Abstract
Background: Feature selection (FS), a crucial preprocessing step in machine learning, greatly reduces the dimension of data and improves model performance. This paper focuses on selecting features for medical data classification. Methods: In this work, a new form of ensemble FS method called weighted rank difference ensemble (WRD-Ensemble) has been put forth. It combines three FS methods to produce a stable and diverse subset of features. The three base FS approaches are Pearson’s correlation coefficient (PCC), reliefF, and gain ratio (GR). These three FS approaches produce three distinct lists of features, and then they order each feature by importance or weight. The final subset of features in this study is chosen using the average weight of each feature and the rank difference of a feature across three ranked lists. Using the average weight and rank difference of each feature, unstable and less significant features are eliminated from the feature space. The WRD-Ensemble method is applied to three medical datasets: chronic kidney disease (CKD), lung cancer, and heart disease. These data samples are classified using logistic regression (LR). Results: The experimental results show that compared to the base FS methods and other ensemble FS methods, the proposed WRD-Ensemble method leads to obtaining the highest accuracy value of 98.97% for CKD, 93.24% for lung cancer, and 83.84% for heart disease. Conclusion: The results indicate that the proposed WRD-Ensemble method can potentially improve the accuracy of disease diagnosis models, contributing to advances in clinical decision-making.
1. Introduction
Artificial intelligence (AI) tries to simulate human intelligence in machines through learning [1,2,3]. Therefore, machine learning (ML) is one of the major branches of AI [1]. It offers many essential tools for insightful data analysis in finance, healthcare, manufacturing, retail, security, etc. ML algorithms can use medical data to find patterns and precisely predict diseases. Data must be prepared for learning and prediction before applying any algorithm to medical datasets. Among various preprocessing steps, feature selection (FS) is an important pre-processing step of ML that prepares data before training any model. FS methods are often used to find the optimal subset of features that maximally increase the performance of the models. The ensemble FS technique has gained importance in the recent literature because of its ability to handle the issue of stability of the results of FS [4]. It integrates the results of two or more FS methods to provide a robust (stable) and diverse subset of important features for classification [4]. With the exception of the combination process, many existing ensemble feature ranking approaches take the same approach to ensemble FS. Several feature selectors are created in articles [5,6,7,8,9,10,11,12] employing different feature ranking techniques, such as information gain, chi-square, symmetric uncertainty, reliefF, etc. Several aggregation functions, such as mean, median, and rank are used by the existing ensemble techniques to combine the lists [12]. The ensemble mean calculates each feature’s score by averaging its weights across all ranking lists [13]. In ensemble median, the combining function for each feature is the median weight across all ranking lists. Even though the feature weightings show how important each feature is in a feature set, they cannot guarantee building a better feature set for classification. The main reason for this is that the features are more similar, making them redundant. The way the features work together is complicated. So, if we try too hard to find features with less redundancy or better purity, we might end up giving up on some good features, which could hurt classification. Consequently, establishing a suitable FS approach still remains a research interest. While there are many research publications on FS algorithms for disease prediction, further research is required for a variety of reasons, including the need for performance improvement, dataset variations, and emerging diseases.
This research focuses on identifying a viable FS strategy for medical data classification. In this paper, mean aggregation is employed for diversity, while absolute rank difference (which has not yet been used in this field) is used for stability in the final feature subset. In this study, three lists of features, generated from three different feature ranking methods, are combined using the mean of the weights or scores for each feature in each ranking list. Pearson’s correlation coefficient (PCC), gain ratio (GR), and reliefF are the three methods to combine in the proposed method. The absolute difference between each feature’s ranks across three ranked lists is used to choose unstable features in the next stage. Finally, if any of these unstable features has rank greater or equal to the last one-fourth of rank in any of the three rank lists, it is removed from the subset of features to obtain the final subset of features. This paper employs datasets of UCI [14] and Kaggle [15] to examine the performance of a new approach that combines mean aggregation with an absolute rank difference. The main contributions of the work are summarized as follows:
- (i)
- This paper proposes a new form of ensemble FS method termed weighted rank difference ensemble (WRD-Ensemble). This WRD-Ensemble method is developed by aggregating the weight of each feature of stand-alone FS methods and then selecting a subset of features by using the rank difference of each feature. In this paper, WRD-Ensembple is shown by combining the PCC, relief, and GR methods;
- (ii)
- The proposed WRD-Ensemble method is tested on chronic kidney disease (CKD), lung cancer, and heart disease datasets, which are widely used datasets for medical data analytics;
- (iii)
- The proposed WRD-Ensemble scheme is compared with the existing stand-alone PCC, GR, and reliefF methods. Finally, its performance is compared with several existing research works on CKD and lung cancer datasets using the proposed method.
Note that the concept of WRD-Ensemble is shown here, combining the well-known PCC, relief, and GR methods; however, this concept of WRD-Ensemble can also be applied by combining other stand-alone FS approaches. The rest of this paper is organized as follows. Section 2 contains related works on ensemble FS methods, and Section 3 describes the existing stand-alone and ensemble FS methods as well as the proposed WRD-Ensemble approach. The performance results for WRD-Ensemble and existing FS methods for the three datasets are included in Section 4. Finally, the conclusion and future scope of the work are presented in Section 5.
2. Related Works
There are several studies that report ensemble FS algorithms. Some work report stand-alone FS, while others apply ensemble or hybrid approaches. These algorithms are applied with different classification methods for the detection of a target in multiple application scenarios.
Firstly, the research on stand-alone FS from the existing studies is reported. SVM classifiers using the filter and wrapper FS approaches are examined by the authors in [16]. It is reported that the SVM classifier, when combined with the best search engine of filter approach, can predict CKD with a maximum accuracy of 98.5%. The study in [17] employs twelve different classifiers to categorize patients as having CKD or not, with the decision tree achieving the highest classification accuracy of 98.6%. The work in [18] analyzes several classifiers along with several FS approaches for ML-based CKD diagnosis. Filter, wrapper, and embedded FS methods are used for feature optimization. In [19], five FS methods, Random forest FS (RF-FS), forward selection, forward exhaustive selection (FES), backward selection (BS), and backward exhaustive (BE), are used to select the most important features of the CKD dataset. Experimental results show that RF with Random Forest FS had achieved the best performance with 98.8% accuracy.
Next, the research on ensemble or hybrid FS methods from the literature is reported. An ensemble of FS methods is introduced in [5] where filter-based FS methods including gain ratio, info gain, chai square, symmetric uncertainty, and reliefF are used to select different subsets of features. Next, these feature subsets are combined by using feature-class and feature–feature mutual information to generate an optimal feature subset. Their proposed FS method, termed as EFS-MI, is evaluated using three classifiers, namely decision trees, random forest, and K-NN. For the colon cancer dataset, EFS-MI yields higher classification accuracy with the decision trees classifier. However, the method gives quite poor classification accuracy with the SVM classifier. The authors in [4] propose an ensemble FS method that combines the ideas of filter and wrapper methods to select the most discriminatory features for the diagnosis of CKD. The density-based FS (DFS) method is used as a filter approach to rank the features of CKD. The results of the DFS method are given to a wrapper-based optimization technique named Improved Teacher Learner Based Optimization (ITLBO) algorithm to find the optimal feature set that contains the most important features for the prediction of CKD. The DFS method computes the density of the features using the probability density function and ranks the features on high-density values to low-density values (i.e., the features with higher density values are prioritized). ITLBO is a variant of the original TLBO algorithm [4]. Using the DFS method on the CKD dataset, 19 features are selected from a total of 24. These features are then input into ITLBO to find an optimal subset containing the most important features for predicting CKD. Finally, these feature subset is evaluated using the ITLBO wrapper method using classification algorithms including SVM and Gradient Boosting. The results of the experiment show that the DFS method can achieve a high classification accuracy of 93% for SVM and 97% for Gradient Boosting, respectively, for the derived optimal feature subset.
One study [11] selects useful features from a dataset using an efficient and comprehensive univariate ensemble-based feature selection (uEFS) method. In uEFS, a unified features scoring algorithm is used to provide a final ranked list of features after a thorough evaluation of a feature set. Next, a threshold value selection approach is applied to choose a subset of features that are deemed important for classifier design excluding unnecessary features. A hybrid wrapper and filter-based FS (HWFFS) is proposed in [20] to reduce the dimensions of the CKD dataset with an SVM classifier. The filter-based FS algorithm is performed based on the three major functions: Information Gain (IG), Correlation Based FS (CFS), and Consistency Based Subset Evaluation (CS) algorithms, respectively. The wrapper-based FS algorithm is performed based on the Enhanced Immune Clonal Selection (EICS) to determine the most relevant features, thus increasing the classifier’s accuracy. The SVM classifier with HWFFS method shows the highest accuracy value of 90.00%.
In short, the topic of ensemble FL algorithms for disease diagnosis is not static, and continual research is required for multiple factors, including adapting to the unique problems associated with different diseases and the changing landscape of medical datasets.
3. Materials and Methods
FS methods can be classified into three different types such as filter, wrapper, and the embedded FS method based on how the supervised learning algorithm is employed in the FS process. Filter methods can be further categorized as univariate and multivariate methods. In univariate approaches, features are evaluated individually based on their relationship with the target variable, whereas multivariate methods consider the interactions between features when determining their relevance. The univariate FS methods PCC, reliefF and GR are described as follows.
3.1. Existing FS Methods
First, some stand-alone FS techniques are briefly described [21,22,23]. One of these is the Pearson’s correlation coefficient (PCC) which is a correlation statistic that measures the strength and direction of the relationship between two features or variables [21]. The gain ratio (GR) method is a normalized version of information gain. It is a non-symmetrical measure that is introduced to compensate for the bias of the IG [24]. ReliefF is a well-known FS method that uses distance measures to evaluate the worth of a feature [25].
The main goal of ensemble FS is to combine the outputs of several base selectors, which should be sufficiently different from one another, to provide a more diverse set of features [26]. Different ensemble FS methods are developed in the context of several health informatics and bioinformatics scenarios [27,28,29,30,31,32].
3.2. The Proposed WRD-Ensemble Method
The proposed ensemble approach is performed in two steps. It starts with creating three different ranking lists of features [11] using the three filter ranking methods, namely PCC, reliefF and GR, then applies the ensemble approach to form a single list of features. The ensemble approach used in this method is the mean of score which is accompanied by absolute rank difference. Each filter ranking method provides a ranking list of features according to the importance of the feature which is also known as the weight of the feature. Suppose L1, L2, and L3 are the three lists from the PCC, reliefF, and GR methods and are the features in a dataset. In the first step, the average of the weight of each feature in three lists is determined. If are the weights of the feature and are the ranks of feature in L1, L2 and L3 lists, then mean of weight, of feature is determined by
Using this formula, the mean weight of each feature is obtained and then the features are sorted according to their mean weight. This will produce a new list of n features each having a mean weight. The second step is to calculate the absolute rank difference of each feature among the three lists. The absolute rank difference, of is determined by
This will create a list of rank differences of n features. To normalize the value of the rank difference of each feature, the following formula is used
where fi is the i-th feature, is the maximum rank, and is the minimum rank in the list of rank differences. The rank difference is used to determine the stability of a feature among three rank lists. A value of zero in the rank difference of a feature means it has the same rank in all three rank lists and it is most consistent in its position in all three rank lists. A threshold [11] Wτ is used to remove those features that have a weight less than or equal to Wτ. Another threshold Rτ is used to remove unstable features from the subset of features and the importance of a feature in any of the three lists is used to select the final list of features. Features which have rank difference greater than or equal to Rτ are considered as unstable features. Before removing these features, the rank of each feature in three rank lists is checked, and if a feature has a rank greater or equal to the last one-fourth of rank in any rank list, it is finally removed from the final subset of features. For example, the CKD dataset has 24 predictive features, and according to the proposed method, the last 6 features of any of the three lists are considered the least important features. The ceiling value of one-fourth of the total number of features is considered. For CKD, if a feature is unstable and has a rank between 19 and 24 in any of the three rank lists, then it will be removed. Algorithm 1 describes the WRD-Ensemble method.
| Algorithm 1 WRD-Ensemble method |
| Input: Dataset D with n features. Output: An array, E, with selected features. 1. Feature Ranking using PCC FS Method Generate L1 using the Pearson Correlation Coefficient Feature Selection (PCC FS) method, and assign ranks and weights to two arrays: Rank and W. for j = 0 to n − 1 do row = L1[j][0] Rank[row][1] = j + 1 W[row][1] = L1[j][1] end for 2. Feature Ranking using ReliefF FS Method Generate L2 using ReliefF Feature Selection (ReliefF FS) method, and assign ranks and weights to two arrays: Rank and W. for j = 0 to n − 1 do row = L2[j][0] Rank[row][2] = j + 1 W[row][2] = L2[j][1] end for 3. Feature Ranking using GR FS Method Generate L3 using Gini Ratio Feature Selection (GR FS) method, and assign ranks and weights to two arrays: Rank and W. for j = 0 to n − 1 do row = L3[j][0] Rank[row][3] = j + 1 W[row][3] = L3[j][1] end for 4. Calculate Mean Weight and Absolute Rank Difference Calculate and assign the mean weight and absolute rank difference of each feature to arrays F and Rank. for j = 0 to n − 1 do F[j][0] = j + 1 F[j][1] = (W[j][1] + W[j][2] + W[j][3])/3 F[j][2] = abs(Rank[j][1] − Rank[j][2]) + abs(Rank[j][2] − Rank[j][3]) + abs(Rank[j][3] − Rank[j][1]) end for 5. Normalize Rank Differences Normalize the rank difference of each feature. for j = 0 to n − 1 do F[j][2] = (F[j][2] − R_min)/(R_max − R_min) // Normalization of rank difference end for 6. Sort the features in F based on their mean weight. 7. Remove features from F with mean weight <= Wτ. 8. Remove features from F with rank difference >= Rτ and do not have a rank equal to the last one-fourth of the rank in any three rank lists. 9. Selected Features_Array, E ← F to E. 10. Return Features_Array, E 11. End |
4. Results and Discussion
The experiments are performed to evaluate the performance of FS approaches. For this, classification models using LR have been built on the three datasets. The 10-fold cross-validation method is utilized to assess the accuracy of the classification models employed. The model has been implemented in Java language. All the FS methods and classification methods are used from WEKA, which is imported to Java as a package. The results reported here are for the cases where the methods are computed on a personal computer having specifications such as, processor: Intel (R) Core (TM) i3-6100 CPU @ 3.70 GHz 3.70 GHz, installed memory (RAM): 4.0 GB (3.88 GB usable), and system type: 64-bit operating system, x64-based processor.
4.1. Results for CKD Dataset
The CKD dataset contains 400 instances and 25 features; among those, 1 feature indicates the decision class. Among the 24 input features, 14 are numerical and 10 are categorical features. The class attribute of CKD is categorical and has two distinct values: ckd, notckd. The dataset is imbalanced because it contains 250 cases of “ckd” class and 150 cases of “notckd”. The CKD dataset contains missing values, and missing values are handled by applying mean for numerical data and mod for nominal data. After applying PCC, reliefF, GR, and WRD-Ensemble on the CKD dataset each method generates a ranked list of features. Table 1 shows the ranked list of features for each method.

Table 1.
Ranked list of features generated by the existing and the proposed FS methods for the CKD dataset.
Table 1 indicates that for PCC, feature 15 is the most important, while feature 14 is the least important feature; for reliefF, feature 19 is the most important, while feature 17 is the least important feature; for GR, feature 12 is the most important, and feature 1 is the least important feature. It is also shown that for WRD-Ensemble, features 19 and 14 are the most important and least important features, respectively. The proposed method’s first phase combines the weights of three base FS approaches to produce a ranked list of features, together with a ranked difference and mean weight for each feature. The following phase of the proposed method involves choosing various Wτ and Rτ values to select multiple feature subsets. Table 2 shows different values of threshold for different numbers of FS.

Table 2.
Different values of thresholds for subsets of features of CKD dataset.
At first, when we choose 0.1062 as the value of Wτ, the WRD-Ensemble algorithm eliminates the lowest-ranked features 9 and 14. Then, when we choose Rτ value 1.0, WRD-Ensemble finds feature 12 as an inconsistent feature. However, feature 12 is still included in the 22 subsets of features because it does not have a rank between 19 and 24 for the total 24 features (i.e., does not have a rank equal to the last one-fourth of the rank) in any of the three methods, namely, PCC, reliefF, and GR methods. The same approach has chosen the top 20, 18, 16, 14, 12, and 10 subsets of features by considering the top ranked features and removing inconsistent features. Lists of features with WRD-Ensemble considering thresholds Wτ and Rτ have been mentioned in Table 3. Note that the list of all the 24 features ranked using WRD-Ensemble is listed in Table 1, while the top 22 features are listed in Table 3. The last 2 features (#9 and #14) of the 24 features in Table 1 are omitted from the 22 features in Table 3.

Table 3.
Lists of features for different subsets of features of the CKD dataset.
On the other hand, the top 22, 20, 18, 16, 14, 12, and 10 features have been selected from each list of features obtained after applying PCC, reliefF, and GR FS methods. Then, LR classifier is applied to build classification models on the dataset with various selected subsets of features. The classification models have been evaluated in terms of accuracy of models. Table 4 shows the experiment results with 22, 20, 18, 16, 14, 12, and 10 features for PCC, reliefF, GR, and WRD-Ensemble, respectively. The best classification accuracy is obtained for the case of WRD-Ensemble when 10 features are used. Table 5 shows the performance comparison of WRD-Ensemble with the stand-alone methods for the case of 10 features. It can be seen that WRD-Ensemble outperforms others in terms of accuracy, recall, and F1-score, while it has comparable performance with others when precision is taken into consideration. The high accuracy value of 98.97% for WRD-Ensemble for 10 features means that the proposed method is suitable for retaining the most relevant features for CKD diseases, leading to high accuracy in disease classification. The high recall values of 98.90% in WRD-Ensemble mean that this method identifies the key factors enabling reliable disease diagnosis in patients, with the possibility that only 1.10% of cases of CKD patients may remain undetected.

Table 4.
Accuracy of models for the CKD dataset for different numbers of features.

Table 5.
Performance comparison of WRD-Ensemble with stand-alone methods for 10 features.
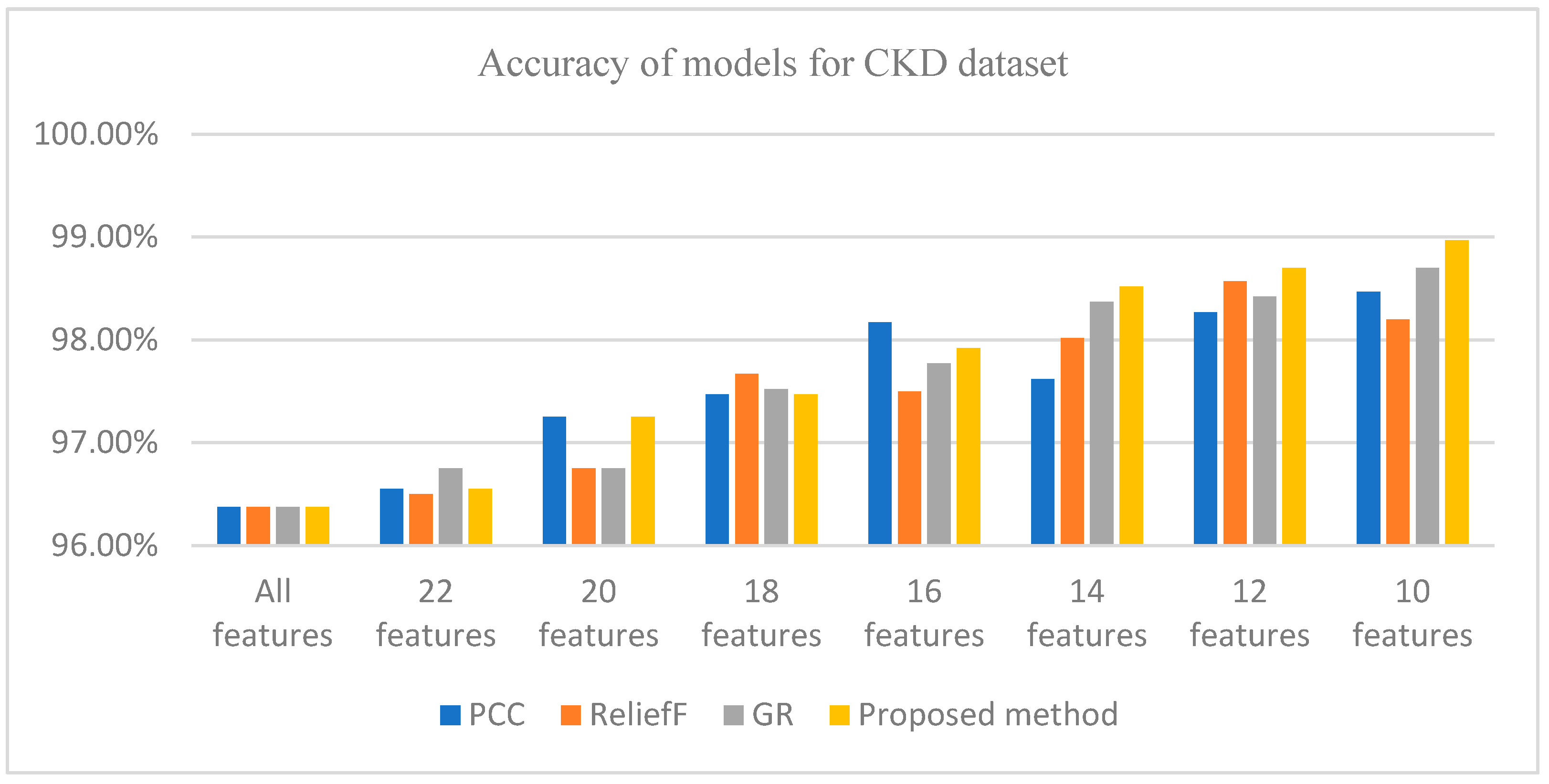
Figure 1 shows that WRD-Ensemble method obtained the highest accuracy value of 98.97% when 10 features were selected; the proposed method WRD-Ensemble has the highest accuracy value compared to PCC, reliefF, and GR. It can be seen that the best accuracy for PCC, reliefF, and GR obtained are 98.47% (10 features), 98.57% (12 features), and 98.7% (10 features), respectively. Table 6 shows a comparative result between WRD-Ensemble and some existing works on the CKD dataset. It can be seen that WRD-Ensemble results in the best accuracy compared to other single and ensemble FS methods reported in the literature.
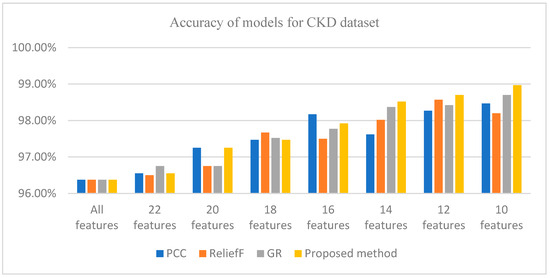
Figure 1.
Accuracy of different methods for the CKD dataset for different numbers of features.

Table 6.
Comparative results between the WRD-Ensemble method and existing works for the CKD dataset.
4.2. Results for Lung Cancer Dataset
The lung cancer dataset of Kaggle contains 309 instances and 16 features; among them one is decision class. Among 15 predictive features, 14 are numerical and 1 is a categorical feature. The class attribute of lung cancer is categorical and has two distinct values: YES and NO. This dataset does not contain any missing values.
After applying PCC, reliefF, GR, and WRD-Ensemble on the lung cancer dataset, each method generates a ranked list of features. Table 7 shows the ranked list of features for each method. Table 8 shows different values of the threshold for different numbers of FS for the lung cancer dataset. Lists of features for various values of Wτ and Rτ have been mentioned in Table 9. Next, Table 10 shows the results of the experiment for the LR classifier with all, 13, 11, 9, and 7 features for PCC, reliefF, GR, and WRD-Ensemble, respectively. The best performance is obtained for WRD-Ensemble when 13 features of the lung cancer dataset are taken into consideration. Table 11 presents the comparative results of these methods for 13 features. Table 11 shows that, in terms of accuracy, precision, recall, and F1-score, WRD-Ensemble surpasses PCC, reliefF, and GR in identifying the key features of the lung cancer dataset.

Table 7.
Ranked list of features generated by three FS methods for lung cancer.

Table 8.
Different values of thresholds for different numbers of subsets of features.

Table 9.
Lists of features for different subsets of features of lung cancer dataset.

Table 10.
Accuracy of models for lung cancer for different numbers of features.

Table 11.
Comparison of WRD-Ensemble with stand-alone methods for lung cancer for 13 features.
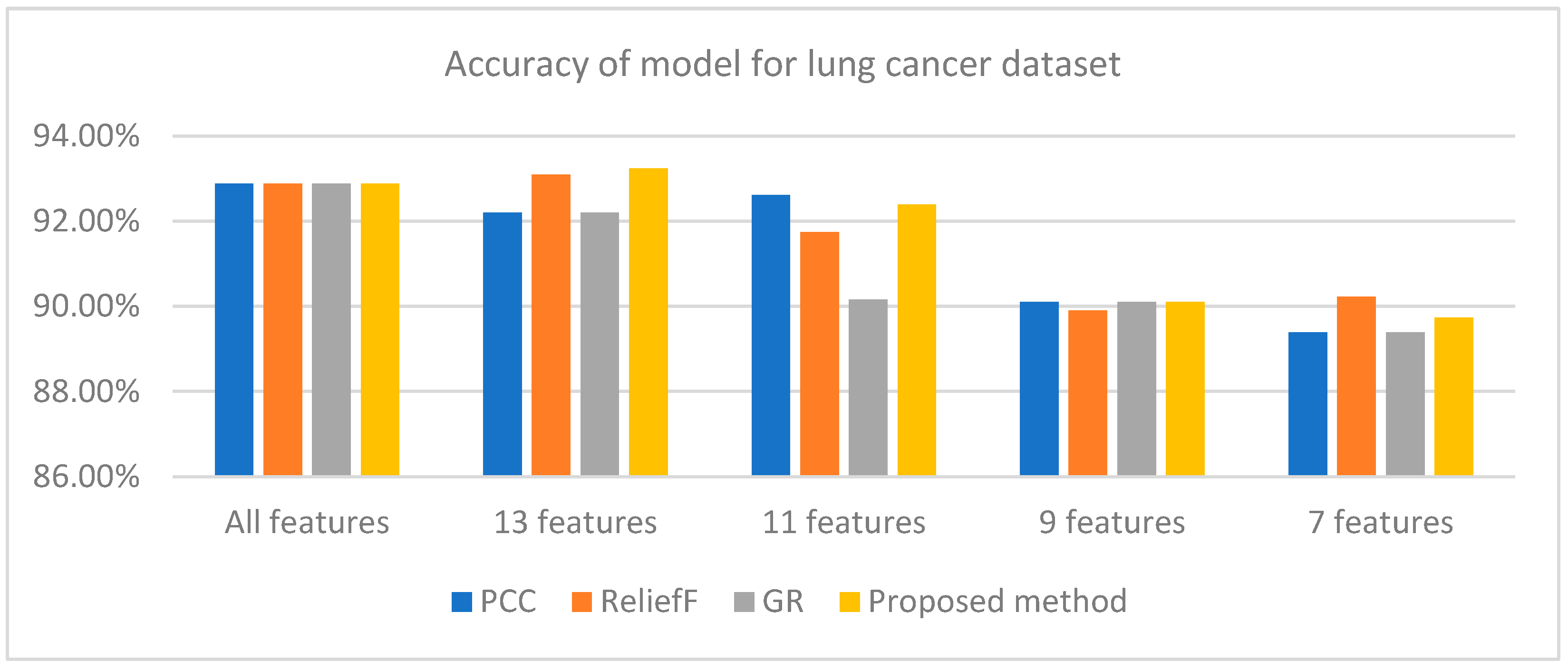
Figure 2 also indicates that the WRD-Ensemble method obtained the highest accuracy value of 93.24% when 13 features are used, where the accuracy value is better than PCC, reliefF, and GR. The highest accuracy values for PCC, reliefF, and GR are 92.88% (15 features), 93.10% (13 features), and 92.88% (15 features). The proposed WRD-Ensemble for lung cancer results is not compared to other existing studies because no published papers have been found to date that employ ensemble FS approaches for lung cancer classification [13]. However, the classification accuracy value of 93.24% and recall value of 96.80% indicate that WRD-Ensemble can be suitable for retaining the critical features of lung cancer which leads to reliable disease detection.
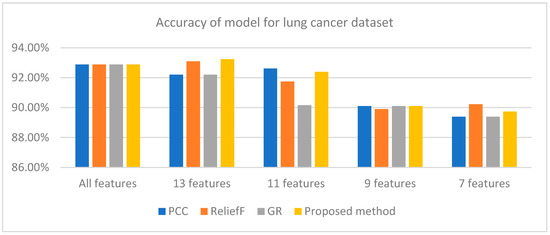
Figure 2.
Accuracy of different methods for lung cancer dataset for different numbers of features.
4.3. Results for Heart Disease Dataset
The heart disease dataset of UCI [33] contains 303 instances and lists 13 features and one target or decision class. The feature selection methods are applied to the heart disease dataset similar to the approaches described in Section 4.1 and Section 4.2. Experimental results show the best performance for the case of the top 7 features with Wτ = 0.0915 and Rτ = 0.4444. When LR is considered as a classification method, the values for the case of WRD-Ensemble, PCC, reliefF, and GR are found to be 83.84%, 82.49%, 82.83%,. and 82.49%, respectively. Hence, the WRD-Ensemble method is also suitable for the heart disease dataset.
5. Conclusions and Future Work
A new form of ensemble FS method termed WRD-Ensemble has been proposed in this paper for the CKD, lung cancer, and heart disease datasets. The proposed ensemble technique integrates PCC, reliefF, and GR schemes by calculating each feature’s mean weight and absolute rank difference. Results from the experiment on the CKD dataset using the LR classifier show the WRD-Ensemble outperforms the other three FS methods, obtaining an accuracy value of 98.97% when 10 features are selected. Similarly, results from the experiment on the lung cancer dataset using the LR classifier show that WRD-Ensemble outperforms the other three FS methods, obtaining the highest accuracy value of 93.24% when 13 features are selected. Finally, WRD-Ensemble is also shown to be effective for the case of heart disease. It should be noted that the results provided in this paper may vary with the variation of the datasets taken into consideration. This is because the effectiveness of FS and machine learning algorithms depend on the data samples considered. Nevertheless, the findings of this paper will contribute to the ongoing data-driven diagnosis of diseases.
The computation time of WRD-Ensemble is longer than that of the three separate FS methods since it integrates them; nevertheless, a thorough analysis of memory usage and computation time is left for further study. In the future, the effectiveness of WRE-Ensemble FS should be compared with other stand-alone and ensemble FS approaches in terms of accuracy and computation time. As additional future work, the concept of WRD-Ensemble can be extended to add more than three univariate methods to increase the diversity. Moreover, other categories of FS methods, such as embedded methods, can also be considered. It is also necessary to assess the proposed WRD-Ensemble method using larger datasets with more features and instances. In the future, the effectiveness of the proposed FS method may be tested for the datasets of other application scenarios as well.
Author Contributions
Conceptualization, A.M.B. and M.R.H.M.; methodology, A.M.B.; software, A.M.B., P.P.; validation, A.M.B. and P.P.; formal analysis, A.M.B.; investigation, A.M.B.; resources, M.R.H.M. and P.P.; data curation, A.M.B. and M.R.H.M.; writing—original draft preparation, A.M.B.; writing—review and editing, M.R.H.M., P.P., and J.K.; visualization, P.P.; supervision, M.R.H.M. and J.K.; project administration, M.R.H.M. and J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding except for the support from Bangladesh University of Engineering and Technology in the form of Basic Research Grant (R60/Ref 5336—M.R.H.M.) in June 2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are available online in repositories [14,15,33], and the details of the algorithm are available at (https://github.com/prajoybuet/WRD-Ensemble.git (accessed on 29 October 2023)).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Abbreviations | Elaboration |
| FS | Feature selection |
| WRD-Ensemble | weighted rank difference ensemble |
| GR | Gain Ratio |
| PCC | Pearson’s correlation coefficient |
| CKD | chronic kidney disease |
| LR | logistic regression |
| DFS | Density-based FS |
| uEFS | univariate ensemble-based feature selection |
| FES | forward exhaustive selection |
| HWFFS | hybrid wrapper and filter-based FS |
| IG | Information Gain |
| CFS | Correlation Based FS |
References
- Kononenko, I. Machine learning for medical diagnosis: History, state of the art and perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef]
- Ahmmed, S.; Podder, P.; Mondal, M.R.H.; Rahman, S.M.A.; Kannan, S.; Hasan, M.J.; Rohan, A.; Prosvirin, A.E. Enhancing Brain Tumor Classification with Transfer Learning across Multiple Classes: An In-Depth Analysis. BioMedInformatics 2023, 3, 1124–1144. [Google Scholar] [CrossRef]
- Rahman, S.M.; Ibtisum, S.; Bazgir, E.; Barai, T. The Significance of Machine Learning in Clinical Disease Diagnosis: A Review. arXiv 2023, arXiv:2310.16978. [Google Scholar] [CrossRef]
- Manonmani, M.; Balakrishnan, S. An Ensemble Feature Selection Method for Prediction of CKD. In Proceedings of the 2020 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 22–24 January 2020; pp. 1–6. [Google Scholar]
- Hoque, N.; Singh, M.; Bhattacharyya, D.K. EFS-MI: An ensemble feature selection method for classification. Complex Intell. Syst. 2018, 4, 105–118. [Google Scholar] [CrossRef]
- Wang, H.; Khoshgoftaar, T.M.; Napolitano, A. Software measurement data reduction using ensemble techniques. Neurocomputing 2012, 92, 124–132. [Google Scholar] [CrossRef]
- Saeys, Y.; Abeel, T.; Peer, Y.V.D. Robust feature selection using ensemble feature selection techniques. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Antwerp, Belgium, 14–18 September 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 313–325. [Google Scholar]
- Osanaiye, O.; Cai, H.; Choo, K.K.R.; Dehghantanha, A.; Xu, Z.; Dlodlo, M. Ensemble-based multi-filter feature selection method for DDoS detection in cloud computing. EURASIP J. Wirel. Commun. Netw. 2016, 1, 130. [Google Scholar] [CrossRef]
- Liu, L.; Tang, S.; Wu, F.X.; Wang, Y.P.; Wang, J. An ensemble hybrid feature selection method for neuropsychiatric disorder classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 1459–1471. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, J.; Zhao, C.; Peng, Y.; Wang, H. An ensemble feature selection method for high-dimensional data based on sort aggregation. Syst. Sci. Control Eng. 2019, 7, 32–39. [Google Scholar] [CrossRef]
- Ali, M.; Ali, S.I.; Kim, D.; Hur, T.; Bang, J.; Lee, S.; Kang, B.H.; Hussain, M. uEFS: An efficient and comprehensive ensemble-based feature selection methodology to select informative features. PLoS ONE 2018, 13, e0202705. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Yuan, W.; Lee, Y.K.; Najeebullah, K.; Rasel, M.K. A review of ensemble learning based feature selection. IETE Tech. Rev. 2014, 31, 190–198. [Google Scholar] [CrossRef]
- Wang, H.; Khoshgoftaar, T.M.; Napolitano, A. A comparative study of ensemble feature selection techniques for software defect prediction. In Proceedings of the 2010 9th International Conference on Machine Learning and Applications, Washington, DC, USA, 12–14 December 2010; pp. 135–140. [Google Scholar]
- Available online: https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease (accessed on 10 October 2022).
- Available online: https://www.kaggle.com/datasets/nancyalaswad90/lung-cancer (accessed on 25 November 2022).
- Polat, H.; Danaei Mehr, H.; Cetin, A. Diagnosis of chronic kidney disease based on support vector machine by feature selection methods. J. Med. Syst. 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, V.; Sharma, A. Performance based evaluation of various machine learning classification techniques for chronic kidney disease diagnosis. arXiv 2016, arXiv:1606.09581. [Google Scholar]
- Chittora, P.; Chaurasia, S.; Chakrabarti, P.; Kumawat, G.; Chakrabarti, T.; Leonowicz, Z.; Bolshev, V. Prediction of chronic kidney disease-a machine learning perspective. IEEE Access 2021, 9, 17312–17334. [Google Scholar] [CrossRef]
- Abdullah, A.A.; Hafidz, S.A.; Khairunizam, W. Performance comparison of machine learning algorithms for classification of chronic kidney disease (CKD). J. Phys. Conf. Ser. 2020, 1529, 052077. [Google Scholar] [CrossRef]
- Sara, S.B.V.; Kalaiselvi, K. Ensemble swarm behaviour based feature selection and support vector machine classifier for chronic kidney disease prediction. Int. J. Eng. Technol. 2018, 7, 190. [Google Scholar] [CrossRef]
- Saidi, R.; Bouaguel, W.; Essoussi, N. Hybrid feature selection method based on the genetic algorithm and pearson correlation coefficient. In Machine Learning Paradigms: Theory and Application; Springer: Cham, Switzerland, 2019; pp. 3–24. [Google Scholar]
- Blessie, E.C.; Karthikeyan, E. Sigmis: A feature selection algorithm using correlation based method. J. Algorithms Comput. Technol. 2012, 6, 385–394. [Google Scholar] [CrossRef]
- Vaghela, V.B.; Vandra, K.H.; Modi, N.K. Information Theory Based Feature Selection for Multi-Relational Naïve Bayesian Classifier. J. Data Min. Genom. Proteom. 2014, 5, 1. [Google Scholar]
- Novaković, J. Toward optimal feature selection using ranking methods and classification algorithms. Yugosl. J. Oper. Res. 2016, 21, 1. [Google Scholar] [CrossRef]
- Yang, F.; Cheng, W.; Dou, R.; Zhou, N. An improved feature selection approach based on ReliefF and Mutual Information. In Proceedings of the International Conference on Information Science and Technology, Nanjing, China, 26–28 March 2011; pp. 246–250. [Google Scholar]
- Afef, B.B.; Mohamed, L. Ensemble feature selection for high dimensional data: A new method and a comparative study. In Advances in Data Analysis and Classification; Springer: Berlin/Heidelberg, Germany, 2017; Volume 12. [Google Scholar]
- Paplomatas, P.; Krokidis, M.G.; Vlamos, P.; Vrahatis, A.G. An ensemble feature selection approach for analysis and modeling of transcriptome data in alzheimer’s disease. Appl. Sci. 2023, 13, 2353. [Google Scholar] [CrossRef]
- Kolukisa, B.; Bakir-Gungor, B. Ensemble feature selection and classification methods for machine learning-based coronary artery disease diagnosis. Comput. Stand. Interfaces 2023, 84, 103706. [Google Scholar] [CrossRef]
- Manzoor, U.; Halim, Z. Protein encoder: An autoencoder-based ensemble feature selection scheme to predict protein secondary structure. Expert Syst. Appl. 2023, 213, 119081. [Google Scholar]
- Wang, A.; Liu, H.; Yang, J.; Chen, G. Ensemble feature selection for stable biomarker identification and cancer classification from microarray expression data. Comput. Biol. Med. 2022, 142, 105208. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Chalise, P.; He, J. Nested cross-validation with ensemble feature selection and classification model for high-dimensional biological data. Commun. Stat.-Simul. Comput. 2023, 52, 110–125. [Google Scholar] [CrossRef]
- Classification Algorithms Logistic Regression. Available online: https://www.tutorialspoint.com/machine_learning_with_python/machine_learning_with_python_classification_algorithms_logistic_regression.htm (accessed on 6 September 2023).
- Available online: https://archive.ics.uci.edu/ml/datasets/Heart+Disease (accessed on 15 January 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).