JavaScript (accessed on 22 January 2024).
- Veeraraghavan, S. 20 Most Popular Programming Languages to Learn in 2023. Available online: https://www.simplilearn.com/best-programming-languages-start-learning-today-article (accessed on 22 January 2024).
- Yang, D. The 9 Best Programming Languages to Learn in 2023. Available online: https://www.fullstackacademy.com/blog/nine-best-programming-languages-to-learn (accessed on 22 January 2024).
- React—A JavaScript Library for Building User Interfaces. Available online: https://reactjs.org (accessed on 22 January 2024).
- D3—Data-Driven Documents. Available online: https://d3js.org (accessed on 22 January 2024).
- Victory—React.js Components for Modular Charting and Data Visualization. Available online: https://formidable.com/open-source/victory (accessed on 22 January 2024).
- C3.js D3-Based Reusable Chart Library. Available online: https://c3js.org (accessed on 22 January 2024).
- Chart.js—Simple Yet Flexible JavaScript Charting Library for the Modern Web. Available online: https://www.chartjs.org (accessed on 22 January 2024).
- Recharts. Recharts—A Composable Charting Library Built on React Components. Available online: https://recharts.org/en-US (accessed on 22 January 2024).
- Plotly—Javascript. Available online: https://plotly.com/javascript (accessed on 22 January 2024).
- Purkayastha, S. Top 15 Python REST API Frameworks in 2022. Available online: https://rapidapi.com/blog/best-python-api-frameworks (accessed on 22 January 2024).
- Kaur, P. Top 5 Python REST API Framework. Available online: https://www.moesif.com/blog/api-product-management/api-analytics/Top-5-Python-REST-API-Frameworks (accessed on 22 January 2024).
- Nicholas, S. Best Python REST API Framework Solutions for 2023. Available online: https://hevodata.com/learn/python-rest-api-framework (accessed on 22 January 2024).
- Manzi, M. How to Build APIs in Python: 8 Popular Frameworks. Available online: https://www.techrepublic.com/article/build-apis-python (accessed on 22 January 2024).
- Flask—Web Development, One Drop at a Time. Available online: https://flask.palletsprojects.com/en/2.2.x (accessed on 22 January 2024).
- Django—The Web Framework for Perfectionists with Deadlines. Available online: https://www.djangoproject.com (accessed on 22 January 2024).
- FastAPI—FastAPI Framework, High Performance, Easy to Learn, Fast to Code, Ready for Production. Available online: https://fastapi.tiangolo.com (accessed on 22 January 2024).
- Pyramid—The Start Small, Finish Big Stay Finished Framework. Available online: https://trypyramid.com (accessed on 22 January 2024).
- React Select. Available online: https://react-select.com/home (accessed on 22 January 2024).
- What Is Application Deployment. Available online: https://www.vmware.com/topics/glossary/content/application-deployment.html (accessed on 22 January 2024).
- Serve-npm. Available online: https://www.npmjs.com/package/serve (accessed on 22 January 2024).
- Nginx. Available online: https://www.nginx.com (accessed on 22 January 2024).
- Apache Tomcat. Available online: https://tomcat.apache.org (accessed on 22 January 2024).
- Docker. Available online: https://www.docker.com (accessed on 22 January 2024).
- IBM. What Is Docker. Available online: https://www.ibm.com/topics/docker (accessed on 22 January 2024).
- Heroku. Available online: https://www.heroku.com (accessed on 22 January 2024).
- Google. App Engine Application Platform. Available online: https://cloud.google.com/appengine (accessed on 22 January 2024).
- Traefik. Traefik Labs: Say Goodbye to Connectivity Chaos. Available online: https://traefik.io (accessed on 22 January 2024).
- Prozan, L.; Shusterman, E.; Ablin, J.; Mitelpunkt, A.; Weiss-Meilik, A.; Adler, A.; Choshen, G.; Kehat, O. Prognostic value of neutrophil-to-lymphocyte ratio in COVID-19 compared with Influenza and respiratory syncytial virus infection. Sci. Rep. 2021, 11, 21519. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.; Whitney, D. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Chan, A.S.; Rout, A. Use of Neutrophil-to-Lymphocyte and Platelet-to-Lymphocyte Ratios in COVID-19. J. Clin. Med. Res. 2020, 12, 448–453. Available online: https://www.jocmr.org/index.php/JOCMR/article/view/4240. (accessed on 27 November 2023). [CrossRef]
- Chen, X.; Gao, W.; Li, J.; You, D.; Yu, Z.; Zhang, M.; Shao, F.; Wei, Y.; Zhang, R.; Lange, T.; et al. A predictive paradigm for COVID-19 prognosis based on the longitudinal measure of biomarkers. Briefings Bioinform. 2021, 22, bbab206. [Google Scholar] [CrossRef]




















Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, R.; Ferreira, A.; Pinto, I.; Geraldes, C.; Von Rekowski, C.; Bento, L. An Interactive Dashboard for Statistical Analysis of Intensive Care Unit COVID-19 Data. BioMedInformatics 2024, 4, 454-476. https://doi.org/10.3390/biomedinformatics4010026
Dias R, Ferreira A, Pinto I, Geraldes C, Von Rekowski C, Bento L. An Interactive Dashboard for Statistical Analysis of Intensive Care Unit COVID-19 Data. BioMedInformatics. 2024; 4(1):454-476. https://doi.org/10.3390/biomedinformatics4010026
Chicago/Turabian StyleDias, Rúben, Artur Ferreira, Iola Pinto, Carlos Geraldes, Cristiana Von Rekowski, and Luís Bento. 2024. "An Interactive Dashboard for Statistical Analysis of Intensive Care Unit COVID-19 Data" BioMedInformatics 4, no. 1: 454-476. https://doi.org/10.3390/biomedinformatics4010026
APA StyleDias, R., Ferreira, A., Pinto, I., Geraldes, C., Von Rekowski, C., & Bento, L. (2024). An Interactive Dashboard for Statistical Analysis of Intensive Care Unit COVID-19 Data. BioMedInformatics, 4(1), 454-476. https://doi.org/10.3390/biomedinformatics4010026









