Phenotypic and Genotypic Characterization of New Kabuli-Type Chickpea Lines in Australia for Resistance to Ascochyta Blight
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Trial Location
2.2. Growing Environment and Experimental Design
2.3. Isolate and Inoculum Preparation
2.4. Inoculation and Disease Assessment
2.5. MAS for AB Resistance
3. Results
3.1. Phenotypic Evaluation for Ascochyta Blight
3.2. MAS for AB Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organisation. FAOSTAT: Crops and Livestock Products. 2022. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 21 March 2024).
- Chickpea Production by Country. 2024. Available online: https://worldpopulationreview.com/country-rankings/chickpea-production-by-country (accessed on 21 March 2024).
- Shahbandeh, M. Volume of Chickpeas Produced Worldwide 2022, by Country. 2024. Available online: https://www.statista.com/statistics/722203/chickpeas-production-volume-by-country-worldwide/#:~:text=Volume%20of%20chickpeas%20produced%20worldwide%202022%2C%20by%20country&text=In%202022%2C%20the%20production%20volume,of%20chickpeas%20were%20produced%20worldwide (accessed on 21 March 2024).
- Arya, M.; Dwivedi, S.; Chaturvedi, S. Management of biotic stresses in chickpea exploiting host plant resistance. Int. J. Agric. Environ. Biotechnol. 2019, 12, 141–149. [Google Scholar] [CrossRef]
- Moore, K.; Ryley, M.; Cumming, G.; Jenkins, L. Chickpea: Ascochyta Blight Management. Australian Pulse Bulletin 2015. Available online: https://www.pulseaus.com.au/growing-pulses/bmp/chickpea/ascochyta-blight (accessed on 15 September 2023).
- Bar, I.; Sambasivam, P.T.; Davidson, J.; Farfan-Caceres, L.M.; Lee, R.C.; Hobson, K.; Moore, K.; Ford, R. Current population structure and pathogenicity patterns of Ascochyta rabiei in Australia. Microb. Genom. 2021, 7, 000627. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, R.; Sandhu, J.S.; Kaur, L.; Gupta, S.K.; Gaur, P.M.; Varshney, R. Genetics of Ascochyta blight resistance in chickpea. Euphytica 2010, 171, 337–343. [Google Scholar] [CrossRef]
- Pulse Australia. Chickpea Fungicide Guide: 2021 Season. Australian Pulse Bulletin 2021. Available online: https://www.pulseaus.com.au/growing-pulses/bmp/chickpea/2021-season-fungicide-guide (accessed on 21 March 2024).
- ABC Rural. Australia the Worlds Largest Exporter of Chickpea as Pulse Council Purses Further Growth. 2023. Available online: https://www.abc.net.au/news/rural/2023-11-05/australia-worlds-largest-chickpea-exporter-pulse-council-growth/103057674 (accessed on 21 March 2024).
- Dang, Y.P.; Dalal, R.C.; Buck, S.R.; Harms, B.R.; Kelly, R.; Hochman, Z.; Schwenke, G.D.; Biggs, A.J.; Ferguson, N.J.; Norrish, S.; et al. Diagnosis, extent, impacts, and management of subsoil constraints in the northern grains cropping region of Australia. Soil Res. 2010, 48, 105–119. [Google Scholar] [CrossRef]
- Javaid, A.; Munir, R.; Khan, I.H.; Shoaib, A. Control of the chickpea blight, Ascochyta rabiei, with the weed plant, Withania somnifera. Egypt. J. Biol. Pest Control. 2020, 30, 114. [Google Scholar] [CrossRef]
- Labdi, M.; Malhotra, R.S.; Benzohra, I.E.; Imtiaz, M. Inheritance of resistance to Ascochyta rabiei in 15 chickpea germplasm accessions. Plant Breed. 2013, 132, 197–199. [Google Scholar] [CrossRef]
- Pande, S.; Siddique, K.H.; Kishore, G.K.; Bayaa, B.; Gaur, P.M.; Gowda, C.L.; Bretag, T.W.; Crouch, J.H. Ascochyta blight of chickpea (Cicer arietinum L.): A review of biology, pathogenicity, and disease management. Aust. J. Agric. Res. 2005, 56, 317–332. [Google Scholar] [CrossRef]
- Mehmood, Y. Evolution of High-Risk Isolates within the Australian Ascochyta rabiei Population and the Differential Defence Responses Instigated in Chickpea; Griffith University: Brisbane, QLD, Australia, 2017. [Google Scholar]
- Berger, J.; Abbo, S.; Turner, N.C. Ecogeography of annual wild Cicer species: The poor state of the world collection. Crop Sci. 2003, 43, 1076–1090. [Google Scholar] [CrossRef]
- Castro, P.; Rubio, J.; Madrid, E.; Fernández-Romero, M.D.; Millán, T.; Gil, J. Efficiency of marker-assisted selection for ascochyta blight in chickpea. J. Agric. Sci. 2013, 153, 56–67. [Google Scholar] [CrossRef]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, K.; Purayannur, S.; Chen, W.; Verma, P.K. Ascochyta rabiei: A threat to global chickpea production. Mol. Plant Pathol. 2022, 23, 1241–1261. [Google Scholar] [CrossRef] [PubMed]
- Iruela, M.; Rubio, J.; Barro, F.; Cubero, J.I.; Millán, T.; Gil, J. Detection of two quantitative trait loci for resistance to ascochyta blight in an intra-specific cross of chickpea (Cicer arietinum L.): Development of SCAR markers associated with resistance. Theor. Appl. Genet. 2006, 112, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Millan, T.; Rubio, J.; Iruela, M.; Daly, K.; Cubero, J.I.; Gil, J. Markers associated with Ascochyta blight resistance in chickpea and their potential in marker-assisted selection. Field Crops Res. 2003, 84, 373–384. [Google Scholar] [CrossRef]
- Tekeoglu, M.; Rajesh, P.; Muehlbauer, F. Integration of sequence tagged microsatellite sites to the chickpea genetic map. Theor. Appl. Genet. 2002, 105, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Udupa, S.M.; Baum, M. Genetic dissection of pathotype-specific resistance to ascochyta blight disease in chickpea (Cicer arietinum L.) using microsatellite markers. Theor. Appl. Genet. 2003, 106, 1196–1202. [Google Scholar] [CrossRef]
- Newman, T.E.; Jacques, S.; Grime, C.; Kamphuis, F.L.; Lee, R.C.; Berger, J.; Kamphuis, L.G. Identification of novel sources of resistance to ascochyta blight in a collection of wild Cicer accessions. Phytopathology 2021, 111, 369–379. [Google Scholar] [CrossRef]
- Isenegger, D.A.; Ford, R.; Taylor, P.W.J. Disease reaction of chickpea (Cicer spp.) genotypes to Botrytis grey mould (Botrytis cinerea). Australas. Plant Pathol. 2011, 40, 583. [Google Scholar] [CrossRef]
- Pande, S.; Sharma, M.; Gaur, P.M.; Tripathi, S.; Kaur, L.; Basandrai, A.; Khan, T.; Gowda, C.L.; Siddique, K.H. Development of screening techniques and identification of new sources of resistance to Ascochyta blight disease of chickpea. Australas. Plant Pathol. 2011, 40, 149–156. [Google Scholar] [CrossRef]
- VSN International. Genstat for Windows, 23rd ed.; Version 23.1.0.651; VSN International: Hemel Hempstead, UK, 2022. [Google Scholar]
- Madrid, E.; Chen, W.; Rajesh, P.N.; Castro, P.; Millán, T.; Gil, J. Allele-specific amplification for the detection of ascochyta blight resistance in chickpea. Euphytica 2013, 189, 183–190. [Google Scholar] [CrossRef]
- Winter, P.; Pfaff, T.; Udupa, S.M.; Hüttel, B.; Sharma, P.C.; Sahi, S.; Arreguin-Espinoza, R.; Weigand, F.; Muehlbauer, F.J.; Kahl, G. Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol. Gen. Genet. MGG 1999, 262, 90–101. [Google Scholar] [CrossRef]
- Madrid, E.; Rajesh, P.N.; Rubio, J.; Gil, J.; Millán, T.; Chen, W. Characterization and genetic analysis of an EIN4-like sequence (CaETR-1) located in QTLAR1 implicated in ascochyta blight resistance in chickpea. Plant Cell Rep. 2012, 31, 1033–1042. [Google Scholar] [CrossRef]
- Powder, K.E. Quantitative trait loci (QTL) mapping. In eQTL Analysis: Methods and Protocols; Humana: New York, NY, USA, 2020; pp. 211–229. [Google Scholar]
- Maheri-Sis, N.; Chamani, M.; Ali-Asghar, S.; Mirza-Aghazadeh, A.; Aghajanzadeh-Golshani, A. Nutritional evaluation of kabuli and desi type chickpeas (Cicer arietinum L.) for ruminants using in vitro gas production technique. Afr. J. Biotechnol. 2008, 7, 2946–2951. [Google Scholar]
- Purushothaman, R.; Upadhyaya, H.D.; Gaur, P.M.; Gowda, C.L.; Krishnamurthy, L. Kabuli and desi chickpeas differ in their requirement for reproductive duration. Field Crops Res. 2014, 163, 24–31. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dwivedi, S.L.; Baum, M.; Varshney, R.K.; Udupa, S.M.; Gowda, C.L.; Hoisington, D.; Singh, S. Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 2008, 8, 1–12. [Google Scholar] [CrossRef]
- Tewari, S.; Pandey, M. Genetics of resistance to ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 1986, 35, 211–215. [Google Scholar] [CrossRef]
- Gil, J.; Castro, P.; Millan, T.; Madrid, E.; Rubio, J. Development of new Kabuli large-seeded chickpea materials with resistance to Ascochyta blight. Crop Pasture Sci. 2017, 68, 967–972. [Google Scholar] [CrossRef]
- Kabakci, H.; Özer, G. Comparison of phenotypic and marker-assisted selection in Turkish cultivars and global genotypes of chickpea for resistance to pathotypes of Ascochyta rabiei (Pass.) Labr. Turk. J. Agric. For. 2021, 45, 1–12. [Google Scholar]
- Bouhadida, M.; Benjannet, R.; Madrid, E.; Amri, M.; Kharrat, M. Efficiency of marker-assisted selection in detection of ascochyta blight resistance in Tunisian chickpea breeding lines. Phytopathol. Mediterr. 2013, 52, 202–211. [Google Scholar]
- Basandrai, A.K.; Basandrai, D.; Pande, S.; Sharma, M.; Thakur, S.K.; Thakur, H.L. Development of ascochyta blight (Ascochyta rabiei) in chickpea as affected by host resistance and plant age. In Ascochyta Blights of Grain Legumes; Tivoli, B., Baranger, A., Muehlbauer, F.J., Cooke, B.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 77–86. [Google Scholar]
- Kaur, L.; Sirari, A.; Kumar, D.; Sandhu, J.S.; Singh, S.; Kapoor, K.; Singh, I.; Gowda, C.L.; Pande, S.; Gaur, P.M.; et al. Combining Ascochyta blight and Botrytis grey mould resistance in chickpea through interspecific hybridization. Phytopathol. Mediterr. 2013, 52, 157–165. [Google Scholar]
- Kimber, R.B.E.; Shtienberg, D.; Ramsey, M.D.; Scott, E.S. The role of seedling infection in epiphytotics of ascochyta blight on chickpea. Eur. J. Plant Pathol. 2007, 117, 141–152. [Google Scholar] [CrossRef]
- Chongo, G.; Gossen, B.D. Effect of plant age on resistance to Ascochyta rabiei in chickpea. Can. J. Plant Pathol. 2001, 23, 358–363. [Google Scholar] [CrossRef]
- Riaz Malik, S.; Iqbal, S.M.; Iqbal, U.; Ahmad, I.; Majeed Haqqani, A. Response of chickpea lines to Ascochyta rabiei at two growing stages. Casp. J. Environ. Sci. 2005, 3, 173–177. [Google Scholar]
- Trapero-Casas, A.; Kaiser, W. Influence of temperature, wetness period, plant age, and inoculum concentration on infection and development of Ascochyta blight of chickpea. Phytopathology 1992, 82, 589–596. [Google Scholar] [CrossRef]

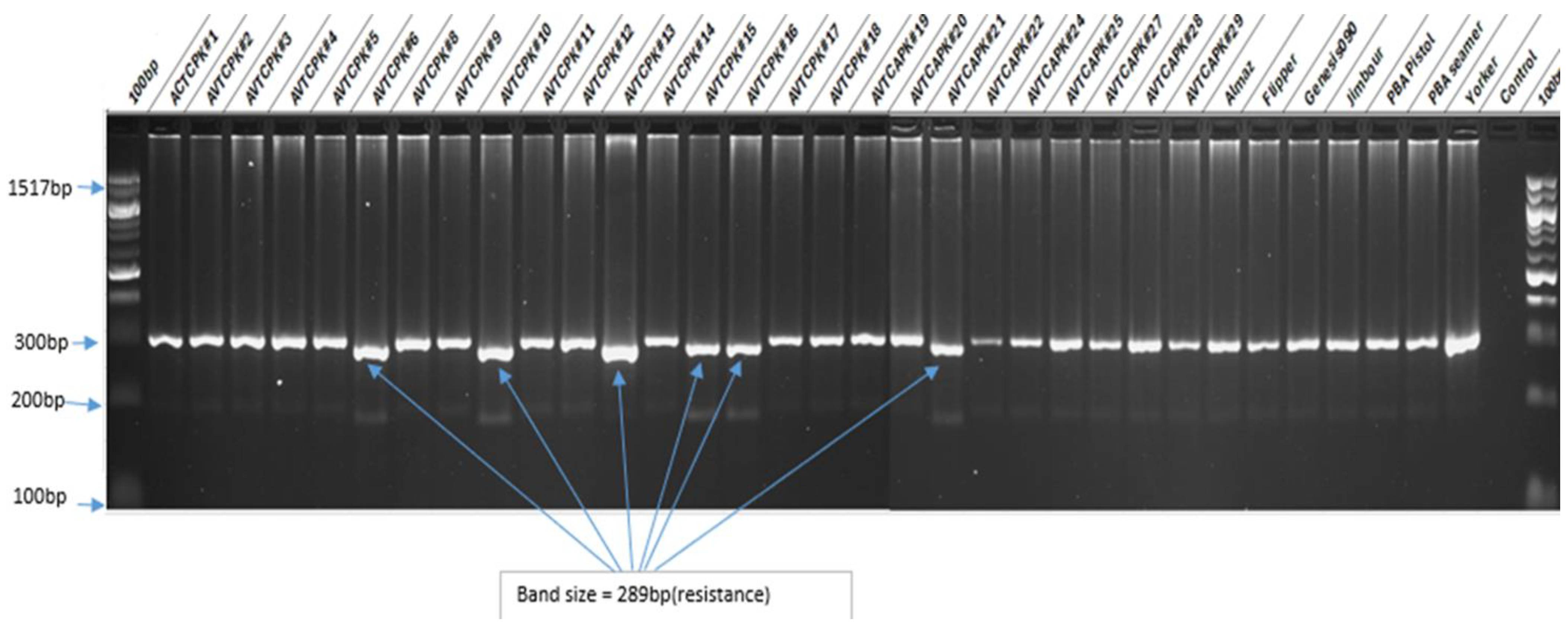
| Line ID | Line ID |
|---|---|
| AVTCPK#1 | AVTCPK#15 |
| AVTCPK#2 | AVTCPK#16 |
| AVTCPK#3 | AVTCPK#17 |
| AVTCPK#4 | AVTCPK#18 |
| AVTCPK#5 | AVTCPK#19 |
| AVTCPK#6 | AVTCPK#20 |
| AVTCPK#8 | AVTCPK#21 |
| AVTCPK#9 | AVTCPK#22 |
| AVTCPK#10 | AVTCPK#24 |
| AVTCPK#11 | AVTCPK#25 |
| AVTCPK#12 | AVTCPK#27 |
| AVTCPK#13 | AVTCPK#28 |
| AVTCPK#14 | AVTCPK#29 |
| Symptoms | Infected Area (%) | Rating Scale | Disease Reaction |
|---|---|---|---|
| No symptoms, immune | 0 | 1 | Asymptomatic |
| Minute lesions/spots on the apical stem | 1–5 | 2 | Resistant (R) |
| Apical stem slightly dropping and lesions up to 5 mm in size. | 6–9 | 3 | Resistant (R) |
| Apical steam-clear dropping and obvious lesions on all the plant parts. | 10–15 | 4 | Moderately Resistant (MR) |
| Obvious lesions on all plant parts, defoliation, and broken branches | 16–20 | 5 | Moderately Resistant (MR) |
| Obvious lesions on all plant parts, defoliation, and broken branches with some plants killed. | 20–40 | 6 | Moderately Susceptible (MR) |
| Same symptoms as 6: up to 25% of the plants killed. | 41–75 | 7 | Susceptible (S) |
| Same symptoms as 6: up to 50% of the plants killed. | 76–100 | 8 | Very Susceptible (VS) |
| Same symptoms as 6: up to 100% of the plants killed. | 100 | 9 | Highly Susceptible (VS) |
| Marker Type | Primer Sequence (5′-3′) | Linkage Group-QTLs | Reference | |
|---|---|---|---|---|
| Allele-specific | CaETR | Fw: CAGGAAGTTCAATGGCCCTA Rev1:TAAGTTGTGACAAAAGACTCAATCG Rev2:TAAGTTGTGACAAAAGACTCAATCG | LG4 QTLAR1 | [27] |
| Sequence-tagged microsatellite (STMS) markers | GAA47 | Fw: CTAAGTTTAATATGTTAGTCCTTAAATTATRev: ACGAACGCAACATTAATTTTATATT | LG4 QTLAR1 | [28] |
| TA146 | Fw: TTTTTGGCTTATTAGACTGACTT Rev: TTGCCATAAAATACAAAATCC | LG4 QTLAR2 | ||
| TA194 | Fw: TTTTTGGCTTATTAGACTGACTT Rev: TTGCCATAAAATACAAAATCC | LG4 QTLAR3 | ||
| Kabuli-Type Chickpea Line ID | Severity Rating 2 | Rank 3 | Disease Reaction 4 |
|---|---|---|---|
| AVTCPK#6 | 4 | a | MR |
| PBA Seamer (R) 1 | 4 | a | MR |
| AVTCPK#14 | 5 | ab | MR |
| Genesis 090 (R) 1 | 5 | ab | MR |
| Yorker (MS/MR) 1 | 5.3 | b | MS |
| Jimbour (S) 1 | 5.5 | b | MS |
| Almaz (MR/MS) 1 | 6 | bc | MS |
| AVTCPK#13 | 6 | bc | MS |
| AVTCPK#28 | 7 | cd | S |
| AVTCPK#5 | 7 | cd | S |
| AVTCPK#1 | 7.4 | de | VS |
| AVTCPK#24 | 7.4 | de | VS |
| AVTCPK#16 | 7.5 | de | VS |
| AVTCPK#21 | 7.5 | de | VS |
| AVTCPK#25 | 7.5 | de | VS |
| AVTCPK#22 | 7.6 | de | VS |
| AVTCPK#12 | 7.7 | de | VS |
| AVTCPK#20 | 7.7 | de | VS |
| AVTCPK#17 | 7.8 | de | VS |
| AVTCPK#8 | 7.8 | de | VS |
| AVTCPK#10 | 8 | de | VS |
| AVTCPK#11 | 8 | de | VS |
| AVTCPK#15 | 8 | de | VS |
| AVTCPK#18 | 8 | de | VS |
| AVTCPK#3 | 8 | de | VS |
| Flipper (MR/MS) 1 | 8 | de | VS |
| PBA Pistol (VS) 1 | 8 | de | VS |
| AVTCPK#19 | 8.2 | de | VS |
| AVTCPK#4 | 8.2 | de | VS |
| AVTCPK#9 | 8.2 | de | VS |
| AVTCPK#2 | 8.4 | e | VS |
| AVTCPK#29 | 8.5 | e | VS |
| AVTCPK#27 | 8.7 | e | VS |
| Line ID | Phenotype 2 | Genotype 3 | |||
|---|---|---|---|---|---|
| CaETR | GAA47 | TA146 | TA194 | ||
| AVTCPK#1 | S | − | − | − | − |
| AVTCPK#2 | VS | − | − | − | − |
| AVTCPK#3 | VS | − | − | − | − |
| AVTCPK#4 | VS | − | − | − | − |
| AVTCPK#5 | S | − | − | − | − |
| AVTCPK#6 | MR | + | + | + | + |
| AVTCPK#8 | S | − | − | − | − |
| AVTCPK#9 | VS | − | − | − | − |
| AVTCPK#10 | MS | + | − | + | − |
| AVTCPK#11 | VS | − | − | − | − |
| AVTCPK#12 | S | − | − | − | − |
| AVTCPK#13 | MS | + | − | − | − |
| AVTCPK#14 | MR | − | − | + | + |
| AVTCPK#15 | VS | + | − | + | − |
| AVTCPK#16 | VS | + | − | − | − |
| AVTCPK#17 | S | − | − | − | − |
| AVTCPK#18 | VS | − | − | − | − |
| AVTCPK#19 | VS | − | − | − | − |
| AVTCPK#20 | S | − | − | − | − |
| AVTCPK#21 | MS | + | − | + | − |
| AVTCPK#22 | S | − | − | − | − |
| AVTCPK#24 | S | − | − | − | − |
| AVTCPK#25 | S | − | − | − | − |
| AVTCPK#27 | VS | − | − | − | − |
| AVTCPK#28 | S | − | − | − | − |
| AVTCPK#29 | VS | − | − | − | − |
| Almaz (MS/MR) 1 | MS | − | − | + | + |
| Flipper (MR) 1 | S | − | − | + | + |
| Genesis 090 (R) 1 | MR | − | − | + | + |
| Jimbour (S) 1 | S | − | − | + | + |
| PBA Pistol (VS) 1 | VS | − | − | − | + |
| PBA Seamer (R) 1 | MR | − | − | − | + |
| Yorker (MS/MR) 1 | MR | − | − | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subedi, M.; Bhattarai, S.; Adorada, D.L. Phenotypic and Genotypic Characterization of New Kabuli-Type Chickpea Lines in Australia for Resistance to Ascochyta Blight. Crops 2024, 4, 400-412. https://doi.org/10.3390/crops4030028
Subedi M, Bhattarai S, Adorada DL. Phenotypic and Genotypic Characterization of New Kabuli-Type Chickpea Lines in Australia for Resistance to Ascochyta Blight. Crops. 2024; 4(3):400-412. https://doi.org/10.3390/crops4030028
Chicago/Turabian StyleSubedi, Megha, Surya Bhattarai, and Dante L. Adorada. 2024. "Phenotypic and Genotypic Characterization of New Kabuli-Type Chickpea Lines in Australia for Resistance to Ascochyta Blight" Crops 4, no. 3: 400-412. https://doi.org/10.3390/crops4030028
APA StyleSubedi, M., Bhattarai, S., & Adorada, D. L. (2024). Phenotypic and Genotypic Characterization of New Kabuli-Type Chickpea Lines in Australia for Resistance to Ascochyta Blight. Crops, 4(3), 400-412. https://doi.org/10.3390/crops4030028







